Abstract
Anti-neuroinflammatory treatment has gained importance in the search for pharmacological treatments of different neurological and psychiatric diseases, such as depression, schizophrenia, Parkinson’s disease, and Alzheimer’s disease. Clinical studies demonstrate a reduction of the mentioned diseases’ symptoms after the administration of anti-inflammatory drugs. Novel coumarin derivates have been shown to elicit anti-neuroinflammatory effects via G-protein coupled receptor GPR55, with possibly reduced side-effects compared to the known anti-inflammatory drugs. In this study, we, therefore, evaluated the anti-inflammatory capacities of the two novel coumarin-based compounds, KIT C and KIT H, in human neuroblastoma cells and primary murine microglia. Both compounds reduced PGE2-concentrations likely via the inhibition of COX-2 synthesis in SK-N-SH cells but only KIT C decreased PGE2-levels in primary microglia. The examination of other pro- and anti-inflammatory parameters showed varying effects of both compounds. Therefore, the differences in the effects of KIT C and KIT H might be explained by functional selectivity as well as tissue- or cell-dependent expression and signal pathways coupled to GPR55. Understanding the role of chemical residues in functional selectivity and specific cell- and tissue-targeting might open new therapeutic options in pharmacological drug development and might improve the treatment of the mentioned diseases by intervening in an early step of their pathogenesis.
1. Introduction
A growing body of research demonstrates the enormous role of neuroinflammation in neurological and psychiatric diseases, such as Alzheimer’s Disease (AD), Parkinson’s Disease (PD), schizophrenia, and depression [1,2,3,4]. Therefore, pharmacological mechanisms targeting neuroinflammation in the treatment of the mentioned diseases might open new therapeutical options. A meta-analysis, covering 36 randomized clinical trials (RCTs) and a total of about 10,000 patients, showed improvement of depressive symptoms after an intervention with specific anti-inflammatory substances, such as non-steroidal anti-inflammatory drugs (NSAIDs) and glucocorticoids, as monotherapy or added to a classic pharmaceutical anti-depressive therapy compared to placebo [5]. For AD, the effects of anti-neuroinflammatory treatment on the progress of neurodegeneration are discussed controversially due to differing results between epidemiological and clinical studies. Nevertheless, new approaches based on statistical modelling using the Alzheimer’s Disease Neuroimaging Initiative showed better cognitive baselines after treatment with NSAIDs and especially diclofenac treatment reduced cognitive decline [6]. PD is associated with neuroinflammation as well, and for this reason, anti-inflammatory treatment with NSAIDs decelerate the disease’s progression and protects remaining dopaminergic neurons [7]. However, these drugs are associated with severe side-effects in their established indications, limiting the period and dose of treatment. A total of 10% of hospitalizations of elderly people are related to preventable drug side-effects, with NSAIDs being responsible for 30% of these avoidable drug reactions. A total of 6% of the patients taking NSAIDs experience side-effects with the need for a consultation of a general doctor within 2 months [8]. Glucocorticoids, for example, impair mitochondrial functions and therefore might foster neurodegeneration due to increased oxidative stress [9]. Therefore, novel anti-inflammatory compounds with fewer potential harmful side-effects might promote the use of anti-inflammatory drugs in diseases such as AD, PD, and depression. The underlying mechanisms of the positive effects of NSAIDs on the mentioned diseases, such as inhibition of mitochondrial Ca2+ overload beside the known cyclooxygenase (COX) inhibition, are still objects of ongoing studies [7,10]. The COX enzymes are mediating the enzymatic conversion of arachidonic acid (AA) to prostaglandin (PG) H2, which is in turn metabolized to PGE2 by different prostaglandin E synthases (PGES), such as microsomal (m)PGES-1 or mPGES-2 [11]. PGE2 is known to act as pro-inflammatory in tissues and the CNS and increased levels are linked to psychiatric disorders such as psychosis [12], and therefore its reduction by NSAID treatment might be beneficial in neuropsychiatric diseases [5] as mentioned above.
Coumarin-based derivates have been shown to elicit anti-neuroinflammatory effects in primary microglia, decreasing lipopolysaccharide (LPS)-induced PGE2 release [13,14]. High levels of PGE2 promote brain injury and therefore act as a pro-inflammatory driver [15]. It has been suggested that the effects of coumarin derivates rely on antagonism with inverse agonistic activity at the G-protein coupled receptor 55 (GPR55) [13,14,16]. A recent study evaluating the effects of the two coumarin derivates KIT C and KIT H, which are investigated in this current study as well, showed anti-oxidative effects of both compounds, which were abolished after GPR55 knockout in SK-N-SH cells [17].
The GPR55 was discovered and cloned in 1999 [18] and is broadly expressed in the CNS, especially in the frontal cortex, putamen, striatum, and caudate [19]. Different studies identified numerous exogenous as well as endogenous ligands of the receptor. Besides ligands known to interact with receptors of the endocannabinoid system, such as delta9-tetrahydrocannabinol (Δ9-THC), L-α-lysophosphatidylinositol (LPI) has been demonstrated to have a strong affinity to GPR55 and is therefore suggested as an endogenous ligand of GPR55 [20]. Besides the endogenous ligands, synthetic selective GPR55 ligands, such as O-1602, acting as an agonist, and the antagonistic ML 193, have been introduced and are commonly used in GPR55 studies, among others [19]. Furthermore, derivates of the coumarin scaffold have been synthesized and suggested to act as antagonists on GPR55 [16,21]. Our group demonstrated the anti-neuroinflammatory effects of different coumarin-based compounds with binding affinities to GPR55 named KIT 10 [14], KIT 3, KIT 17, and KIT 21 [13], showing anti-neuroinflammatory properties in primary microglial cell cultures.
The GPR55-mediated biological effects remain the focus of current research. However, GPR55 agonism might be associated with negative effects for the cells, tissues, and, consequently, the organism itself. Agonism and overexpression of GPR55 are associated with cancer proliferation [22], metabolic diseases, such as obesity and diabetes [23], and decreased osteoclast formation [24]. Therefore, GPR55 antagonists might reverse negative GPR55-mediated effects and open new therapeutical options in the treatment of several diseases. Various in vivo and in vitro studies with central nervous cells or tissues and model organisms are focusing on the effects of GPR55 expression and antagonists in different conditions and diseases. A model for AD, 5xFAD-mice, showed a higher expression of GPR55 in the hippocampus compared to heterozygotic and wildtype mice with impairments in novel object recognition [25]. In the chemically-induced murine PD model, chronic abnormal cannabidiol (GPR55 agonist) treatment improved motoric functions and acted neuroprotectively [26]. GPR55 agonists, as well as antagonists, enfolded beneficial effects on motor coordination and sensorimotor deficits on 6-hydroxydopamine-induced PD symptoms in rats [27], suggesting a more complex role of GPR55 in PD. Furthermore, intrahippocampal administration of the GPR55 agonist O-1602 protected against LPS-induced inflammatory insults of neural stem cells [28]. In another study, intracerebroventricular injection of O-1602 induced anxiolytic effects in an elevated plus-maze test in rats, whereas ML 193 led to increased anxiety-like behavior [29]. In the corticosterone-induced depressive-like behavior of rats, O-1602 reversed depressive-like behavior and normalized increased levels of interleukin (IL)-1β and tumor necrosis factor (TNF)α [30]. GPR55-knockout mice repealed hyperalgesia to mechanical stimuli suggesting GPR55 to be a promising target for treating inflammatory and neuropathic pain [31]. The featured studies indicate a complex role of GPR55 in neurological and psychiatric diseases, with the agonism, as well as the antagonism, being beneficial dependent on the concrete situation. Association of a GPR55-mutation with psychiatric diseases has been shown in human clinical trials as well. In suicide victims without any diagnosed mental illness, decreased GPR55 and CB2 gene expression with increased GPR55-CB2 heteromers were found in the dorsolateral prefrontal cortex (DLPFC), eliciting a potential involvement of GPR55 in impulsivity and decision-making in suicide [32]. The single nucleotide polymorphism Gly195Val of the GPR55 is associated with an increased risk of Anorexia nervosa (AN) in a study comparing Japanese AN-patients with an age-unmatched control group [33].
GPR55 transduces extracellular signals via Gα12/13 [34] and Gαq [35], resulting in the phosphorylation and activation of phospholipase C, protein kinase C (PKC), mitogen-activated protein kinases (MAPK) such as p38 MAPK, and extracellular signal-regulated kinase (ERK), followed by the activation of transcriptional factors [19]. The activation of the different pathways is complexly regulated and might differ between various ligands [36]. These phenomena might be explained by different primary active states of one receptor as a response to different ligands resulting in distinct conformations responsible for the selective pathway activation, also referred to as functional selectivity [37,38]. As shown in a previous study [13], comparing the three coumarin-based compounds, KIT 3, KIT 17, and KIT 21, the effects on PGE2-reduction of the compounds show enormous differences probably dependent on the chemical residues, which might be explained by functional selectivity for antagonists. In contrast to GPR55 agonists, GPR55 antagonists are defined by a head region with the most electronegative region near the end of the central portion, whereas agonists have the electronegativity in the head region. Furthermore, GPR55 antagonists show an aromatic or heterocyclic ring that protrudes out of the binding pocket of GPR55, potentially preventing any conformational change [19]. Therefore, the different residues of the tested coumarin-based compounds [13,14] might determine how deep the compounds fit in the binding pocket and therefore, how potent the compounds might change the receptors state following the extent of the biological effects.
For this current study, two novel synthesized coumarin derivates named KIT C and KIT H were investigated in human neuroblastoma cells and in primary microglial cell cultures of mice. The effects of KIT C and KIT H on the COX-2/PGE2 pathway and pro- and anti-inflammatory mediators were evaluated in comparison to the commercial GPR55 agonist O-1602 and antagonist ML 193.
