Transcriptomic and Metabolomic Analysis of a Pseudomonas-Resistant versus a Susceptible Arabidopsis Accession
Abstract
1. Introduction
2. Results
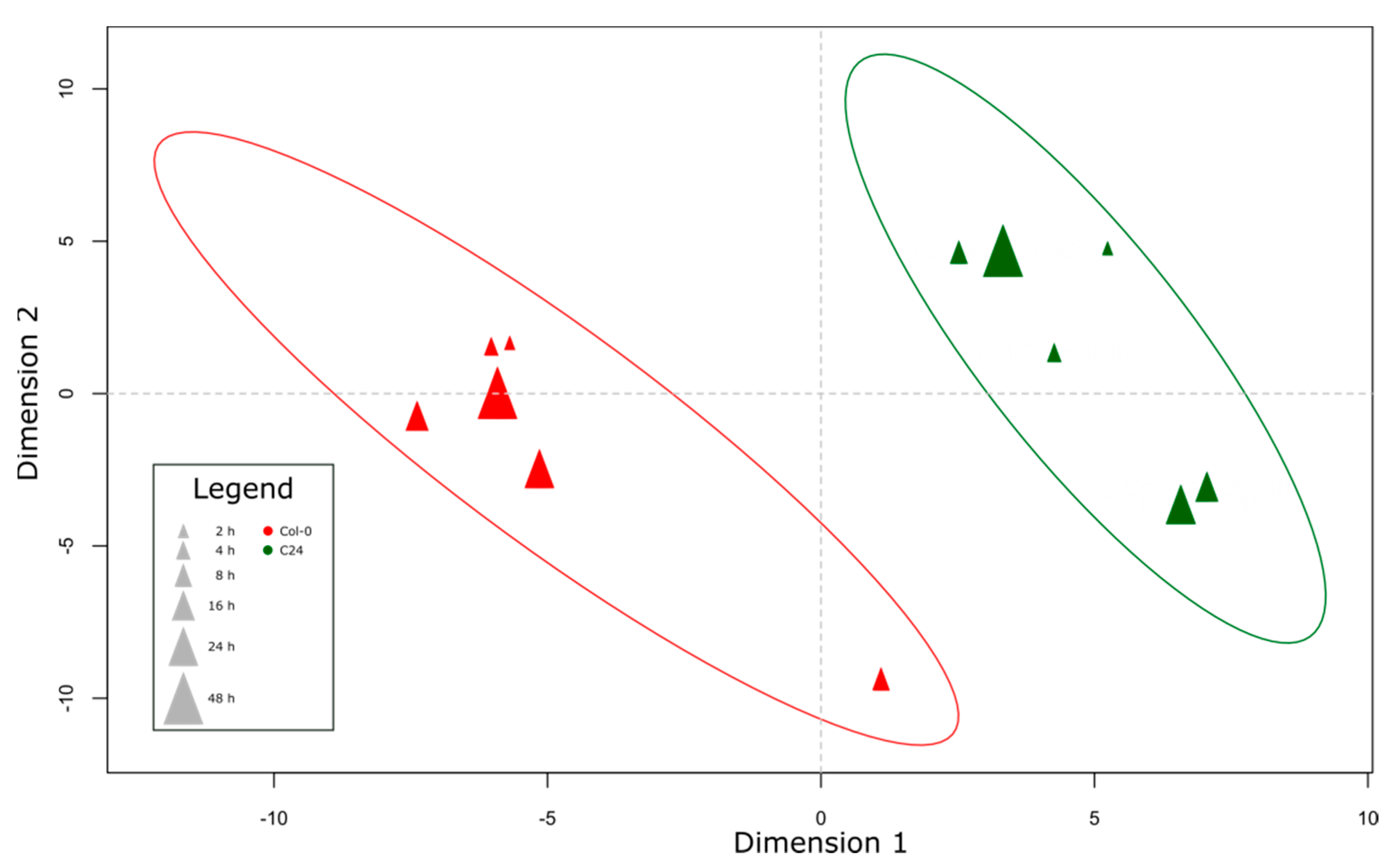
2.1. Appropriate Experimental Study to Dissect the Arabidopsis–Pseudomonas Pathosystem
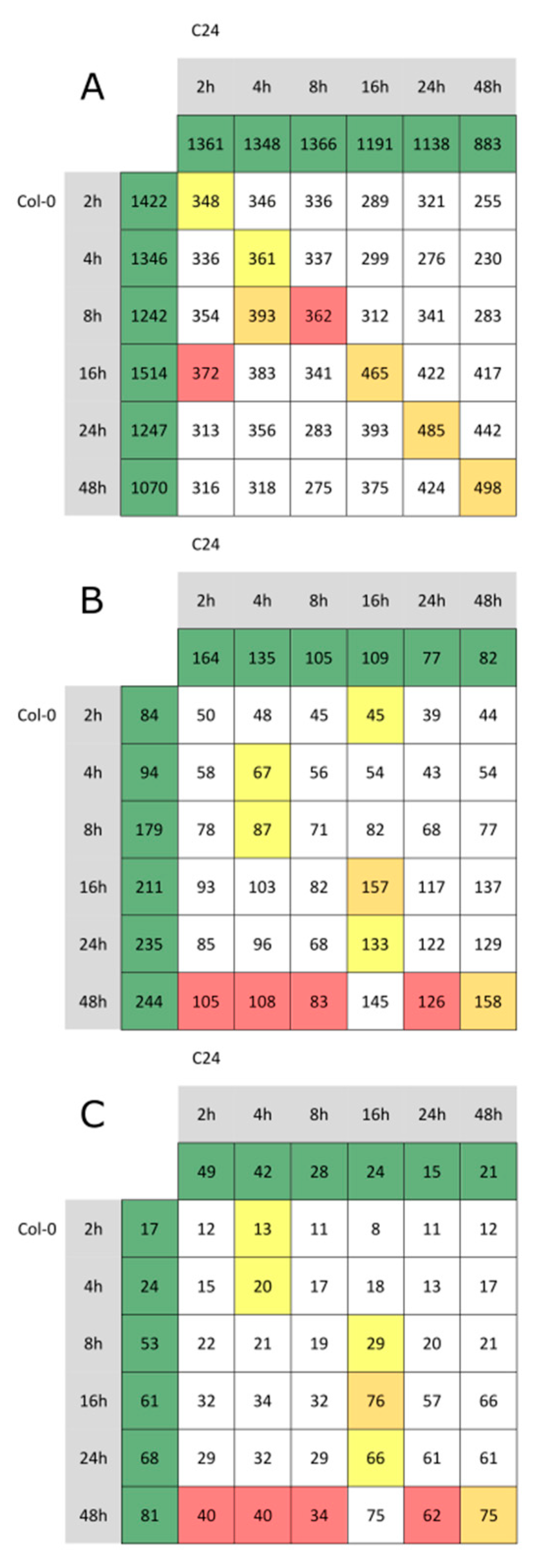
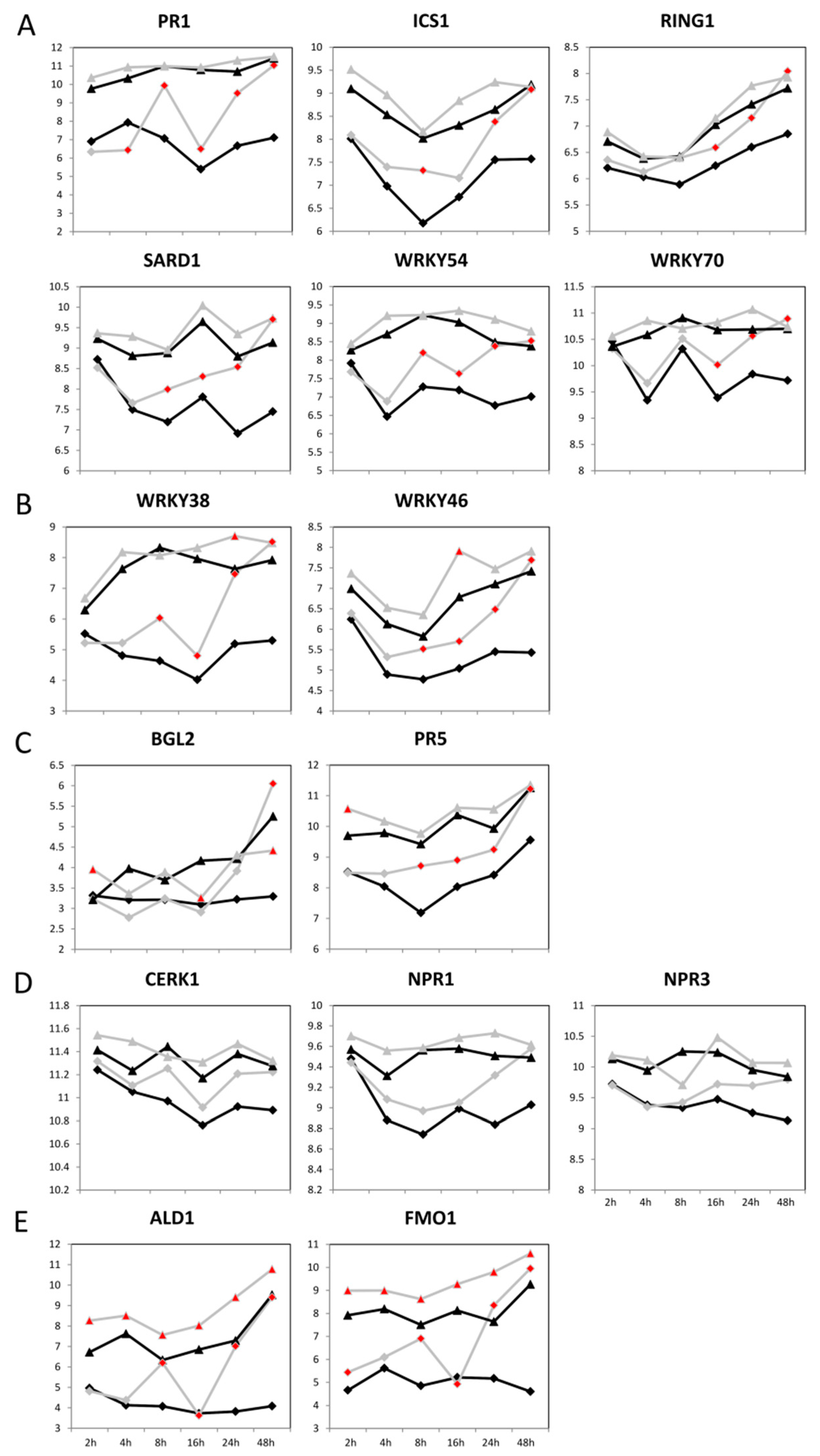
2.2. Significant Transcriptomic Differences in the Basal Gene Expression of SA-Dependent SAR in Col-0 and C24
2.3. The Basal Expression of SA-Related Genes in C24 under Axenic Conditions
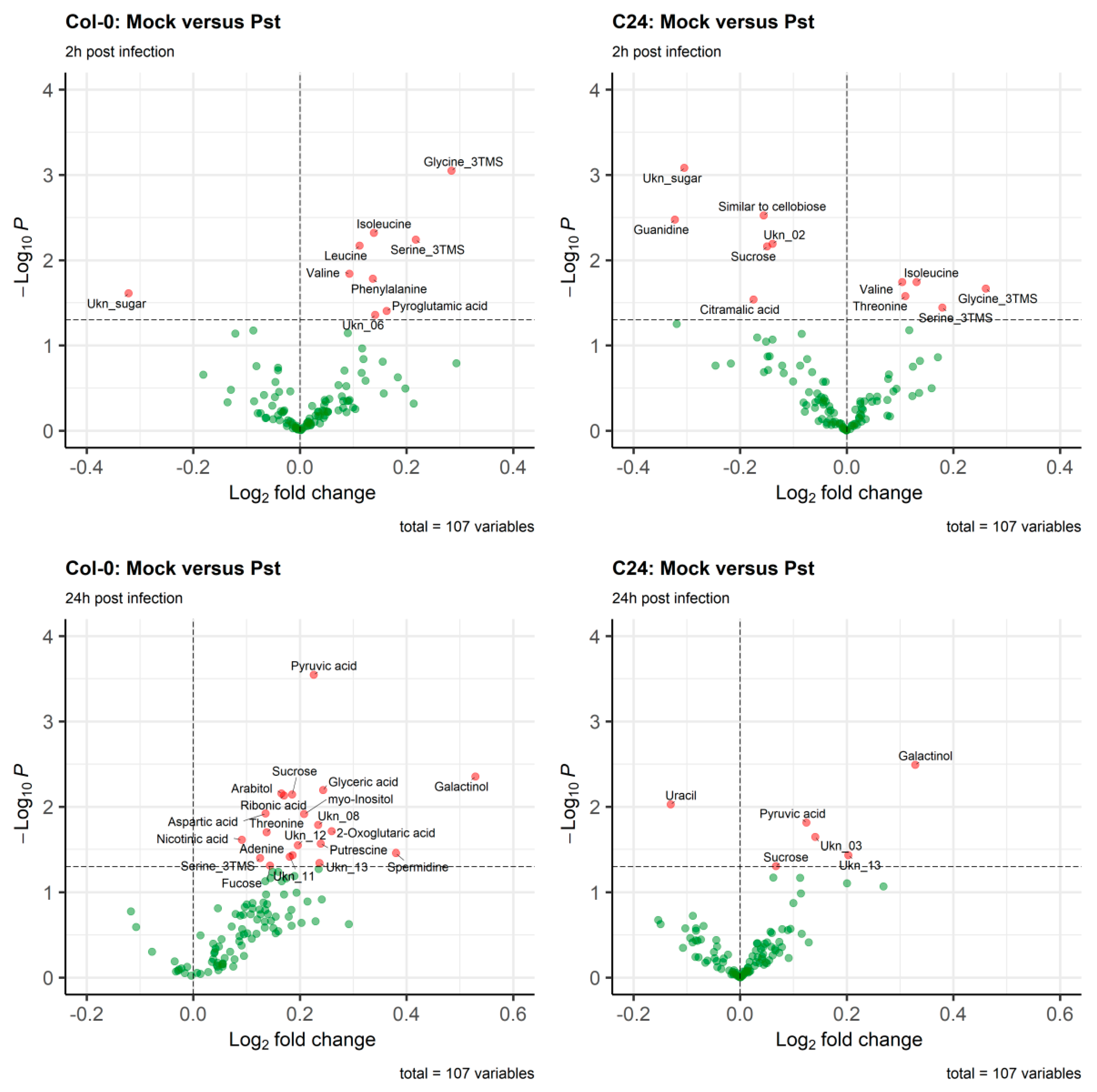
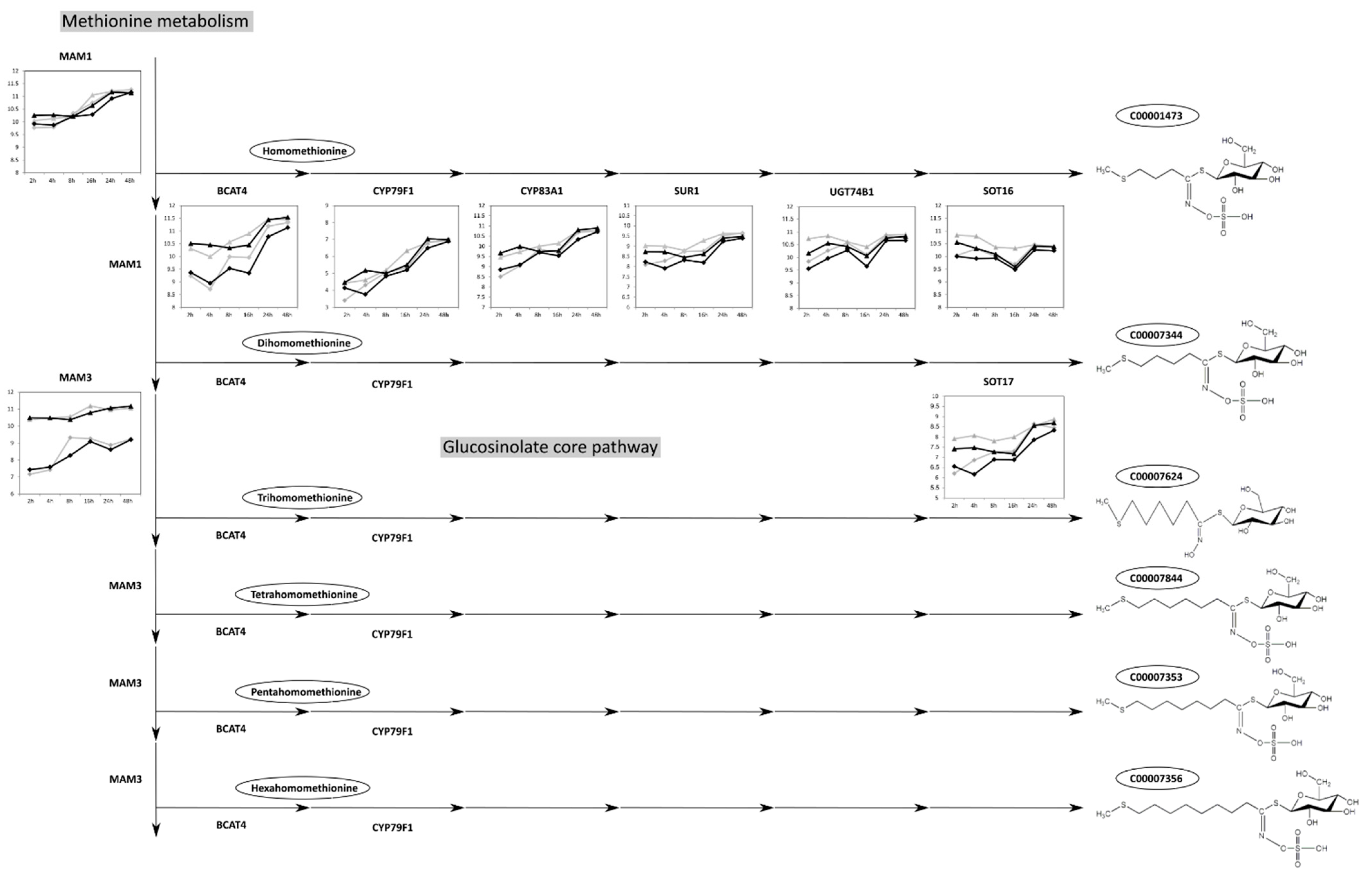
2.4. The Aliphatic Glucosinolate Biosynthesis Pathway Is Differential between Col-0 and C24
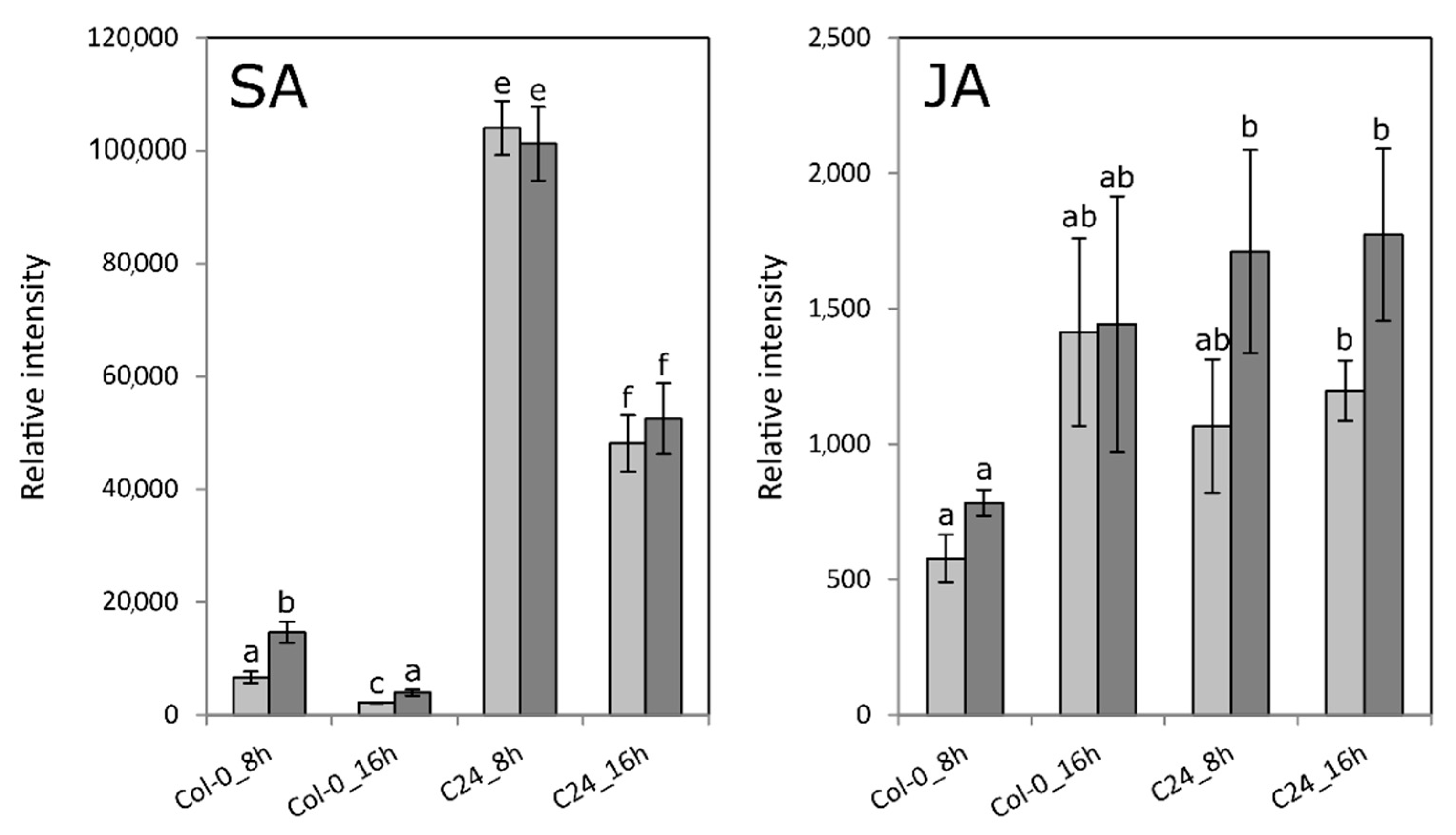
2.5. Col-0 and C24 Share JA-Dependent Responses upon Infection with Pst
2.6. JA-Related Responses after Infection with Pst Are Delayed in Col-0
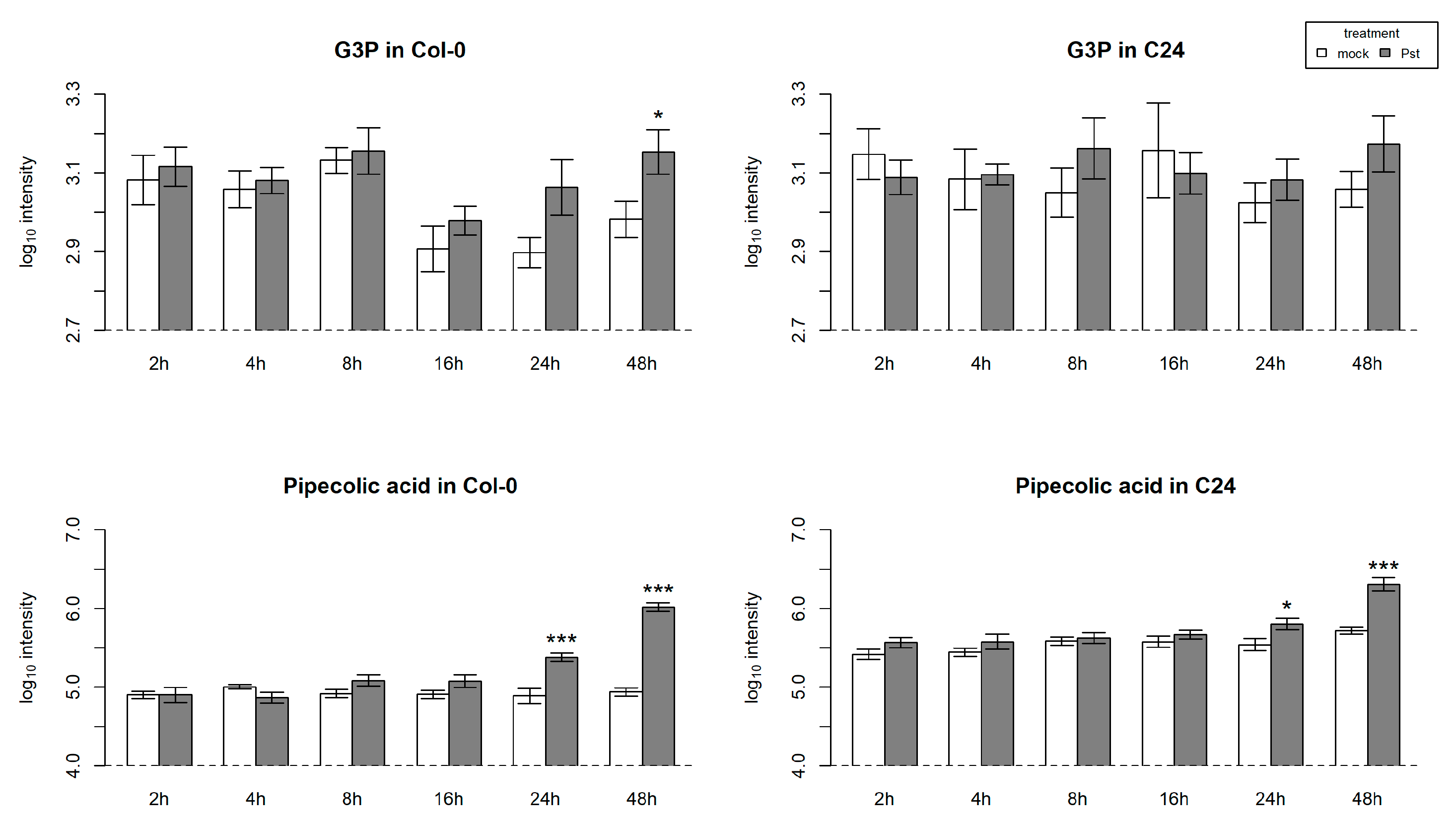
2.7. Pst Induces SA-Dependent SAR-Related Gene Expression in Col-0
3. Discussion
3.1. SAR-Related Differences between Col-0 and C24 Play a Major Role in Determining Resistance to Pst
3.2. Nitrogen Metabolism and Aliphatic Glucosinolates
3.3. JA-Related Responses against Pst Are Shared between Accessions, though Are Delayed in Col-0
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Bacterial Growth
4.3. Inoculation and Sampling
4.4. Transcriptomics
4.5. Metabolomics
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-Pseudomonas syringae Interaction. Arabidopsis B 2002, 1, e0039. [Google Scholar] [CrossRef] [PubMed]
- Ritter, C.; Dangl, J.L. Interference between Two Specific Pathogen Recognition Events Mediated by Distinct Plant Disease Resistance Genes. Plant Cell 1996, 8, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Felix, G.; Boller, T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999, 18, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Mindrinos, M.; Davis, K.; Ausubel, F.M. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 1991, 3, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shah, J.; Klessig, D.F. Signal perception and transduction in plant defense responses. Genes Dev. 1997, 11, 1621–1639. [Google Scholar] [CrossRef] [PubMed]
- Lindeberg, M.; Cunnac, S.; Collmer, A. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol. Plant Pathol. 2009, 10, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Lindeberg, M.; Cunnac, S.; Collmer, A. Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends Microbiol. 2012, 20, 199–208. [Google Scholar] [CrossRef]
- Maleck, K.; Levine, A.; Eulgem, T.; Morgan, A.; Schmid, J.; Lawton, K.A.; Dangl, J.L.; Dietrich, R.A. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 2000, 26, 403–410. [Google Scholar] [CrossRef]
- Tao, Y.; Xie, Z.; Chen, W.; Glazebrook, J.; Chang, H.-S.; Han, B.; Zhu, T.; Zou, G.; Katagiri, F. Quantitative Nature of Arabidopsis Responses during Compatible and Incompatible Interactions with the Bacterial Pathogen Pseudomonas syringae [W]. Plant Cell 2003, 15, 317–330. [Google Scholar] [CrossRef]
- Navarro, L.; Zipfel, C.; Rowland, O.; Keller, I.; Robatzek, S.; Boller, T.; Jones, J.D.G. The Transcriptional Innate Immune Response to flg22. Interplay and Overlap with Avr Gene-Dependent Defense Responses and Bacterial Pathogenesis. Plant Physiol. 2004, 135, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Thilmony, R.; Underwood, W.; He, S.Y. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringaepv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006, 46, 34–53. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Dong, X.; Li, Z.-G.; He, F.; Zhang, Z. Differential Coexpression Analysis Reveals Extensive Rewiring of Arabidopsis Gene Coexpression in Response to Pseudomonas syringae Infection. Sci. Rep. 2016, 6, 35064. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010, 63, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.A.; Polanski, K.; de Torres-Zabala, M.; Jayaraman, S.; Bowden, L.; Moore, J.; Penfold, C.A.; Jenkins, D.J.; Hill, C.; Baxter, L.; et al. Transcriptional Dynamics Driving MAMP-Triggered Immunity and Pathogen Effector-Mediated Immunosuppression in Arabidopsis Leaves Following Infection with Pseudomonas syringae pv tomato DC3000. Plant Cell 2015, 27, 3038–3064. [Google Scholar] [CrossRef]