2. Results
2.1. Effects of the Compounds on Cell Viability
Results of the performed MTT cell viability assay for the used compounds are presented in Figure 1. Neither KIT C (light grey bars) or KIT H (dark grey bars), as shown before [17], nor O-1602 (light blue bar) or ML 193 (blue bar) showed cytotoxic effects in IL-1β-stimulated SK-N-SH cells compared to untreated cells. KIT C in concentrations of 5 and 10 µM, 1 µM KIT H, and 25 µM ML 193, on the contrary, significantly increased cell viability or metabolism. Ethanol, used as positive control, strongly induced cell death as expected. Since none of the compounds in the concentrations tested elicited cytotoxic effects, we proceeded with further experiments.
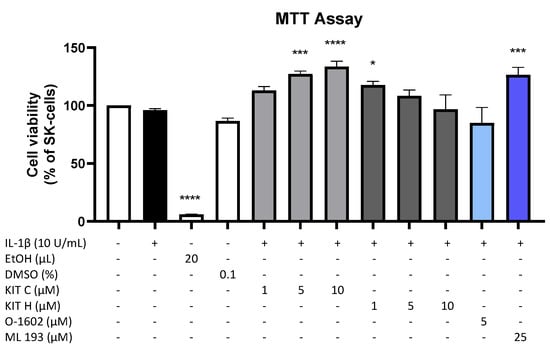
Figure 1.
Effects of KIT C (light grey bars), KIT H (dark grey bars), O-1602 (light blue bar), and ML 193 (blue bar) on cell viability in IL-1β-stimulated SK-N-SH cells (24 h treatment). Cell viability was measured by change in color due to MTT-oxidation, and absorbance was measured at 595 nm using an ELISA-reader. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunett’s post hoc tests with * p < 0.05, *** p < 0.001, **** p < 0.0001 compared to untreated cells. The figure is derived from our previous publication [17].
2.2. Effects of the Compounds on IL-1β-Induced PGE2-Release
Since PGE2 is the central molecule in the AA/COX-2/PGE2 pathway and acts pro-inflammatory, next we investigated the effects of KIT C, KIT H, O-1602, and ML 193 on PGE2-release in IL-1β-stimulated SK-N-SH cells. KIT C (light grey bars), as well as KIT H (dark grey bars), showed a significant and concentration-dependent reduction of IL-1β-induced PGE2-levels (Figure 2) starting at concentrations of 5 µM. KIT H elicited a more potent PGE2-reduction than KIT C, reaching basal PGE2-concentrations of untreated cells in the concentration of 25 µM. ML 193 (blue bar), a GPR55 antagonist, also showed significant inhibition of IL-1β-mediated PGE2-release, with an effect size between KIT C and KIT H. O-1602 (light blue bar), a potent GPR55 agonist, did not significantly inhibit IL-1β-induced PGE2-synthesis.
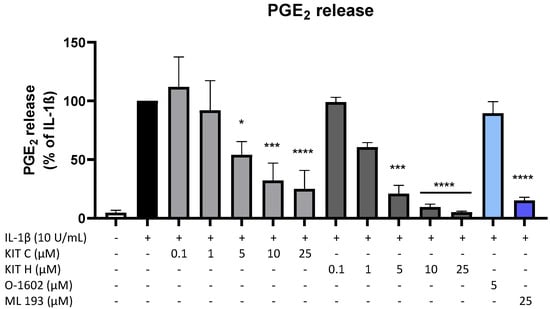
Figure 2.
Effects of KIT C (light grey bars), KIT H (dark grey bars), O-1602 (light blue bar), and ML 193 (blue bar) on PGE2-release in IL-1β-stimulated SK-N-SH cells. Cells were stimulated as described under material and methods. After 24 h of stimulation, supernatants were collected and the release of PGE2 was measured by EIA. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc tests with * p < 0.05, *** p < 0.001, **** p < 0.0001 compared to IL-1β.
2.3. GPR55 Activity of KIT C and KIT H
To prove whether the observed anti-inflammatory effects of KIT C and KIT H are mediated via GPR55, a GPR55 activation assay was performed (Figure 3). AM251 (1 µM), a GPR55 agonist with additional activities at CB1- and CB2-receptors, and LPI (10 µM), the physiological agonist of GPR55, were used as positive controls. KIT C in concentrations of 5 and 10 µM showed about a 4-fold higher GPR55 activation than 1 µM AM251 without reaching significance but showing a clear trend. KIT H revealed about a 2-fold but not significantly higher GPR55 activation than 1 µM AM251 in all tested concentrations, comparable to the GPR55 activation capacity of 10 µM LPI.
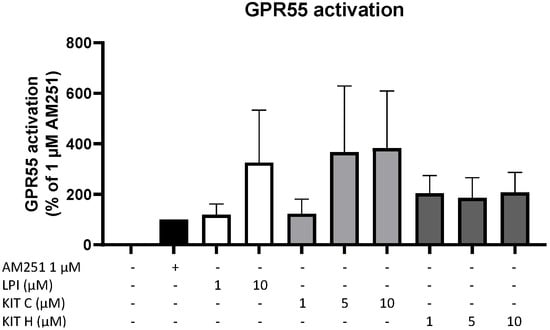
Figure 3.
GPR55 activation by AM251 (black bar), LPI (white bars), KIT C (light grey bars), and KIT H (dark grey bars) in HEK293T-GPR55 cells. Cells were treated as described under material and methods. After 6 h of stimulation, cells were lysed, and the luciferase activity was measured. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc tests compared to 1 µM AM251.
2.4. Effects of KIT C and KIT H on COX-2 mRNA and Protein Levels
To investigate the underlying mechanisms of the strong PGE2-reduction, COX-2 expression and synthesis were evaluated using Western Blot (Figure 4A) and qPCR (Figure 4B). COX-2 protein synthesis was potently increased by IL-1β if compared to the untreated control. Pre-treatment with exclusively 5 µM KIT C (light grey bars) reduced IL-1β-stimulated COX-2 levels in SK-N-SH cells. KIT H (dark grey bars) significantly reduced IL-1β-mediated COX-2 synthesis starting with the concentration of 1 µM. As shown in Figure 4B, COX-2 mRNA expression was potently induced by IL-1β-stimulation for 4 h. Whereas KIT H (dark grey bars) did not affect IL-1β-induced COX-2 expression, KIT C (light grey bars) significantly enhanced IL-1β-induced COX-2 expression in concentrations of 0.1, 5, and 10 µM which contrasts with the Western Blot results. Treatment with KIT C for different time points (2, 4, 8, 12, 24 h) followed by the analysis of COX-2 protein synthesis and mRNA expression did not explain the observed diverging effects on COX-2 synthesis and expression (Supplementary Figure S1), so we can exclude effects based on differences in the IL-1β-stimulation time course. COX-2 mRNA expression was higher or at least comparable to the IL-1β positive control at all time points, whereas COX-2 protein levels were first detectable after 8 h at higher levels than IL-1β-stimulated cells and started to decrease after 12 h of stimulation. After 24 h, COX-2 expression remained comparable to or higher than IL-1β-treated cells, while protein levels were significantly reduced as shown in Figure 4.
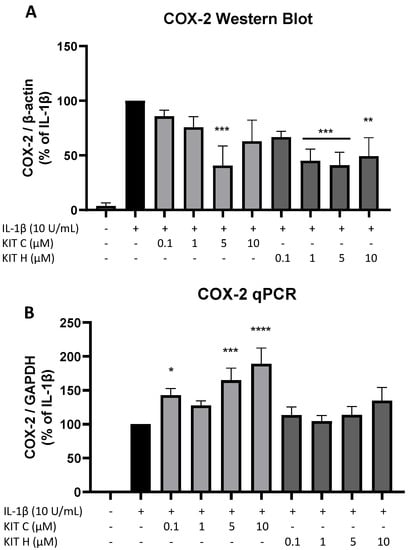
Figure 4.
Effects of KIT C (light grey bars) and KIT H (dark grey bars) on COX-2 protein levels (A) and COX-2 mRNA expression (B) in IL-1β-stimulated SK-N-SH cells. Cells were stimulated and analyzed as described under material and methods. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc tests with * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 compared to IL-1β.
2.5. Effects of KIT C and KIT H on COX-Activity
Besides COX synthesis and expression, enzyme activities of COX-1 (Figure 5A) and COX-2 (Figure 5B) were examined as another possible mechanism of PGE2-reduction independent of COX synthesis and expression. Neither KIT C (light grey bars) nor KIT H (dark grey bars), the GPR55 agonist O-1602, and antagonist ML 193 affected COX-1 or COX-2 activities in concentrations between 0.1 and 10 µM. Both COX inhibitor controls potently decreased COX activities. The selective COX-1 inhibitor SC-560 decreased COX-1 activity by about 70% but did not reach significance. The COX-1 and COX-2 inhibitor diclofenac in concentrations of 0.1 and 1 µM significantly reduced COX-2 activity.
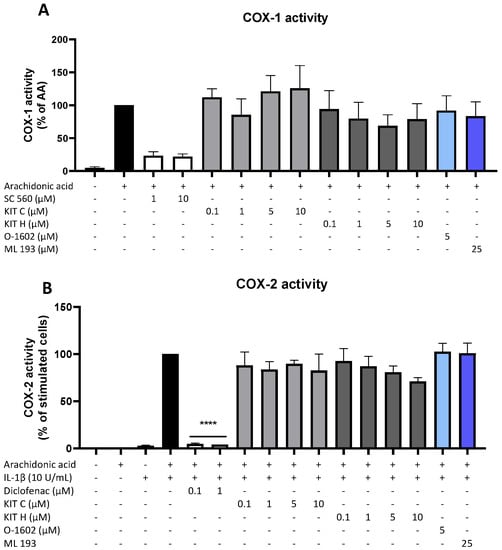
Figure 5.
Effects of KIT C (light grey bars), KIT H (dark grey bars), O-1602 (light blue bar), and ML 193 (blue bar) on COX-1 (A) and COX-2 enzyme activities (B) in SK-N-SH cells. (A) COX-1-activity was measured after 15 min of incubation with arachidonic acid (AA). Levels of PGE2 in the supernatants were quantified by EIA. (B) After 24 h of IL-1β pre-stimulation, 15 µM of arachidonic acid (AA) was added and PGE2-release was measured by EIA. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was carried out by using one-way ANOVA with Dunnett’s post hoc tests with **** p < 0.0001 compared to AA (A) or IL-1β with AA (B).
2.6. Effects of KIT C and KIT H on COX-1 and mPGES-1 Expression
Next, we studied the effects of KIT C (light grey bars) and KIT H (dark grey bars) on two other important enzymes involved in the AA/PGE2 pathway, COX-1 and mPGES-1. The expression of both enzymes was evaluated using qPCR. The expression of mPGES-1 (Figure 6A) was strongly induced by IL-1β-treatment for 4 h and 10 µM of KIT C slightly but significantly increased mPGES-1 expression compared to the IL-1β positive control. KIT H did not affect IL-1β-stimulated mPGES-1 expression. COX-1 expression (Figure 6B) was decreased by stimulation with IL-1β, and KIT C, as well as KIT H, partially ameliorated the IL-1β-induced reduction of COX-1 expression.
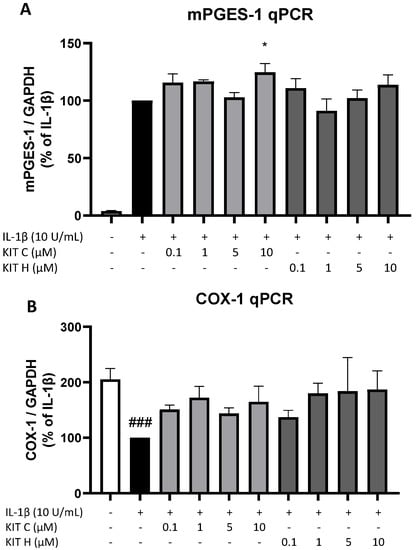
Figure 6.
Effects of KIT C (light grey bars) and KIT H (dark grey bars) on mPGES-1 (A) and COX-1 mRNA expression (B) in IL-1β-stimulated SK-N-SH cells. Cells were stimulated as described under material and methods. After 4 h of stimulation, RNA was isolated and mRNA levels of the shown target genes were measured using qPCR. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc tests with * p < 0.05 and ### p < 0.001 compared to IL-1β (A) or to untreated cells (B).
2.7. Effects of KIT C and KIT H on IL-1β–Induced Cytokine Release
Besides the AA/PGE2 pathway, the effects of KIT C (light grey bars) and KIT H (dark grey bars) on IL-1β-induced IL-6 as pro-inflammatory and IL-10 as anti-inflammatory cytokines were investigated (Figure 7). Stimulation with IL-1β for 24 h potently induced IL-6 release in SK-N-SH cells but neither KIT C nor KIT H nor O-1602 affected IL-6 production (Figure 7A). The GPR55 antagonist ML 193, however, significantly reduced IL-1β and increased IL-6 release by about 60% in IL-1β-treated SK-N-SH cells.
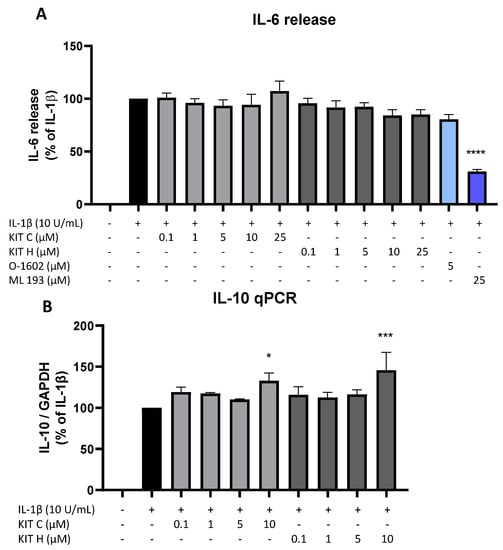
Figure 7.
Effects of KIT C (light grey bars), KIT H (dark grey bars), O-1602 (light blue bar), and ML 193 (blue bar) on IL-6 release (A) and effects of KIT C and KIT H on IL-10 mRNA expression (B) in IL-1β-stimulated SK-N-SH cells. Cells were stimulated as described under material and methods. After 24 h of stimulation, supernatants were collected and the release of IL-6 was measured by ELISA (A). After 4 h of stimulation, RNA was isolated and the mRNA levels of the shown target genes were measured using qPCR. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc tests with * p < 0.05, *** p < 0.001 and **** p < 0.0001 compared to IL-1β.
IL-1β reliably induced IL-10 mRNA expression in SK-N-SH cells as shown by qPCR, and KIT C as well as KIT H both enhanced IL-1β-stimulated IL-10 mRNA levels in concentrations of 10 µM compared to IL-1β the positive control (Figure 7B).
2.8. Effects of KIT C and KIT H on PGE2- and IL-6 Release in LPS-Stimulated Primary Mouse Microglia
The promising results of KIT C and KIT H in SK-N-SH cells were re-evaluated in primary mouse microglia as preliminary results for follow-up studies. Only KIT C (light grey bars) but not KIT H (dark grey bars) significantly reduced PGE2-levels after LPS-induction in primary microglia (Figure 8A). A total of 10 µM KIT C reduced PGE2 levels to concentrations compared to untreated primary microglia.
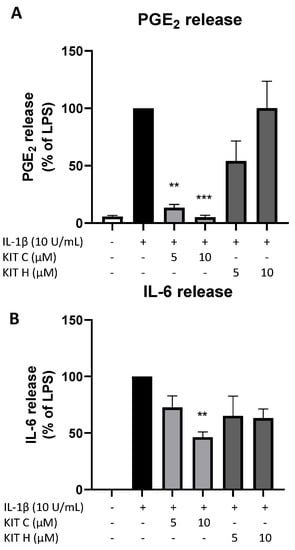
Figure 8.
Effects of KIT C (light grey bars) and KIT H (dark grey bars) on PGE2- (A) and IL-6-synthesis (B) in LPS-stimulated primary mice microglia. Cells were stimulated as described under material and methods. After 24 h of stimulation, supernatants were collected and the release of PGE2 or IL-6 was measured by EIA/ELISA. Values are presented as the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc tests with ** p < 0.01 and *** p < 0.001 compared to LPS.
LPS-stimulation for 24 h potently induced IL-6 production (Figure 8B) in primary mice microglia as well. KIT C (light grey bars) significantly decreased LPS-induced IL-6-synthesis by about 50% and KIT H (dark grey bars) showed a non-significant trend of reducing IL-6 release in primary mice microglial cells.
3. Discussion
The current study investigates the anti-neuroinflammatory effects of two novel coumarin derivates, KIT C and KIT H, in SK-N-SH cells as well as in primary microglial cell cultures. Selected results of KIT C and KIT H were compared to the commercial GPR55 agonist O-1602 and the GPR55 antagonist ML 193. None of the tested compounds showed toxic effects in SK-N-SH cells. KIT H demonstrated the most potent reduction of IL-1β-induced PGE2-levels compared to KIT C and ML 193, whereas O-1602 did not affect PGE2-levels. In primary mice microglia, KIT C decreased LPS-induced PGE2-release, whereas KIT H did not affect PGE2-levels significantly. Although KIT C increased IL-1β-induced COX-2 expression in SK-N-SH cells, both coumarin derivates decreased COX-2 protein synthesis. Neither COX-1 nor COX-2 enzymatic activity was affected by the tested compounds and thus, the PGE2 decreasing effects are not due to direct inhibition of those enzymes as shown for classical NSAIDs. Furthermore, the IL-1β-mediated reduction of COX-1 expression was partially reversed by KIT C and KIT H. In SK-N-SH cells, ML 193 significantly decreased IL-1β induced IL-6 levels, but neither KIT C nor KIT H or O-1602 affected IL-6 release. In primary microglia, however, KIT C but not KIT H significantly inhibited LPS-induced IL-6 release. IL-1β-mediated IL-10 expression was significantly increased by KIT C and KIT H in SK-N-SH cells.
The anti-neuroinflammatory effects of other coumarin derivates have been shown using different coumarin derivates (KITs) in primary rat microglia, potently inhibiting LPS-induced PGE2-release [13,14]. Our group showed a strong reduction of PGE2-synthesis, COX-2 gene expression, and protein synthesis as well as mPGES-1 protein synthesis after treatment with KIT 17 [13]. KIT 10 also reduced COX-2 and mPGES-1 protein synthesis [14]. Neither KIT 17 nor KIT 10 affected COX-2 enzymatic activity, whereas KIT 17 but not KIT 10 increased COX-1 activity [13,14]. The demonstrated reduction of COX-2 protein synthesis by KIT 17 was replicated in the current study for KIT C and KIT H. Since COX-2 is a key step in the enzymatic transformation of arachidonic acid to PGH2 [39], the reduction of COX-2 protein levels might be at least partially responsible for the observed PGE2-reduction. mPGES-1, the enzyme responsible for the final step in the synthesis of PGE2 out of PGH2 [40], could not be investigated on protein levels in the current study, since SK-N-SH cells showed a high basal mPGES-1 signal (data not shown). This might be explained by the cross reaction of the available antibodies, potentially binding to cPGES or mPGES-2 as well and, therefore, pretending high basal mPGES-1 protein levels. Thus, the observed reduction of PGE2 in SK-N-SH cells is likely and at least in part mediated by decreased COX-2 protein levels. The role of mPGES-1 in the observed decrease of PGE2 after KIT C and KIT H pretreatment, however, needs to be evaluated in future studies. Since COX-1 expression was not significantly affected by KIT C or KIT H compared to the untreated control, gastrointestinal ulcers as a side effect of COX-1 inhibition in vivo may be prevented. However, COX-1 protein levels were not examined in the current studies. The observed effects of KIT C and KIT H in LPS-stimulated primary mice microglia are likely to be mediated by similar mechanisms as in IL-1β-stimulated SK-N-SH cells. Therefore, further studies are necessary to investigate the effects of coumarin derivates on COX-1 protein synthesis and gastrointestinal effects as well as the responsible pathways and mechanisms in primary microglia. The reduction of the pro-inflammatory IL-6 in LPS-stimulated primary microglia and the enhanced expression of the anti-inflammatory IL-10 in IL-1β-stimulated SK-N-SH cells after treatment with KIT C and KIT H supports the anti-inflammatory potential of these coumarin compounds. Since the results of KIT C and KIT H in SK-N-SH cells as well as the preliminary data in primary mice microglia are promising, future experiments with both compounds using animals are ethically justifiable.