- Frerigmann, H.; Glawischnig, E.; Gigolashvili, T. The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 654. [Google Scholar] [CrossRef][Green Version]
- Fan, J.; Crooks, C.; Lamb, C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2007, 53, 393–399. [Google Scholar] [CrossRef]
- Vetter, M.M.; Kronholm, I.; He, F.; Haweker, H.; Reymond, M.; Bergelson, J.; Robatzek, S.; de Meaux, J. Flagellin Perception Varies Quantitatively in Arabidopsis thaliana and Its Relatives. Mol. Biol. Evol. 2012, 29, 1655–1667. [Google Scholar] [CrossRef]
- Baudin, M.; Martin, E.C.; Sass, C.; Hassan, J.A.; Bendix, C.; Sauceda, R.; Diplock, N.; Specht, C.D.; Petrescu, A.J.; Lewis, J.D. A natural diversity screen in Arabidopsis thaliana reveals determinants for recognition in the ZAR1-ZED1 immune complex. Plant. Cell Environ. 2021, 44, 629–644. [Google Scholar] [CrossRef]
- Van de Weyer, A.-L.; Monteiro, F.; Furzer, O.J.; Nishimura, M.T.; Cevik, V.; Witek, K.; Jones, J.D.G.; Dangl, J.L.; Weigel, D.; Bemm, F. A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana. Cell 2019, 178, 1260–1272.e14. [Google Scholar] [CrossRef]
- Mishina, T.E.; Zeier, J. Bacterial non-host resistance: Interactions of Arabidopsis with non-adapted Pseudomonas syringae strains. Physiol. Plant. 2007, 131, 448–461. [Google Scholar] [CrossRef]
- Hu, X.; Reddy, A.S. Cloning and expression of a PR5-like protein from Arabidopsis: Inhibition of fungal growth by bacterially expressed protein. Plant Mol. Biol. 1997, 34, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Weigel, R.R.; Bäuscher, C.; Pfitzner, A.J.P.; Pfitzner, U.M. NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 2001, 46, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. The Arabidopsis Flavin-Dependent Monooxygenase FMO1 Is an Essential Component of Biologically Induced Systemic Acquired Resistance. Plant Physiol. 2006, 141, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Loake, G.; Grant, M. Salicylic acid in plant defence—The players and protagonists. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef]
- Heidel, A.J.; Clauss, M.J.; Kroymann, J.; Savolainen, O.; Mitchell-Olds, T. Natural Variation in MAM Within and Between Populations of Arabidopsis lyrata Determines Glucosinolate Phenotype. Genetics 2006, 173, 1629–1636. [Google Scholar] [CrossRef]
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911. [Google Scholar] [CrossRef]
- Stenzel, I.; Otto, M.; Delker, C.; Kirmse, N.; Schmidt, D.; Miersch, O.; Hause, B.; Wasternack, C. ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: Tissue- and organ-specific promoter activities and in vivo heteromerization. J. Exp. Bot. 2012, 63, 6125–6138. [Google Scholar] [CrossRef]
- Seo, H.S.; Song, J.T.; Cheong, J.-J.; Lee, Y.-H.; Lee, Y.-W.; Hwang, I.; Lee, J.S.; Choi, Y. Do Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef]
- Schaller, F.; Biesgen, C.; Müssig, C.; Altmann, T.; Weiler, E.W. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 2000, 210, 979–984. [Google Scholar] [CrossRef]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef] [PubMed]
- Guranowski, A.; Miersch, O.; Staswick, P.E.; Suza, W.; Wasternack, C. Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 2007, 581, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Marquis, V.; Smirnova, E.; Poirier, L.; Zumsteg, J.; Schweizer, F.; Reymond, P.; Heitz, T. Stress- and pathway-specific impacts of impaired jasmonoyl-isoleucine (JA-Ile) catabolism on defense signalling and biotic stress resistance. Plant. Cell Environ. 2020, 43, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Bowling, S.A.; Guo, A.; Cao, H.; Gordon, A.S.; Klessig, D.F.; Dong, X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 1994, 6, 1845–1857. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Shi, Z.; Maximova, S.; Liu, Y.; Verica, J.; Guiltinan, M.J. The Salicylic Acid Receptor NPR3 Is a Negative Regulator of the Transcriptional Defense Response during Early Flower Development in Arabidopsis. Mol. Plant 2013, 6, 802–816. [Google Scholar] [CrossRef]