The signal transduction induced by GPR55 activation and targeted by KIT C and H resulting in the shown COX-2 protein reduction might involve different downstream pathways associated with the GPR55. We analyzed different downstream pathways of the GPR55 using KIT 10 in primary microglia demonstrating no effects on the mitogen activated protein kinases (MAPK)-pathway but a reduced LPS-induced IκB-α phosphorylation [14]. As shown in previous publications, PGE2-suppression as well as decreasing COX-2 protein levels can be achieved by inhibiting NF-κB- and MAPK-pathways [41]. For the current study, different pathways, such as MAP-kinases (phosphor-Erk1/2, p38 MAPK, SAPK/Jnk), IκB-α, NF-κB, protein kinase C (PKC), nuclear factor of activated t-cells (NFAT), nuclear growth factor (NGF), and brain-derived neurotrophic factor (BDNF) were examined, but neither KIT C nor KIT H significantly affected the investigated pathways (data not shown).
The inhibition of oxidative stress/isoprostane pathways independent of COX-2 might be responsible for the reduction of PGE2 by KIT C and KIT H as well, as discussed in our previous study [42] showing that COX enzymatic activity is enhanced by antioxidative stress [43]. In this and a previous study, KIT C as well as KIT H decreased oxidative stress and 8-Iso-PGF2α synthesis via GPR55 coupled to the inhibition of PGE2 and therefore possibly affecting the AA pathway by reduced COX-activity [17]. However, COX enzymatic activity is not significantly affected by treatment with KIT C or KIT H in the current study. Inhibition of phospholipase A2 (PLA2) reduces the concentration of available AA in the cells; therefore, PGE2-levels are reduced as a consequence [44]. In the current study, we did not assess PLA2 expression, protein levels, or activity, so we are not able to exclude a PLA2 dependent mechanism of the PGE2-suppression. For this reason, the molecular targets of KIT C and KIT H leading to PGE2/COX-2 reduction need to be further investigated in future studies.
In contrast to the decrease of COX-2 protein levels in 24-h IL-1β-stimulated SK-N-SH cells, mRNA concentrations were significantly increased due to the treatment with KIT C and as tendence after treatment with KIT H in 4-h stimulated cells. We conducted a time curve with 10 µM of both compounds, investigating protein as well as mRNA levels after 2, 4, 8, 12 and 24 h of IL-1β-stimulation. After 2 and 4 h of stimulation, we were not able to detect COX-2 by Western Blot. Protein levels of KIT C and KIT H pretreated cells were increased compared to IL-1β only treated cells at 8 (KIT C and KIT H) and 12 h (KIT C only). KIT H pretreated cells showed less COX-2 protein after 12 h of stimulation. COX-2 mRNA levels were significantly increased by both KITs after only 4 h of stimulation but were higher or at least comparable to the IL-1β positive control at all time points. Therefore, the difference between mRNA levels and protein levels of COX-2 cannot be explained by the dynamic over time. It has been shown that COX-2 mRNA stability is mediated by Erk1/2 and can be affected by G-protein coupled receptors (GPCRs). The Kaposi sarcoma virus oncogenic protein (vGPCR), for example, is associated with COX-2 overexpression and enhances mRNA stability of COX-2 [45]. Reduced mRNA-stability, however, leads to an earlier degradation of COX-2 mRNA, so even higher levels of mRNA might not usher high levels of COX-2 protein, because the mRNA is shorter and available for translation. Since the inhibition of pathways such as Erk1/2 coupled to the GPR55 has been shown to affect mRNA stability, this could be a possible explanation for increased COX-2 mRNA levels not leading to increased protein levels. However, phosphorylation of Erk1/2 is not affected by treatment with KIT C or KIT H in SK-N-SH-cells (not shown). Another mechanism leading to reduced COX-2 protein levels with increased mRNA concentrations might be a ubiquitin-mediated degradation of COX-2 protein. It has been shown that Centromere Protein U (CENPU) reduces ubiquitination as well as degradation of COX-2 in breast cancer [46]. Even if CENPU-dependent affection of COX-2 degradation is not likely to be responsible for the observed effects of KIT C and KIT H on COX-2 protein and mRNA levels, the mechanism of enhanced COX-2 degradation might be triggered by other pathways such as protein kinase C. Further studies are needed to elucidate the underlying mechanisms of the observed effects of KIT C and H on the AA pathway.
The observed COX-2 protein levels as well as PGE2 reduction after 24 h of IL-1β-stimulation is not accompanied by a reduction of COX-1 or COX-2 enzymatic activity. The enzymatic activity was measured after 30 min of treatment with KIT C and KIT H, so the enzymatic activity is independent of changes to protein levels of COX-1 or COX-2, as demonstrated in the time curve (supplementary Figure S1). The time curves demonstrate that changes of COX-2 levels can be found earliest after 8 h. The results show that KIT C and KIT H do not specifically bind and inhibit COX-1 or COX-2 enzymes, but rather affect the enzyme synthesis via pathways coupled to the GPR55.
It has been shown and suggested that coumarin derivates (KITs) enfold their biological effects by GPR55 antagonisms with inverse agonistic activity at the GPR55 [13,14,16]. However, neither for KIT C nor for KIT H radioligand assays have been implemented. Instead, a GPR55 activation assay was performed demonstrating a biological activity of KIT C and KIT H at the receptor, comparable to the known agonists LPI and AM251. The GPR55 activation data support the hypothesis that KIT C and KIT H enfold their effects by acting via direct activation of GPR55. Therefore, we further studied the effects of the commercially available GPR55 agonist O-1602 as well as the GPR55 antagonist ML 193. Interestingly, the GPR55 antagonist ML 193, but not the agonist O-1602, showed comparable biological effects to KIT C and KIT H in most experiments, suggesting an antagonistic activity of ML 193 with additional inverse agonistic activity like KIT C and KIT H at the GPR55 as shown with the previously studied coumarin derivates [16,21]. In line with this hypothesis, the decrease of IL-1β-induced PGE2-levels after ML 193 treatment remains between the effect size of PGE2-inhibition due to KIT C and KIT H administration. Our previous study, investigating the anti-oxidative effects of KIT C and KIT H in SK-N-SH cells, furthermore demonstrated the GPR55-dependency of the effects by both compounds since the antioxidative effects of KIT C and KIT H were abolished after GPR55 knockout in SK-N-SH cells [17]. The molecular structure of ligands determines either agonistic or antagonistic (respective with inverse agonistic activity) effects at the receptor [19]. All KITs share the coumarin scaffold with different chemical residues, which might momentously change the biological effects via GPR55. This might be an explanation of the observed differences between the effect size of KIT C and KIT H in different experiments. In a previous study, the efficacy of KIT 3, KIT 17, and KIT 21 showed enormous differences in the magnitude of inhibition of LPS-induced PGE2-release [13], underlining the hypothesized importance of the chemical residues. The chemical residues and the distribution of electronegativity in the molecules determine the position of the molecule and the deepness of binding in the GPR55 binding pocket and, therefore, the triggered effects [19]. Furthermore, heterocyclic or aromatic residues are described as a common characteristic of GPR55 antagonists, protruding out of the binding pocket and stabilizing the receptors “Off”-conformation [19], which at least KIT H exhibits. Since we observe effects that suggest an inverse agonistic activity of the substances, the lack of the heterocyclic or aromatic ring of KIT C basically does not question its GPR55-affinity as antagonist with inverse agonistic activity. On a biomolecular level, functional selectivity and differences in conformational change [37,38] after the binding of the compounds to GPR55 due to the differences in the chemical residues might be the explanation for the different effects of the tested coumarin derivates. However, the underlying mechanisms of the observed differences of the effects of KIT C and KIT H cannot be answered by the recent study. Therefore, future research is necessary to identify the exact role of the chemical residues in the biological effectiveness. This might open new options in the pharmacological drug design based on the coumarin scaffold.
Interestingly, the effects of KIT C and KIT H vary dependent on the cell type used as well. For example, KIT C but not KIT H showed a significant reduction of IL-1β-induced PGE2 in primary mice microglia. In SK-N-SH cells, however, KIT H showed a stronger inhibition of PGE2 than KIT C. Since we compared two different types of cells, namely neuroblastoma (neuronal) and microglial cells, the observed digressing effects might be explained by differences in GPR55 density on the cell surface, coupled Gα proteins, and associated downstream pathways as shown for other GPCRs. For example, for muscarinic agonists such as carbachol, weak efficacy in combination with differing levels of receptor affinity leads to cell- or tissue-dependent selectivity at the muscarinic GPCRs [37]. Furthermore, the proposed differences in receptor density and G-protein coupling dependent on the cell cycle phase has been shown for calcitonin before [47]. Since the investigated cells take over different functions in the CNS, with microglia fulfilling a key function in immune defense and neuroinflammation [48], the different effects might be explained by the different physiological cell functions. Neurons are primarily affected by the microglial inflammatory response [48] and physiologically building neuronal networks for signal transduction and processing [49].