- Lee, D.H.; Choi, H.W.; Hwang, B.K. The Pepper E3 Ubiquitin Ligase RING1 Gene, CaRING1, Is Required for Cell Death and the Salicylic Acid-Dependent Defense Response. Plant Physiol. 2011, 156, 2011–2025. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Kim, K.-C.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 Transcription Factors Interact with Histone Deacetylase 19 in Basal Defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.K.; Dixon, R.A.; Lamb, C.J. Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J. 1994, 5, 715–725. [Google Scholar] [CrossRef]
- Shah, J.; Chaturvedi, R.; Chowdhury, Z.; Venables, B.; Petros, R.A. Signaling by small metabolites in systemic acquired resistance. Plant J. 2014, 79, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Holmes, E.C.; Rajniak, J.; Kim, J.-G.; Tang, S.; Fischer, C.R.; Mudgett, M.B.; Sattely, E.S. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4920–E4929. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Mekonnen, D.W.; Hartmann, M.; Yildiz, I.; Janowski, R.; Lange, B.; Geist, B.; Zeier, J.; Schäffner, A.R. UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid, and balances plant immunity. Plant Cell 2021, 33, 714–734. [Google Scholar] [CrossRef]
- Yu, K.; Soares, J.M.; Mandal, M.K.; Wang, C.; Chanda, B.; Gifford, A.N.; Fowler, J.S.; Navarre, D.; Kachroo, A.; Kachroo, P. A Feedback Regulatory Loop between G3P and Lipid Transfer Proteins DIR1 and AZI1 Mediates Azelaic-Acid-Induced Systemic Immunity. Cell Rep. 2013, 3, 1266–1278. [Google Scholar] [CrossRef]
- Bechtold, U.; Lawson, T.; Mejia-Carranza, J.; Meyer, R.C.; Brown, I.R.; Altmann, T.; Ton, J.; Mullineaux, P.M. Constitutive salicylic acid defences do not compromise seed yield, drought tolerance and water productivity in the Arabidopsis accession C24. Plant. Cell Environ. 2010, 33, 1959–1973. [Google Scholar] [CrossRef]
- Durrant, W.; Dong, X. SYSTEMIC ACQUIRED RESISTANCE. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Shah, J.; Zeier, J. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 2013, 4, 30. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic Acquired Resistance: Turning Local Infection into Global Defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Whalen, M.C.; Innes, R.W.; Bent, A.F.; Staskawicz, B.J. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 1991, 3, 49–59. [Google Scholar] [CrossRef]
- Ishihara, T.; Sakurai, N.; Sekine, K.-T.; Hase, S.; Ikegami, M.; Shibata, D.; Takahashi, H. Comparative Analysis of Expressed Sequence Tags in Resistant and Susceptible Ecotypes of Arabidopsis thaliana Infected with Cucumber Mosaic Virus. Plant Cell Physiol. 2004, 45, 470–480. [Google Scholar] [CrossRef][Green Version]
- Lapin, D.; Meyer, R.C.; Takahashi, H.; Bechtold, U.; Ackerveken, G. Broad-spectrum resistance of Arabidopsis C24 to downy mildew is mediated by different combinations of isolate-specific loci. New Phytol. 2012, 196, 1171–1181. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Carver, T.L.W.; Prats, E. NO way to live; the various roles of nitric oxide in plant–pathogen interactions. J. Exp. Bot. 2006, 57, 489–505. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Kim, S.-H.; Kim, S.-H.; Yoo, S.-J.; Min, K.-H.; Nam, S.-H.; Cho, B.H.; Yang, K.-Y. Putrescine regulating by stress-responsive MAPK cascade contributes to bacterial pathogen defense in Arabidopsis. Biochem. Biophys. Res. Commun. 2013, 437, 502–508. [Google Scholar] [CrossRef]
- Liu, C.; Atanasov, K.E.; Tiburcio, A.F.; Alcázar, R. The Polyamine Putrescine Contributes to H2O2 and RbohD/F-Dependent Positive Feedback Loop in Arabidopsis PAMP-Triggered Immunity. Front. Plant Sci. 2019, 10, 894. [Google Scholar] [CrossRef]
- Schuhegger, R.; Nafisi, M.; Mansourova, M.; Petersen, B.L.; Olsen, C.E.; Svatoš, A.; Halkier, B.A.; Glawischnig, E. CYP71B15 (PAD3) Catalyzes the Final Step in Camalexin Biosynthesis. Plant Physiol. 2006, 141, 1248–1254. [Google Scholar] [CrossRef]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Levrel, A.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Front. Plant Sci. 2015, 6, 539. [Google Scholar] [CrossRef]
- Cheval, C.; Perez, M.; Leba, L.J.; Ranty, B.; Perochon, A.; Reichelt, M.; Mithöfer, A.; Robe, E.; Mazars, C.; Galaud, J.P.; et al. PRR2, a pseudo-response regulator, promotes salicylic acid and camalexin accumulation during plant immunity. Sci. Rep. 2017, 7, 6979. [Google Scholar] [CrossRef]
- Müller, T.M.; Böttcher, C.; Glawischnig, E. Dissection of the network of indolic defence compounds in Arabidopsis thaliana by multiple mutant analysis. Phytochemistry 2019, 161, 11–20. [Google Scholar] [CrossRef]
- Graser, G.; Oldham, N.J.; Brown, P.D.; Temp, U.; Gershenzon, J. The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 2001, 57, 23–32. [Google Scholar] [CrossRef]
- Textor, S.; de Kraker, J.-W.; Hause, B.; Gershenzon, J.; Tokuhisa, J.G. MAM3 Catalyzes the Formation of All Aliphatic Glucosinolate Chain Lengths in Arabidopsis. Plant Physiol. 2007, 144, 60–71. [Google Scholar] [CrossRef]
- Stotz, H.U.; Sawada, Y.; Shimada, Y.; Hirai, M.Y.; Sasaki, E.; Krischke, M.; Brown, P.D.; Saito, K.; Kamiya, Y. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotiniasclerotiorum. Plant J. 2011, 67, 81–93. [Google Scholar] [CrossRef]
- Giamoustaris, A.; Mithen, R. The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica napus ssp. oleifera) on its interaction with specialist and generalist pests. Ann. Appl. Biol. 1995, 126, 347–363. [Google Scholar] [CrossRef]
- Traw, M.B.; Kniskern, J.M.; Bergelson, J. SAR increases fitness of Arabidopsis thaliana in the presence of natural bacterial pathogens. Evolution. 2007, 61, 2444–2449. [Google Scholar] [CrossRef]
- Mewis, I.; Appel, H.M.; Hom, A.; Raina, R.; Schultz, J.C. Major Signaling Pathways Modulate Arabidopsis Glucosinolate Accumulation and Response to Both Phloem-Feeding and Chewing Insects. Plant Physiol. 2005, 138, 1149–1162. [Google Scholar] [CrossRef]
- von Saint Paul, V.; Zhang, W.; Kanawati, B.; Geist, B.; Faus-Keßler, T.; Schmitt-Kopplin, P.; Schäffner, A.R. The Arabidopsis Glucosyltransferase UGT76B1 Conjugates Isoleucic Acid and Modulates Plant Defense and Senescence. Plant Cell 2011, 23, 4124–4145. [Google Scholar] [CrossRef]
- Maksym, R.P.; Ghirardo, A.; Zhang, W.; von Saint Paul, V.; Lange, B.; Geist, B.; Hajirezaei, M.-R.; Schnitzler, J.-P.; Schäffner, A.R. The Defense-Related Isoleucic Acid Differentially Accumulates in Arabidopsis Among Branched-Chain Amino Acid-Related 2-Hydroxy Carboxylic Acids. Front. Plant Sci. 2018, 9, 766. [Google Scholar] [CrossRef]
- Bauer, S.; Mekonnen, D.W.; Geist, B.; Lange, B.; Ghirardo, A.; Zhang, W.; Schäffner, A.R. The isoleucic acid triad: Distinct impacts on plant defense, root growth, and formation of reactive oxygen species. J. Exp. Bot. 2020, 71, 4258–4270. [Google Scholar] [CrossRef]
- Yang, L.; Teixeira, P.J.P.L.; Biswas, S.; Finkel, O.M.; He, Y.; Salas-Gonzalez, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J.L. Pseudomonas syringae Type III Effector HopBB1 Promotes Host Transcriptional Repressor Degradation to Regulate Phytohormone Responses and Virulence. Cell Host Microbe 2017, 21, 156–168. [Google Scholar] [CrossRef]
- Hesse, H.; Nikiforova, V.; Gakiere, B.; Hoefgen, R. Molecular analysis and control of cysteine biosynthesis: Integration of nitrogen and sulphur metabolism. J. Exp. Bot. 2004, 55, 1283–1292. [Google Scholar] [CrossRef]
- Benstein, R.M.; Ludewig, K.; Wulfert, S.; Wittek, S.; Gigolashvili, T.; Frerigmann, H.; Gierth, M.; Flügge, U.-I.; Krueger, S. Arabidopsis Phosphoglycerate Dehydrogenase1 of the Phosphoserine Pathway Is Essential for Development and Required for Ammonium Assimilation and Tryptophan Biosynthesis. Plant Cell 2014, 25, 5011–5029. [Google Scholar] [CrossRef]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 Complete Salicylic Acid Biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef]