Another possible explanation for the observed cell-dependent effects of KIT C and KIT H might be the cells’ donor species. In this study, we compared human neuroblastoma cells with primary microglia cells of C57B1/6 WT mice. The cloning of the GPR55 showed 4.3, 7, and 10 kilobase (kb) mRNA transcripts in human brain tissue whereas the examination of the rat spleen revealed a 3.5 kb mRNA transcript [18]. A search in the NCBI Protein Database comparing GPR55-sequences evinced small differences in the number of amino acids. In humans, 319 amino acids form the GPR55, and in mice, the receptor is built by 327 amino acids. Therefore, the receptor most likely reveals differences between humans and mice, and for this reason, the same compound might elicit different effects depending on the species investigated. Species-selective effects in the endocannabinoid system have been shown for rats and mice investigating the cannabinoid receptor (CB)2. Rats and mice were treated with intranasal JWH-133, a selective CB2 agonist, and mice but not rats showed a reduction in the self-administration of intravenous cocaine as a consequence [50]. A third possible mechanism explaining the observed species-differences might be the expression and protein synthesis of the GPR55 as well as the coupled Gα proteins and pathways that may vary between species, as mentioned for cell-type selective effects earlier [37]. Further studies are necessary to distinguish the proposed mechanisms of the cell-/species-selective effects of KIT C and KIT H. Since studies in model organisms are often required before clinical studies can be implemented, differences between species are important to draw the right conclusions about doses and expected effects in humans.
The used concentrations of KIT C and KIT H as well as O-1602 and ML 193 were chosen based on previous experiences with similar coumarin derivates [13,14,17] as well as publications investigating the commercial GPR55 agonist and antagonist [51,52,53,54]. O-1602 is a potent agonist at the GPR55 with an EC50 of 13 nm [55], and ML 193 is a potent antagonist of the GPR55 with a GPR55-potency of 221 nm [29]. IC50 of other related coumarin-derivates such as KIT 10 [14] and KIT 17 [13] was between approximately 5 µM and 10 µM in radioligand-assays. The IC50 data of KIT 10 and Kit 17 were determined using a cellular system and was therefore in the µM range when compared to ML 193 that was determined using membrane preparation. Unfortunately, for KIT C and KIT H, no IC50 values are available, and for the GPR55 and the investigated compounds no Bmax or KD values are available. However, the used concentrations of O-1602 and ML-193 are much higher than the reported EC50 and IC50 values. Therefore, the observed effects for at least those two compounds might not only be GPR55-dependent but mediated by specific intermolecular actions. Specific interactions are defined as ligand-receptor-binding, whereas non-specific interactions can occur between ligands and other enzymes or proteins of the cells that are not the targeted receptor [56,57]. The used concentrations of KIT C and KIT H, in contrast, are at sufficient levels compared to the IC50 values of other coumarin based compounds. Moreover, for KIT C and KIT H, knockout of the GPR55 in SK-N-SH-cells, as described in a previously published paper, fully abolished observed effects of both coumarin compounds on oxidative stress-induced cell death [17]. This, and the data obtained from our GPR55 activity assay, support the hypothesis that effects of KIT C and KIT H are GPR55 dependent and not non-specific receptor-independent interactions, even if they have lower affinity to the GPR55 than the commercially available ligands used.
A growing body of research focuses on anti-inflammatory treatment in numerous neurological and psychiatric diseases. For depression, positive effects of NSAIDs and glucocorticoids were observed [5], but NSAIDs are associated with gastrointestinal ulcers, and long glucocorticoid treatments may cause numerous side effects, such as osteoporosis and an increase of infections [58]. Coumarin derivates with anti-inflammatory characteristics and possibly milder side effects might be an alternative to the already known anti-inflammatory drugs. Some authors support the hypothesis of the inflammatory genesis of depression by the fact that autoimmune diseases, as well as depression, are observed at about two times higher prevalence in women than in men [2]. Besides depression, anti-inflammatory drugs such as COX-2 inhibitors ameliorated psychiatric symptoms in schizophrenia, Borderline Personality Disorder, and obsessive-compulsive disorders, whereas NSAID treatment reduced the anti-depressive effects of selective serotonin reuptake inhibitors (SSRI) [4]. Furthermore, neuroinflammation is associated with neurological diseases, such as PD and AD, as well as neuroinflammation possibly disturbing the balanced interaction of different CNS cells and leading to neurodegeneration [3].
Anti-neuroinflammatory capacities have been shown for coumarin derivates in previous studies [13,14] as well as for KIT C and KIT H in the current study. Therefore, coumarin derivates such as KIT C as well as KIT H might be promising novel compounds in the treatment of diseases with (neuro)inflammatory etiologies and pathomechanisms. Further understanding of functional selectivity [37,38] at the GPR55 based on different chemical residues as well as species differences might improve pharmaceutical drug design for these diseases. The evaluation of KIT C and KIT H in animal disease models might improve the understanding of the potential of both compounds in the future treatment of neurological and psychiatric diseases and demonstrate possible side-effects of coumarin derivates.
4. Materials and Methods
4.1. Chemicals
KIT C and KIT H were synthesized as described previously [17] by the Karlsruher Institute for Technology (KIT), Institute of Organic Chemistry, dissolved in DMSO (Merck KGaA, Darmstadt, Germany) and used in final concentrations of 0.1–25 µM. The GPR55 agonist O-1602 and GPR55 antagonist ML 193 (both from Cayman Chemicals, Ann Arbor, MI, USA, distributed by BioMol, Hamburg, Germany) were dissolved in DMSO and used in final concentrations of 5 µM (O-1602) and 25 µM (ML 193). Human interleukin (IL)-1β (100,000 U/mL in phosphate buffered saline (PBS)) was purchased from Roche Diagnostics (Manheim, Germany) and was used at a final concentration of 10 U/mL for the experiments. 5 mg/mL lipopolysaccharide (LPS) from Salmonella typhimurium (Sigma-Aldrich GmbH, Taufkirchen, Germany) was dissolved in PBS as stock and diluted with distilled water for a final concentration of 10 ng/mL in primary microglia cultures.
Figure 9 shows the chemical structure of KIT C and KIT H that were already introduced in a previous paper [17], as well as the structures of O-1602 and ML 193 obtained from the manufacturer’s (Cayman Chemicals, Ann Arbor, MI, USA) website.
Figure 9.
Chemical structures of KIT C, KIT H, O-1602, and ML 193 as previously published [17], respectively, as provided by the Cayman Chemicals website (https://www.caymanchem.com (accessed on 6 January 2022)).
4.2. Human Neuroblastoma (SK-N-SH) Cell Culture
SK-N-SH neuronal cells were obtained from the American Type Culture Collection (HTB-11, Rockville, MD, USA) and approximately 2 × 106 cells were grown in 1× minimum essential medium (MEM) containing Earl’s salts, 10% fetal bovine serum (Bio & SELL GmbH, Feucht/Nürnberg, Germany), 1 mM l-glutamine, 1 mM sodium pyruvate, 2 mL of 100× MEM vitamin solution, 40 units/mL penicillin, 40 µg/mL streptomycin and 0.1 µg/mL fungizone® (all obtained from Gibco, Thermo Fisher Scientific, Bonn, Germany). Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2 in 75 cm2 cell culture flasks (Falcon, Heidelberg, Germany) with approximately 2 × 105 cells per cm2 when splitting the sub-cultures and grown to about 90% confluency. Confluent monolayers were passaged routinely by trypsinization. After trypsinization, cells were harvested and re-seeded into 6-, 12-, 24- or 96-well plates (between 2.8 × 105 and 2 × 104 cells depending on the plate size). On the next day, the medium was changed after 1 h, and cells were stimulated for respective experiments.
4.3. Primary Mouse Microglial Cell Cultures
4.3.1. Ethics Statement
Animals were obtained from the Center for Experimental Models and Transgenic Services-Freiburg (CEMT-FR). All the experiments were approved and conducted according to the guidelines of the ethics committee of the University of Freiburg Medical School under protocol No. X-19/06R and the study was carefully planned to minimize the number of animals used and their suffering.
4.3.2. Primary Mouse Microglia Cultures
Primary mouse mixed glia cultures were prepared from 2 to 3 days old C57B1/6 WT mice as described before [44,59,60,61]. Briefly, brains were carefully taken under sterile conditions and meninges were removed. The cortices were dissociated and filtered through a 70 µm nylon cell strainer (BD biosciences, Heidelberg, Germany). After centrifugation at 1000 rpm for 10 min the cells were resuspended in LPS-free Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS; Bio & SELL GmbH, Nürnberg/Feucht, Germany) and antibiotics (DMEM and anti-anti obtained from Gibco, Thermo Fisher Scientific, Bonn, Germany) and cultured in 10 cm cell culture dishes at a density of 5 × 105 cells/plate (Falcon, Heidelberg, Germany) in the humified atmosphere at 10% CO2 and 37 °C. After 12 days in vitro, floating microglia were harvested by gently shaking on an orbital shaker and re-seeded into 6-well plates with approximately 3 × 105 cells per well and an 80% survival rate to obtain pure microglial cell cultures. The protocol for preparing pure primary microglial cultures was established by Gebicke-Härter et al. in our laboratory and published in 1989. This publication demonstrated a purity of >98% for microglia using morphological features, immunofluorescence (monocyte/macrophage marker anti-CD68 = ED1), and cytochemical analysis [60]. On the next day, the medium was changed to remove non-adherent cells, and after 1 h, cells were stimulated for the experiments.
4.4. Cell Viability Assay
MTT assay (Sigma-Aldrich GmbH, Taufkirchen, Germany) was used for measuring the viability of SK-N-SH neuronal cells after treatment with KIT C (1, 5 and 10 µM), KIT H (1, 5 and 10 µM), O-1602 (5µM), or ML 193 (25 µM). This assay determines the number of metabolically active cells and allows conclusions about viable cells in the culture based on the reduction of a yellow tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide or MTT) to purple formazan in the cells. Briefly, cells were cultured in 96-well plates at a density of 25 × 103 cells/well for 24 h. The medium was changed and after at least 1 h, cells were pre-treated with different concentrations of the compounds for 30 min. Cells were then incubated with or without IL-1β for the next 20 h. 20 µL Ethanol (approximately 20% end conc.) was used to induce cell death as a positive control. Next, 20 µL of MTT-solution (working concentration 5 mg/mL) were added to all wells and incubated for another 4 h at 37 °C. Afterwards, the medium was removed and replaced with 200 µL of DMSO. The colorimetric reaction was measured using the MRXe Microplate reader (Dynex Technologies, Denkerdorf, Germany) at 595 nm.