- Okrent, R.A.; Brooks, M.D.; Wildermuth, M.C. Arabidopsis GH3.12 (PBS3) Conjugates Amino Acids to 4-Substituted Benzoates and Is Inhibited by Salicylate. J. Biol. Chem. 2009, 284, 9742–9754. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural Basis for Signaling by Exclusive EDS1 Heteromeric Complexes with SAG101 or PAD4 in Plant Innate Immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef]
- Wittek, F.; Hoffmann, T.; Kanawati, B.; Bichlmeier, M.; Knappe, C.; Wenig, M.; Schmitt-Kopplin, P.; Parker, J.E.; Schwab, W.; Vlot, A.C. Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistance via azelaic acid and its precursor 9-oxo nonanoic acid. J. Exp. Bot. 2014, 65, 5919–5931. [Google Scholar] [CrossRef]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469.e16. [Google Scholar] [CrossRef]
- Mohnike, L.; Rekhter, D.; Huang, W.; Feussner, K.; Tian, H.; Herrfurth, C.; Zhang, Y.; Feussner, I. The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 2021, 33, 735–749. [Google Scholar] [CrossRef]
- Noutoshi, Y.; Okazaki, M.; Kida, T.; Nishina, Y.; Morishita, Y.; Ogawa, T.; Suzuki, H.; Shibata, D.; Jikumaru, Y.; Hanada, A.; et al. Novel Plant Immune-Priming Compounds Identified via High-Throughput Chemical Screening Target Salicylic Acid Glucosyltransferases in Arabidopsis. Plant Cell 2012, 24, 3795–3804. [Google Scholar] [CrossRef]
- AbuQamar, S.; Chen, X.; Dhawan, R.; Bluhm, B.; Salmeron, J.; Lam, S.; Dietrich, R.A.; Mengiste, T. Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 2006, 48, 28–44. [Google Scholar] [CrossRef]
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis Defense against Botrytis cinerea: Chronology and Regulation Deciphered by High-Resolution Temporal Transcriptomic Analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic Interaction between Abscisic Acid and Jasmonate-Ethylene Signaling Pathways Modulates Defense Gene Expression and Disease Resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis [W]. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, G.; Wang, X.; Miao, J.; Liu, X.; Chen, Z.; Qu, L.-J.; Gu, H. Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep. 2007, 27, 125–135. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, J.; Gao, X.; Tong, J.; Xiao, L.; Li, W.; Zhang, H. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 2010, 457, 1–12. [Google Scholar] [CrossRef]
- Krishnaswamy, S.; Verma, S.; Rahman, M.H.; Kav, N.N. V Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 2011, 75, 107–127. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef]
- Thaler, J.S.; Owen, B.; Higgins, V.J. The Role of the Jasmonate Response in Plant Susceptibility to Diverse Pathogens with a Range of Lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef]
- Weiler, E.W.; Kutchan, T.M.; Gorba, T.; Brodschelm, W.; Niesel, U.; Bublitz, F. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 1994, 345, 9–13. [Google Scholar] [CrossRef]
- Geng, X.; Jin, L.; Shimada, M.; Kim, M.G.; Mackey, D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta. 2014, 240, 1149–1165. [Google Scholar] [CrossRef]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Kloek, A.P.; Verbsky, M.L.; Sharma, S.B.; Schoelz, J.E.; Vogel, J.; Klessig, D.F.; Kunkel, B.N. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001, 26, 509–522. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent Defense Pathways through a Novel Function in the Cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Irizarry, R.A. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef]
- Irizarry, R.A. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Horváth, L.; Rice, G. Extensions of some classical methods in change point analysis. TEST 2014, 23, 219–255. [Google Scholar] [CrossRef]
- Omranian, N.; Mueller-Roeber, B.; Nikoloski, Z. Segmentation of biological multivariate time-series data. Sci. Rep. 2015, 5, 8937. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer-Verlag: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Cuadros-Inostroza, Á.; Caldana, C.; Redestig, H.; Kusano, M.; Lisec, J.; Peña-Cortés, H.; Willmitzer, L.; Hannah, M.A. TargetSearch-a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinform. 2009, 10, 428. [Google Scholar] [CrossRef]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dormann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef]
- Steinfath, M.; Groth, D.; Lisec, J.; Selbig, J. Metabolite profile analysis: From raw data to regression and classification. Physiol. Plant. 2007, 132, 150–161. [Google Scholar] [CrossRef]
- Hummel, J.; Segu, S.; Li, Y.; Irgang, S.; Jueppner, J.; Giavalisco, P. Ultra Performance Liquid Chromatography and High Resolution Mass Spectrometry for the Analysis of Plant Lipids. Front. Plant Sci. 2011, 2, 54. [Google Scholar] [CrossRef]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef]
| GO BP ID | p-Value | Count | Size | Term | |
|---|---|---|---|---|---|
| 1 | GO:0009627 | 6.35 × 10−53 | 128 | 442 | systemic acquired resistance |
| 2 | GO:0009697 | 5.88 × 10−49 | 85 | 207 | salicylic acid biosynthetic process |
| 3 | GO:0009814 | 1.16 × 10−47 | 134 | 532 | defense response, incompatible interaction |
| 4 | GO:0009696 | 1.90 × 10−47 | 86 | 220 | salicylic acid metabolic process |
| 5 | GO:0042537 | 8.94 × 10−45 | 90 | 258 | benzene-containing compound metabolic process |
| 6 | GO:0006952 | 4.69 × 10−44 | 249 | 1644 | defense response |
| 7 | GO:0002376 | 1.78 × 10−42 | 179 | 977 | immune system process |
| 8 | GO:0045087 | 1.90 × 10−42 | 166 | 859 | innate immune response |
| 9 | GO:0006955 | 4.89 × 10−42 | 166 | 865 | immune response |
| 10 | GO:0071446 | 6.40 × 10−35 | 93 | 353 | cellular response to salicylic acid stimulus |
| 11 | GO:0009751 | 1.12 × 10−34 | 108 | 469 | response to salicylic acid stimulus |
| 12 | GO:0009607 | 1.15 × 10−34 | 208 | 1413 | response to biotic stimulus |
| 13 | GO:0009863 | 2.31 × 10−34 | 92 | 351 | salicylic acid mediated signaling pathway |
| 14 | GO:0051707 | 3.09 × 10−34 | 207 | 1412 | response to other organism |
| 15 | GO:0031348 | 6.87 × 10−33 | 79 | 273 | negative regulation of defense response |
| 16 | GO:0031347 | 1.27 × 10−31 | 112 | 539 | regulation of defense response |
| 17 | GO:0080134 | 1.07 × 10−30 | 113 | 560 | regulation of response to stress |
| 18 | GO:0009862 | 6.30 × 10−30 | 72 | 250 | systemic acquired resistance, salicylic acid mediated signaling pathway |
| 19 | GO:0051704 | 2.42 × 10−29 | 231 | 1802 | multi-organism process |
| 20 | GO:0050832 | 4.46 × 10−29 | 84 | 344 | defense response to fungus |
| GO BP ID | p-Value | Count | Size | Term | |
|---|---|---|---|---|---|
| 1 | GO:0006952 | 2.79 × 10−4 | 63 | 1644 | defense response |
| 2 | GO:0007165 | 4.80 × 10−4 | 71 | 1948 | signal transduction |
| 3 | GO:0044700 | 8.90 × 10−4 | 72 | 2027 | single organism signaling |
| 4 | GO:0023052 | 9.02 × 10−4 | 72 | 2028 | signaling |
| 5 | GO:0007154 | 4.15 × 10−3 | 78 | 2367 | cell communication |
| 6 | GO:0016145 | 4.35 × 10−3 | 3 | 14 | S-glycoside catabolic process |
| 7 | GO:0080028 | 5.70 × 10−3 | 2 | 5 | nitrile biosynthetic process |
| 8 | GO:0006591 | 8.41 × 10−3 | 2 | 6 | ornithine metabolic process |
| 9 | GO:0015824 | 9.54 × 10−3 | 6 | 74 | proline transport |
| 10 | GO:0015804 | 1.15 × 10−2 | 6 | 77 | neutral amino acid transport |
| 11 | GO:0032890 | 1.16 × 10−2 | 2 | 7 | regulation of organic acid transport |
| 12 | GO:0050898 | 1.16 × 10−2 | 2 | 7 | nitrile metabolic process |
| 13 | GO:0051952 | 1.16 × 10−2 | 2 | 7 | regulation of amine transport |
| 14 | GO:0070417 | 1.22 × 10−2 | 3 | 20 | cellular response to cold |
| 15 | GO:0000304 | 1.52 × 10−2 | 2 | 8 | response to singlet oxygen |
| 16 | GO:0019674 | 1.52 × 10−2 | 2 | 8 | NAD metabolic process |
| Content Fold Change after 48 h | ||
|---|---|---|
| KNApSAcK ID: Name(s) | Col-0 | C24 |
| Mock vs. Pst | Mock vs. Pst | |
| C00007850: 5-Oxooctyl glucosinolate; Glucocappasalin | n.d. | 1.63 |
| C00007855: 4-Phenylbutyl glucosinolate | n.d. | 1.53 |
| C00007801: 5-Hexenyl glucosinolate | n.d. | 1.37 |
| 3-Hydroxy-5-(methylthio)pentyl glucosinolate | n.d. | 1.21 |
| C00001463: Glucobrassicanapin; 4-Pentenyl glucosinolate | n.d. | 1.19 |