4.5. Determination of PGE2-Release
SK-N-SH neuronal cells were pretreated with KIT C (0.1–25 µM), KIT H (0.1–25 µM), O-1602 (5 µM), or ML 193 (25 µM) for 30 min. Afterwards, cells were incubated for the next 24 h with or without IL-1β (10 U/mL) and supernatants were collected. The levels of PGE2 were measured using a commercially available enzyme immunoassay (EIA) kit (from Cayman Chemicals, Ann Arbor, MI, USA, distributed by BioMol, Hamburg, Germany) following the manufacturer’s protocol. The results were normalized to IL-1β and presented as a percentage of change in PGE2 levels of at least three independent experiments.
PGE2-release in primary microglia cultures was measured as described for SK-N-SH cells. Only KIT C (5 and 10 µM) and KIT H (5 and 10 µM) were evaluated in primary microglia and LPS (10 ng/mL) was used for stimulation instead of IL-1β. Supernatants were collected after 24 h and used in the EIA following the protocol. The results were normalized to LPS and presented as percentage of change in prostaglandin levels of at least three independent experiments.
4.6. Determination of GPR55 Agonistic Activity
The determination of GPR55 agonistic activity was carried out using HEK293T-GPR55 cells that overexpress the human GPR55 [62]. Briefly, HEK293T-GPR55 cells were cultured in 24-well plates (105 cells/well) and transiently transfected with 0.2 µg of the reporter plasmid CRE-Luc that contains six consensus cAMP responsive elements (CRE) linked to firefly luciferase reporter gene using Roti©-Fect (Carl Roth, Karlsruhe, Germany). Transfected cells were treated with increasing concentrations of either the test compounds KIT C and KIT H or the positive controls AM251 and LPI for 6 h. Then, the cells were washed twice with PBS 1× and lysed in 100 µL lysis buffer containing 25 mM Tris-phosphate (pH 7.8), 8 mM MgCl2, 1 mM DTT, 1% Triton X-100, and 7% glycerol for 15 min at room temperature in a horizontal shaker. Luciferase activity was measured using a TriStar2 Berthold/LB942 multimode reader (Berthold Technologies, Bad Wildbad, Germany) following the instructions of the luciferase assay kit (Promega, Madison, WI, USA). The relative light units (RLUs) were calculated, and the results were expressed as the percentage of activation over the control. The experiment was performed three times.
4.7. RNA Isolation and Quantitative PCR
For quantification of the mRNA of the enzymes of the COX-2/PGE2 pathway, we performed quantitative real-time PCR (qPCR) in SK-N-SH cells. Cultured cells were pretreated with KIT C or KIT H (0.1–10 µM) for 30 min, followed by stimulation with IL-1β (10 U/mL) for 4 h. For the COX-2 time curve, cells were incubated with IL-1β for 2, 8, 12 and 24 h, respectively. Total RNA was extracted using the GeneMATRIX Universal RNA Purification Kit (Roboklon GmbH, Berlin, Deutschland) according to the manufacturer’s protocol. Then, cDNA was reverse transcribed from 500 ng of total RNA in a 30 μL total reaction volume with an initial denaturation at 70 °C (10 min) with the following amplification cycle after the addition of the master mix. The qPCR amplification was carried out by the CFX96 real-time PCR detection system (Bio-Rad Laboratories GmbH, Feldkirchen, Germany). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an internal control for sample normalization. The primer sequences were GAPDH: Forward (Fwd): 5′-TGGGAAGCTGGTCATCAAC-3′/Reverse (Rev): 5′-GCATCACCCCATTTGATGTT-3′, COX-2: Fwd: 5′-CTTCACGCATTTCAAG-3′/Rev: 5′-TCACCGTAAAGTCCAC-3′, mPGES-1: Fwd 5′-TGCAGCACGCTGCTGGTCAT-3′/Rev 5′-GTCGTTGCGGTGGGCTCTGAG-3′, COX-1: Fw: 5′-TCCATGTTGGCTATGG-3′/Rv: 5′-GTGGTGGTCCTTCCTG-3′ and IL-10: Fwd: 5′-GATGCCTTCAAGTGAA-3′/Rev: 5′-GCAACCCAGGCTTAAA-3′. Primers were designed using Universal ProbeLibrary Assay Design Center (Roche Diagnostics, Mannheim, Germany) and obtained from biomers.net GmbH (Ulm, Germany).
4.8. Immunoblotting
SK-N-SH neuronal cells were pre-treated with KIT C and KIT H (0.1–10 µM) for 30 min. After 24 h (COX-2) of IL-1β-stimulation (10 U/mL), cells were washed with cold PBS and lysed mechanically in lysis buffer (42 mM Tris–HCl, 1.3% sodium dodecyl sulfate, 6.5% glycerin, 100 μM sodium orthovanadate, and 2% phosphatase and 0.2% protease inhibitors). For the COX-2 time curve, cells were incubated with IL-1β for 2, 4, 8 and 12 h, respectively. Protein concentrations of the samples were measured using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Bonn, Germany). For Western blotting, 20 μg of total protein from each sample were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions. Proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Darmstadt, Germany) by semi-dry blotting. After blocking with Roti-Block (Roth, Karlsruhe, Germany), membranes were incubated overnight with primary antibodies (mouse anti-COX-2 (MAB-4198, 1:1000; R&D systems, Wiesbaden, Germany), mouse anti-actin (sc-47778, 1:5000, Santa Cruz Biotechnology Inc., Heidelberg, Germany)). The proteins were detected with horseradish peroxidase-coupled sheep anti-mouse IgG (1:20,000 dilution; Amersham Biosciences GmbH, Freiburg, Germany) using enhanced chemiluminescence (ECL) reagents (Biozym, Hessisch Oldendorf, Germany) and taking pictures with the ChemiDoc MP imaging system (Bio-Rad Laboratories GmbH, Feldkirchen, Germany). Densitometric analysis was performed using ImageJ software (V1.48t, NIH, Bethesda, MD, USA).
4.9. Cyclooxygenase Activity Assay
COX enzymatic activity was investigated using the arachidonic acid assay, as described previously [63]. For COX-1 activity, neuroblastoma cells were plated in 24-well plates and after 24 h, the medium was removed and replaced with a serum-free medium. KIT C or KIT H (0.1–10 µM) or the selective inhibitor of COX-1 SC560 [(1 and 10 μM); Sigma-Aldrich GmbH, Taufkirchen, Germany] was added and left for 15 min. Then, arachidonic acid (15 µM; Sigma-Aldrich GmbH, Taufkirchen, Germany) was applied for another 15 min. Finally, supernatants were collected and used for the determination of PGE2 as described above.
For COX-2 enzymatic activity, the assay was conducted as for COX-1, but with pre-incubation of IL-1β (10 U/mL) for 24 h to induce COX-2 synthesis and using diclofenac sodium ((0.1 and 1 μM); Sigma-Aldrich GmbH, Taufkirchen, Germany) as commercial COX-2 preferential inhibitor.
4.10. Determination of IL-6 Release
Effects of KIT C and KIT H (0.1–25 µM) on IL-6 release in IL-1β-stimulated SK-N-SH cells and (concentrations 5 and 10 µM) in LPS-stimulated primary microglia were evaluated using ELISA. Commercially available InvitrogenTM eBioscienceTM human or mouse IL-6 ELISA Ready-SET-Go!TM Kits (Thermo Fisher Scientific, Bonn, Germany) were used following the manufacturer’s protocol. Briefly, cells were stimulated after pretreatment with KIT C or KIT H for 24 h with IL-1β (SK-N-SH cells) or LPS (primary microglial cells). Supernatants were collected and stored at −80 °C for further experiments. ELISA-plates (Nunc MaxiSorpTM; Thermo Fisher Scientific, Bonn, Germany) were coated with IL-6 capture antibody overnight. The next day, samples were added followed by the addition of an IL-6 determination antibody after removing supernatants and washing the plate. The amount of bound IL-6 determination antibody was quantified using an HRP-dependent colorimetric reaction. The absorbance of the wells was read at 450 nm using the MRXe Microplate reader and calculated as % of IL-1β or LPS after blank substraction.
4.11. Statistical Analysis
Raw values were converted to percentage and IL-1β (10 U/mL), LPS (10 ng/mL), or the appropriate negative control, such as untreated cells for MTT-assay, were considered as 100%. Data are represented as mean ± SEM of at least three independent experiments. The statistical comparisons were performed using one-way ANOVA with Dunett’s post hoc test (Prism 8 software, GraphPad software Inc., San Diego, CA, USA). The level of significance was set at * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 and is indicated in the figures.
5. Conclusions
Anti-inflammatory treatment of neurological and psychiatric diseases has been shown to abate disease’s symptoms in murine models and clinical studies. However, many available anti-inflammatory drugs are associated with momentous side effects. Therefore, the development of new pharmacological therapeutics might improve the treatment of these CNS diseases. KIT C and KIT H, likely enfolding their effects via GPR55, might be promising novel anti-inflammatory strategies for future interventions by decreasing pro-inflammatory PGE2, IL-6, and increasing anti-inflammatory IL-10. Future research is necessary to fully understand the involved mechanisms and effects of KIT C and KIT H in in vivo models and following clinical studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23020959/s1.
Author Contributions
B.L.F., S.H., C.N., J.S., S.W.S., T.H., S.B., E.M. and M.A. participated in research design. The experiments were performed by M.A., J.S., S.W.S., J.A.C., E.M. and T.H. Data were analyzed by M.A., J.S., E.M., J.A.C. and T.H., M.A., E.M. and B.L.F. wrote or contributed to the writing of the manuscript. In addition, M.A., J.S., S.W.S., S.G., T.H., E.M., J.A.C., S.B., C.N., S.H. and B.L.F. reviewed the data and discussed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The article processing charge was funded by the Baden-Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg Library in the funding program (Open Access Publishing).