| C00007340: 3-Hydroxypropyl glucosinolate | n.d. | 1.16 |
| C00007817: 2-Hydroxypropyl glucosinolate | ||
| C00007826: 1-Methyl-2-hydroxyetyl glucosinolate; Glucosisymbrin | ||
| C00001488: Sinigrin; 2-Phenylethyl glucosinolate; | n.d. | 1.11 |
| Allyl glucosinolate | ||
| C00007845: 9-Methylthiononyl glucosinolate | n.d. | 1.01 |
| C00007800: 6-Heptenyl glucosinolate | 2.92 ** | 1.28 |
| C00001473: Glucoiberverin; | 2.59 *** | 1.26 |
| 3-(Methylthio)propyl glucosinolate | ||
| C00000125: Glucobrassicin; Glucobrassicine; 3-Indolylmethylglucosinolate | 1.71 *** | 2.17 * |
| C00007856: 3-Phenylpropyl glucosinolate | 1.67 ** | 1.26 |
| C00000130: 5-Hydroxyglucobrassicin; 5-Hydroxy-3-indolylmethylglucosinolate | 1.63 | 2.03 * |
| C00007348: 6-Methylsulfinylhexyl glucosinolate; Glucohesperin | 1.53 * | 1.24 |
| C00007813: 3-Hydroxy-6-(methylthio)hexyl glucosinolate | ||
| C00007352: 7-(Methylsulfinyl)heptyl glucosinolate; Glucoibarin | 1.52 * | 1.29 |
| C00007624: 5-Methylthiopentyldesulfoglucosinolate | 1.47 * | 1.21 |
| C00007355: 8-Methylsulfinyloctyl glucosinolate; Glucohirsutin | 1.41 * | 1.13 |
| C00007593: 5-Methylthiopentylglucosinolate; Glucoberteroin | 1.32 * | 1.22 |
| C00007353: 7-Methylthioheptyl glucosinolate | 1.31 * | 1.19 |
| C00007356: 8-Methylthio-octyl glucosinolate | 1.28 * | 1.07 |
| C00001474: Glucolepidiin; Ethyl glucosinolate | 1.16 | n.d. |
| Gene Annotation | Group | 2 h Col-0 Pst—Mock | 4 h Col-0 Pst—Mock | 8 h Col-0 Pst—Mock | 16 h Col-0 Pst—Mock | 24 h Col-0 Pst—Mock | 48 h Col-0 Pst—Mock | 2 h C24 Pst—Mock | 4 h C24 Pst—Mock | 8 h C24 Pst—Mock | 16 h C24 Pst—Mock | 24 h C24 Pst—Mock | 48 h C24 Pst—Mock |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interval 1 | Interval 2 | Interval 1 | Interval 2 | ||||||||||
| ABO3 | I | 0.103 | −0.012 | 0.366 | −0.181 | 1.630 | 1.265 | 0.971 | 0.393 | 0.764 | 0.071 | 0.589 | 0.536 |
| ACT11 | I | −0.226 | −0.402 | −0.731 | −0.359 | −0.850 | −1.324 | −0.627 | −0.724 | −0.560 | −0.053 | −0.024 | −0.773 |
| AOC3 | II | 0.328 | 0.529 | 0.892 | 0.897 | 1.573 | 2.488 | 0.967 | 0.540 | 0.347 | 1.386 | 1.594 | 1.313 |
| ASB1 | I | 0.298 | −0.187 | 0.006 | 0.799 | 1.482 | 1.588 | 0.966 | 0.865 | 0.963 | −0.168 | 1.132 | −0.674 |
| AT1G26390 | II | 0.685 | 0.642 | 1.478 | 0.340 | 0.852 | 3.186 | 1.371 | 0.819 | 0.858 | 0.868 | 0.676 | 2.555 |
| AT1G53885 | II | 0.449 | 0.307 | 0.441 | 2.016 | 2.292 | 3.491 | 0.609 | 0.437 | 1.076 | 2.354 | 2.846 | 3.425 |
| AT1G53885 | II | 0.449 | 0.307 | 0.441 | 2.016 | 2.292 | 3.491 | 0.609 | 0.437 | 1.076 | 2.354 | 2.846 | 3.425 |
| AT1G60730 | II | 0.321 | 0.378 | 0.520 | 0.452 | 0.524 | 1.035 | 0.684 | 0.747 | 0.406 | 0.690 | 0.510 | 0.788 |
| AT1G66310 | I | −0.296 | −0.121 | −0.794 | −0.718 | −1.015 | −0.734 | −0.016 | −0.885 | −0.933 | 0.352 | −0.472 | 0.443 |
| AT2G45220 | II | 0.376 | 0.369 | 2.129 | 0.467 | 0.965 | 3.166 | 0.987 | 1.035 | 1.201 | 0.818 | 0.546 | 1.904 |
| AT3G04000 | II | 0.529 | 0.432 | 0.664 | 0.359 | 0.487 | 1.505 | 0.574 | 0.721 | 0.735 | 0.686 | 1.116 | 1.529 |
| AT3G11340 | II | 0.311 | 1.576 | 2.301 | 1.246 | 3.784 | 4.448 | 1.297 | 1.275 | 1.021 | 1.130 | 2.061 | 0.305 |
| AT3G22600 | II | 0.324 | 0.524 | 2.631 | 1.069 | 1.626 | 1.798 | 1.040 | 0.859 | 0.477 | 0.387 | 0.883 | 1.069 |
| AT3G46080 | I | 0.082 | −0.065 | 0.103 | −0.019 | 1.822 | 2.888 | 1.302 | 0.915 | 1.227 | 0.472 | 0.883 | 1.278 |
| AT4G03130 | I | 0.383 | −0.324 | −0.452 | −0.726 | −0.209 | −1.041 | −0.513 | −0.414 | −1.063 | 0.567 | 0.210 | 0.324 |
| AT4G11890 | I | 0.251 | 0.398 | 1.576 | 0.830 | 1.309 | 1.781 | 0.466 | 0.915 | 0.396 | 0.404 | 0.188 | 0.098 |
| AT4G19970 | II | −0.096 | −0.208 | 1.366 | 0.529 | 0.393 | 1.702 | 0.977 | 0.687 | 1.105 | 0.142 | 0.742 | 1.559 |
| AT5G05600 | II | 0.111 | 0.545 | 0.909 | 1.480 | 1.805 | 3.007 | 0.852 | 0.593 | 1.127 | 1.921 | 1.887 | 1.873 |
| AT5G08240 | II | 0.644 | 0.272 | 0.675 | 0.497 | 0.655 | 1.840 | 0.949 | 0.698 | 0.572 | 0.485 | 0.566 | 0.800 |
| AT5G39050 | I | 0.168 | 0.613 | 0.472 | 0.551 | 0.919 | 1.364 | 0.808 | 0.598 | 0.581 | 0.255 | 0.576 | 0.674 |