Institutional Review Board Statement
All experiments using animals for the generation of microglia were approved and conducted according to the guidelines of the ethics committee of the University of Freiburg under protocol Nr. X-19/06A (approved on 9 October 2019).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this manuscript are available from the corresponding author upon request. The compounds used in this study were described before [17] and the data were submitted to the repository chemotion under the submission numbers CRS-9508 (KIT C) and CRS-9514 (KIT H). For a direct link to the data please refer to: KIT C (8-Isopropyl-3,5-dimethyl-2H-chromen-2-one): https://dx.doi.org/10.14272/LSYXPDGXGOSFCW-UHFFFAOYSA-N.1 (accessed on 14 January 2022) and KIT H (3-Benzyl-8-isopropyl-5-methyl-2H-chromen-2-one): https://dx.doi.org/10.14272/WNXYNMKVCJSJKO-UHFFFAOYSA-N.1 (accessed on 14 January 2022).
Acknowledgments
The skillful technical assistance of Ulrike Götzinger-Berger and Brigitte Günter in cell culture and EIA is greatly acknowledged. This project was supported by the core facility “Molecule Archive” of the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG project number: 284178167) and the Repository Chemotion (DFG, grant number 266379491). The authors thank the Baden-Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg Library for their support via the funding program « Open Access Publishing ».
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Craft, J.M.; Watterson, D.M.; Van Eldik, L.J. Neuroinflammation: A potential therapeutic target. Expert Opin. Ther. Targets 2005, 9, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Troubat, R.; Barone, P.; Leman, S.; Desmidt, T.; Cressant, A.; Atanasova, B.; Brizard, B.; El Hage, W.; Surget, A.; Belzung, C.; et al. Neuroinflammation and depression: A review. Eur. J. Neurosci. 2021, 53, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Coelho-Junior, H.J.; Landi, F.; Bernabei, R.; Marzetti, E. Mitochondrial Dysfunction, Oxidative Stress, and Neuroinflammation: Intertwined Roads to Neurodegeneration. Antioxidants 2020, 9, 647. [Google Scholar] [CrossRef] [PubMed]
- Najjar, S.; Pearlman, D.M.; Alper, K.; Najjar, A.; Devinsky, O. Neuroinflammation and psychiatric illness. J. Neuroinflamm. 2013, 10, 816. [Google Scholar] [CrossRef] [Green Version]
- Köhler-Forsberg, O.N.; Lydholm, C.; Hjorthøj, C.; Nordentoft, M.; Mors, O.; Benros, M.E. Efficacy of anti-inflammatory treatment on major depressive disorder or depressive symptoms: Meta-analysis of clinical trials. Acta Psychiatr. Scand. 2019, 139, 404–419. [Google Scholar] [CrossRef]
- Rivers-Auty, J.; Mather, A.E.; Peters, R.; Lawrence, C.B.; Brough, D. Anti-inflammatories in Alzheimer’s disease—Potential therapy or spurious correlate? Brain Commun. 2020, 2, fcaa109. [Google Scholar] [CrossRef] [PubMed]
- Esposito, E.; Di Matteo, V.; Benigno, A.; Pierucci, M.; Crescimanno, G.; Di Giovanni, G. Non-steroidal anti-inflammatory drugs in Parkinson’s disease. Exp. Neurol. 2007, 205, 295–312. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.; Robson, J. The dangers of NSAIDs: Look both ways. Br. J. Gen. Pract. J. R. Coll. Gen. Pract. 2016, 66, 172–173. [Google Scholar] [CrossRef]
- Choi, G.E.; Han, H.J. Glucocorticoid impairs mitochondrial quality control in neurons. Neurobiol. Dis. 2021, 152, 105301. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Blasco, S.; Valero, R.A.; Rodríguez-Crespo, I.; Villalobos, C.; Núñez, L. Mitochondrial Ca2+ Overload Underlies Aβ Oligomers Neurotoxicity Providing an Unexpected Mechanism of Neuroprotection by NSAIDs. PLoS ONE 2008, 3, e2718. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Trott, J.S.; Haque, S.; McCormick, S.; Chiorazzi, N.; Mongini, P.K.A. A Cyclooxygenase-2/Prostaglandin E Pathway Augments Activation-Induced Cytosine Deaminase Expression within Replicating Human B Cells. J. Immunol. 2010, 185, 5300–5314. [Google Scholar] [CrossRef] [Green Version]
- Moreno, C.; Parellada, M.; MacDowell, K.S.; García-Bueno, B.; Cabrera, B.; González-Pinto, A.; Saiz, P.; Lobo, A.; Rodriguez-Jimenez, R.; Berrocoso, E.; et al. Differences in the regulation of inflammatory pathways in adolescent- and adult-onset first-episode psychosis. Eur. Child Adolesc. Psychiatry 2019, 28, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Saliba, S.W.; Jauch, H.; Gargouri, B.; Keil, A.; Hurrle, T.; Volz, N.; Mohr, F.; van der Stelt, M.; Bräse, S.; Fiebich, B.L. Anti-neuroinflammatory effects of GPR55 antagonists in LPS-activated primary microglial cells. J. Neuroinflamm. 2018, 15, 322. [Google Scholar] [CrossRef] [PubMed]
- Saliba, S.W.; Gläser, F.; Deckers, A.; Keil, A.; Hurrle, T.; Apweiler, M.; Ferver, F.; Volz, N.; Endres, D.; Bräse, S.; et al. Effects of a Novel GPR55 Antagonist on the Arachidonic Acid Cascade in LPS-Activated Primary Microglial Cells. Int. J. Mol. Sci. 2021, 22, 2503. [Google Scholar] [CrossRef]
- Liu, Q.; Liang, X.; Wang, Q.; Wilson, E.N.; Lam, R.; Wang, J.; Kong, W.; Tsai, C.; Pan, T.; Larkin, P.B.; et al. PGE signaling via the neuronal EP2 receptor increases injury in a model of cerebral ischemia. Proc. Natl. Acad. Sci. USA 2019, 116, 10019–10024. [Google Scholar] [CrossRef] [Green Version]
- Rempel, V.; Volz, N.; Gläser, F.; Nieger, M.; Bräse, S.; Müller, C.E. Antagonists for the Orphan G-Protein-Coupled Receptor GPR55 Based on a Coumarin Scaffold. J. Med. Chem. 2013, 56, 4798–4810. [Google Scholar] [CrossRef]
- Apweiler, M.; Saliba, S.W.; Streyczek, J.; Hurrle, T.; Gräßle, S.; Bräse, S.; Fiebich, B.L. Targeting Oxidative Stress: Novel Coumarin-Based Inverse Agonists of GPR55. Int. J. Mol. Sci. 2021, 22, 11665. [Google Scholar] [CrossRef] [PubMed]
- Sawzdargo, M.; Nguyen, T.; Lee, D.K.; Lynch, K.R.; Cheng, R.; Heng, H.H.Q.; George, S.R.; O’Dowd, B.F. Identification and cloning of three novel human G protein-coupled receptor genes GPR52, ΨGPR53 and GPR55: GPR55 is extensively expressed in human brain. Mol. Brain Res. 1999, 64, 193–198. [Google Scholar] [CrossRef]
- Shore, D.M.; Reggio, P.H. The therapeutic potential of orphan GPCRs, GPR35 and GPR55. Front. Pharmacol. 2015, 6, 69. [Google Scholar] [CrossRef] [Green Version]
- Oka, S.; Nakajima, K.; Yamashita, A.; Kishimoto, S.; Sugiura, T. Identification of GPR55 as a lysophosphatidylinositol receptor. Biochem. Biophys. Res. Commun. 2007, 362, 928–934. [Google Scholar] [CrossRef]
- Rempel, V.; Volz, N.; Hinz, S.; Karcz, T.; Meliciani, I.; Nieger, M.; Wenzel, W.; Bräse, S.; Müller, C.E. 7-Alkyl-3-benzylcoumarins: A Versatile Scaffold for the Development of Potent and Selective Cannabinoid Receptor Agonists and Antagonists. J. Med. Chem. 2012, 55, 7967–7977. [Google Scholar] [CrossRef] [PubMed]
- Falasca, M.; Ferro, R. Role of the lysophosphatidylinositol/GPR55 axis in cancer. Adv. Biol. Regul. 2016, 60, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Navarrete, J.M.; Catalan, V.; Whyte, L.; Diaz-Arteaga, A.; Vazquez-Martinez, R.; Rotellar, F.; Guzman, R.; Gomez-Ambrosi, J.; Pulido, M.R.; Russell, W.R.; et al. The L-α-Lysophosphatidylinositol/GPR55 System and Its Potential Role in Human Obesity. Diabetes 2012, 61, 281–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whyte, L.S.; Ryberg, E.; Sims, N.A.; Ridge, S.A.; Mackie, K.; Greasley, P.J.; Ross, R.A.; Rogers, M.J. The putative cannabinoid receptor GPR55 affects osteoclast function in vitro and bone mass in vivo. Proc. Natl. Acad. Sci. USA 2009, 106, 16511–16516. [Google Scholar] [CrossRef] [Green Version]
- Medina-Vera, D.; Rosell-Valle, C.; López-Gambero, A.J.; Navarro, J.A.; Zambrana-Infantes, E.N.; Rivera, P.; Santín, L.J.; Suarez, J.; Rodríguez de Fonseca, F. Imbalance of Endocannabinoid/Lysophosphatidylinositol Receptors Marks the Severity of Alzheimer’s Disease in a Preclinical Model: A Therapeutic Opportunity. Biology 2020, 9, 377. [Google Scholar] [CrossRef] [PubMed]
- Celorrio, M.; Rojo-Bustamante, E.; Fernández-Suárez, D.; Sáez, E.; de Mendoza, A.E.-H.; Müller, C.E.; Ramírez, M.J.; Oyarzábal, J.; Franco, R.; Aymerich, M.S. GPR55: A therapeutic target for Parkinson’s disease? Neuropharmacology 2017, 125, 319–332. [Google Scholar] [CrossRef]