| AT5G57510 | II | 0.984 | 0.034 | 0.666 | 0.159 | 0.914 | 3.287 | 1.357 | 0.702 | 0.672 | 0.654 | 1.405 | 2.171 |
| CEN2 | II | −0.228 | 0.843 | 1.387 | 0.228 | 2.256 | 2.978 | 0.562 | 1.072 | 0.688 | 0.771 | 1.000 | 1.479 |
| COBL5 | I | 0.023 | 0.641 | 0.809 | 0.533 | 0.778 | 1.461 | 0.567 | 0.681 | 0.553 | 0.448 | 0.525 | 0.302 |
| DUR | II | 0.534 | 0.791 | 1.592 | 0.092 | 1.595 | 3.018 | 0.785 | 0.803 | 0.729 | 0.730 | 0.979 | 1.989 |
| EDS1 | I | −0.275 | −0.142 | 0.000 | 0.429 | 0.690 | 1.035 | 1.260 | 0.528 | 0.346 | 0.326 | −0.263 | −0.433 |
| FMO1 | II | 0.779 | 0.480 | 2.049 | −0.297 | 3.178 | 5.345 | 1.072 | 0.803 | 1.119 | 1.146 | 2.160 | 1.341 |
| GRX480 | II | −0.088 | 0.007 | 1.703 | 1.759 | 2.910 | 3.277 | 0.987 | 0.613 | 0.316 | 0.846 | 1.046 | 0.796 |
| GSTU11 | II | 0.029 | 0.257 | 0.254 | 0.333 | 1.101 | 3.677 | 1.264 | 0.997 | 0.128 | 2.348 | 2.694 | 2.566 |
| GSTU4 | II | 0.999 | 0.931 | 1.966 | 1.391 | 1.722 | 4.467 | 1.929 | 1.435 | 1.548 | 1.520 | 1.509 | 0.904 |
| GSTU8 | I | −0.586 | −0.769 | 1.098 | 0.937 | 1.166 | 1.823 | 1.408 | 1.644 | 1.064 | 1.143 | 0.616 | 0.585 |
| IAR3 | II | −0.053 | 0.397 | 0.932 | 0.884 | 1.251 | 2.349 | 0.860 | 0.533 | 0.611 | 0.727 | 0.986 | 1.138 |
| KCO2 | I | −0.318 | 0.669 | −1.301 | −1.217 | −0.333 | −0.673 | −0.033 | −1.172 | −0.582 | −0.079 | −0.793 | −0.726 |
| LHT7 | II | −0.610 | 0.736 | 2.174 | 1.331 | 3.433 | 3.421 | 0.126 | 1.278 | 0.406 | 1.260 | 1.436 | 0.584 |
| MYB13 | II | 0.059 | 0.560 | 0.600 | 0.328 | 0.797 | 1.428 | 0.569 | 0.875 | 0.743 | 0.386 | 0.958 | 0.935 |
| MYC2 | II | −0.030 | 0.155 | 0.367 | 1.191 | 1.256 | 1.525 | 0.791 | 0.782 | 0.190 | 1.131 | 0.931 | 0.983 |
| PDF1.2 | II | −0.351 | 0.428 | 2.226 | 2.240 | 1.757 | 2.317 | 1.654 | 1.651 | 2.028 | 1.620 | 1.277 | 0.082 |
| RAP2.6 | II | −0.201 | −0.194 | 0.100 | 0.302 | 1.627 | 3.460 | 1.092 | 0.340 | 0.895 | 0.679 | 0.745 | 1.586 |
| Rap2.6L | II | 0.595 | 0.889 | 0.897 | 0.431 | 1.205 | 2.564 | 0.747 | 1.115 | 0.810 | 0.579 | 0.975 | 1.987 |
| RLP43 | I | 0.503 | 0.117 | 1.137 | 0.388 | 1.122 | 1.656 | 0.829 | 0.937 | 0.422 | 0.084 | 0.557 | 0.447 |
| SBT3.3 | II | −0.498 | 0.952 | 1.016 | 0.524 | 0.655 | 2.469 | 1.394 | 0.949 | 1.221 | 1.041 | 1.584 | 1.582 |
| TPS04 | II | 0.253 | 0.916 | 0.453 | 1.528 | 2.856 | 5.219 | 1.063 | 1.163 | 0.861 | 2.564 | 1.961 | 1.249 |
| UGT73D1 | II | 0.123 | −0.107 | 0.044 | 0.770 | 0.723 | 2.176 | 0.469 | 1.061 | 0.903 | 0.544 | 1.313 | 0.924 |
| WAK3 | II | 0.359 | 0.272 | 1.733 | 1.003 | 1.688 | 2.445 | 0.824 | 0.669 | 0.489 | 0.276 | 0.767 | 0.962 |
| WRKY75 | II | 0.759 | 0.181 | 1.547 | 0.269 | 1.214 | 4.190 | 1.208 | 1.271 | 0.733 | 0.886 | 0.674 | 1.676 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orf, I.; Tenenboim, H.; Omranian, N.; Nikoloski, Z.; Fernie, A.R.; Lisec, J.; Brotman, Y.; Bromke, M.A. Transcriptomic and Metabolomic Analysis of a Pseudomonas-Resistant versus a Susceptible Arabidopsis Accession. Int. J. Mol. Sci. 2022, 23, 12087. https://doi.org/10.3390/ijms232012087
Orf I, Tenenboim H, Omranian N, Nikoloski Z, Fernie AR, Lisec J, Brotman Y, Bromke MA. Transcriptomic and Metabolomic Analysis of a Pseudomonas-Resistant versus a Susceptible Arabidopsis Accession. International Journal of Molecular Sciences. 2022; 23(20):12087. https://doi.org/10.3390/ijms232012087
Chicago/Turabian StyleOrf, Isabel, Hezi Tenenboim, Nooshin Omranian, Zoran Nikoloski, Alisdair R. Fernie, Jan Lisec, Yariv Brotman, and Mariusz A. Bromke. 2022. "Transcriptomic and Metabolomic Analysis of a Pseudomonas-Resistant versus a Susceptible Arabidopsis Accession" International Journal of Molecular Sciences 23, no. 20: 12087. https://doi.org/10.3390/ijms232012087
APA StyleOrf, I., Tenenboim, H., Omranian, N., Nikoloski, Z., Fernie, A. R., Lisec, J., Brotman, Y., & Bromke, M. A. (2022). Transcriptomic and Metabolomic Analysis of a Pseudomonas-Resistant versus a Susceptible Arabidopsis Accession. International Journal of Molecular Sciences, 23(20), 12087. https://doi.org/10.3390/ijms232012087