- Fatemi, I.; Abdollahi, A.; Shamsizadeh, A.; Allahtavakoli, M.; Roohbakhsh, A. The effect of intra-striatal administration of GPR55 agonist (LPI) and antagonist (ML193) on sensorimotor and motor functions in a Parkinson’s disease rat model. Acta Neuropsychiatr. 2021, 33, 15–21. [Google Scholar] [CrossRef]
- Hill, J.D.; Zuluaga-Ramirez, V.; Gajghate, S.; Winfield, M.; Sriram, U.; Rom, S.; Persidsky, Y. Activation of GPR55 induces neuroprotection of hippocampal neurogenesis and immune responses of neural stem cells following chronic, systemic inflammation. Brain. Behav. Immun. 2019, 76, 165–181. [Google Scholar] [CrossRef]
- Rahimi, A.; Hajizadeh Moghaddam, A.; Roohbakhsh, A. Central administration of GPR55 receptor agonist and antagonist modulates anxiety-related behaviors in rats. Fundam. Clin. Pharmacol. 2015, 29, 185–190. [Google Scholar] [CrossRef]
- Wróbel, A.; Serefko, A.; Szopa, A.; Ulrich, D.; Poleszak, E.; Rechberger, T. O-1602, an Agonist of Atypical Cannabinoid Receptors GPR55, Reverses the Symptoms of Depression and Detrusor Overactivity in Rats Subjected to Corticosterone Treatment. Front. Pharmacol. 2020, 11, 1002. [Google Scholar] [CrossRef]
- Staton, P.C.; Hatcher, J.P.; Walker, D.J.; Morrison, A.D.; Shapland, E.M.; Hughes, J.P.; Chong, E.; Mander, P.K.; Green, P.J.; Billinton, A.; et al. The putative cannabinoid receptor GPR55 plays a role in mechanical hyperalgesia associated with inflammatory and neuropathic pain. Pain 2008, 139, 225–236. [Google Scholar] [CrossRef]
- García-Gutiérrez, M.S.; Navarrete, F.; Navarro, G.; Reyes-Resina, I.; Franco, R.; Lanciego, J.L.; Giner, S.; Manzanares, J. Alterations in Gene and Protein Expression of Cannabinoid CB2 and GPR55 Receptors in the Dorsolateral Prefrontal Cortex of Suicide Victims. Neurotherapeutics 2018, 15, 796–806. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, H.; Onaivi, E.S.; Horiuchi, Y.; Imai, K.; Komaki, G.; Ishikawa, T.; Suzuki, M.; Watanabe, Y.; Ando, T.; Higuchi, S.; et al. Functional polymorphism in the GPR55 gene is associated with anorexia nervosa. Synapse 2011, 65, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Ryberg, E.; Larsson, N.; Sjögren, S.; Hjorth, S.; Hermansson, N.-O.; Leonova, J.; Elebring, T.; Nilsson, K.; Drmota, T.; Greasley, P.J. The orphan receptor GPR55 is a novel cannabinoid receptor: GPR55, a novel cannabinoid receptor. Br. J. Pharmacol. 2007, 152, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Lauckner, J.E.; Jensen, J.B.; Chen, H.-Y.; Lu, H.-C.; Hille, B.; Mackie, K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc. Natl. Acad. Sci. USA 2008, 105, 2699–2704. [Google Scholar] [CrossRef] [Green Version]
- Henstridge, C.M.; Balenga, N.A.; Schröder, R.; Kargl, J.K.; Platzer, W.; Martini, L.; Arthur, S.; Penman, J.; Whistler, J.L.; Kostenis, E.; et al. GPR55 ligands promote receptor coupling to multiple signalling pathways. Br. J. Pharmacol. 2010, 160, 604–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenakin, T. Functional Selectivity through Protean and Biased Agonism: Who Steers the Ship? Mol. Pharmacol. 2007, 72, 1393–1401. [Google Scholar] [CrossRef] [Green Version]
- Kenakin, T.; Christopoulos, A. Signalling bias in new drug discovery: Detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 2013, 12, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Nørregaard, R.; Kwon, T.-H.; Frøkiær, J. Physiology and pathophysiology of cyclooxygenase-2 and prostaglandin E2 in the kidney. Kidney Res. Clin. Pract. 2015, 34, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Siljehav, V.; Olsson Hofstetter, A.; Jakobsson, P.-J.; Herlenius, E. mPGES-1 and prostaglandin E2: Vital role in inflammation, hypoxic response, and survival. Pediatr. Res. 2012, 72, 460–467. [Google Scholar] [CrossRef] [Green Version]
- Olajide, O.A.; Velagapudi, R.; Okorji, U.P.; Sarker, S.D.; Fiebich, B.L. Picralima nitida seeds suppress PGE2 production by interfering with multiple signalling pathways in IL-1β-stimulated SK-N-SH neuronal cells. J. Ethnopharmacol. 2014, 152, 377–383. [Google Scholar] [CrossRef]
- Apweiler, M.; Streyczek, J.; Saliba, S.W.; Ditrich, J.; Muñoz, E.; Fiebich, B.L. Anti-Inflammatory and Anti-Oxidative Effects of AM404 in IL-1β-Stimulated SK-N-SH Neuroblastoma Cells. Front. Pharmacol. 2021, 12, 789074. [Google Scholar] [CrossRef]
- Candelario-Jalil, E.; Akundi, R.S.; Bhatia, H.S.; Lieb, K.; Appel, K.; Muñoz, E.; Hüll, M.; Fiebich, B.L. Ascorbic acid enhances the inhibitory effect of aspirin on neuronal cyclooxygenase-2-mediated prostaglandin E2 production. J. Neuroimmunol. 2006, 174, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Akundi, R.S.; Candelario-Jalil, E.; Hess, S.; Hüll, M.; Lieb, K.; Gebicke-Haerter, P.J.; Fiebich, B.L. Signal transduction pathways regulating cyclooxygenase-2 in lipopolysaccharide-activated primary rat microglia. Glia 2005, 51, 199–208. [Google Scholar] [CrossRef]
- Medina, M.V.; D Agostino, A.; Ma, Q.; Eroles, P.; Cavallin, L.; Chiozzini, C.; Sapochnik, D.; Cymeryng, C.; Hyjek, E.; Cesarman, E.; et al. KSHV G-protein coupled receptor vGPCR oncogenic signaling upregulation of Cyclooxygenase-2 expression mediates angiogenesis and tumorigenesis in Kaposi’s sarcoma. PLoS Pathog. 2020, 16, e1009006. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Zhou, D.; Shi, Z.; Qiu, Y.; Zhou, G.; Liu, J.; Yang, Q.; Cao, L.; Zhang, J. Centromere protein U (CENPU) enhances angiogenesis in triple-negative breast cancer by inhibiting ubiquitin–proteasomal degradation of COX-2. Cancer Lett. 2020, 482, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Kenakin, T. The Classification of Seven Transmembrane Receptors in Recombinant Expression Systems. Pharmacol. Rev. 1996, 48, 413–463. [Google Scholar] [PubMed]
- Yap, J.K.Y.; Pickard, B.S.; Chan, E.W.L.; Gan, S.Y. The Role of Neuronal NLRP1 Inflammasome in Alzheimer’s Disease: Bringing Neurons into the Neuroinflammation Game. Mol. Neurobiol. 2019, 56, 7741–7753. [Google Scholar] [CrossRef]
- Huang, Y.; Thathiah, A. Regulation of Neuronal Communication by G Protein-Coupled Receptors. FEBS Lett. 2015, 589, 1607–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.-Y.; Bi, G.-H.; Li, X.; Li, J.; Qu, H.; Zhang, S.-J.; Li, C.-Y.; Onaivi, E.S.; Gardner, E.L.; Xi, Z.-X.; et al. Species Differences in Cannabinoid Receptor 2 and Receptor Responses to Cocaine Self-Administration in Mice and Rats. Neuropsychopharmacology 2015, 40, 1037–1051. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Lee, A.-Y.; Park, S.-Y.; Liu, K.-H.; Im, D.-S. O-1602 Promotes Hepatic Steatosis through GPR55 and PI3 Kinase/Akt/SREBP-1c Signaling in Mice. Int. J. Mol. Sci. 2021, 22, 3091. [Google Scholar] [CrossRef]
- Lee, S.-J.; Im, D.-S. GPR55 Antagonist CID16020046 Protects against Atherosclerosis Development in Mice by Inhibiting Monocyte Adhesion and Mac-1 Expression. Int. J. Mol. Sci. 2021, 22, 13084. [Google Scholar] [CrossRef] [PubMed]
- Janefjord, E.; Mååg, J.L.V.; Harvey, B.S.; Smid, S.D. Cannabinoid Effects on β Amyloid Fibril and Aggregate Formation, Neuronal and Microglial-Activated Neurotoxicity In Vitro. Cell. Mol. Neurobiol. 2014, 34, 31–42. [Google Scholar] [CrossRef]
- Lipina, C.; Walsh, S.K.; Mitchell, S.E.; Speakman, J.R.; Wainwright, C.L.; Hundal, H.S. GPR55 Defi-ciency Is Associated with Increased Adiposity and Impaired Insulin Signaling in Peripheral Metabolic Tissues. FASEB J. 2019, 33, 1299–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schicho, R.; Bashashati, M.; Bawa, M.; McHugh, D.; Saur, D.; Hu, H.-M.; Zimmer, A.; Lutz, B.; Mackie, K.; Bradshaw, H.B.; et al. The Atypical Cannabinoid O-1602 Protects against Experimental Colitis and Inhibits Neutrophil Recruitment. Inflamm. Bowel. Dis. 2011, 17, 1651–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decuzzi, P.; Ferrari, M. The Role of Specific and Non-Specific Interactions in Receptor-Mediated En-docytosis of Nanoparticles. Biomaterials 2007, 28, 2915–2922. [Google Scholar] [CrossRef]
- Helm, C.A.; Knoll, W.; Israelachvili, J.N. Measurement of Ligand-Receptor Interactions. Proc. Natl. Acad. Sci. USA 1991, 88, 8169–8173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashti-Khavidaki, S.; Saidi, R.; Lu, H. Current status of glucocorticoid usage in solid organ transplantation. World J. Transplant. 2021, 11, 443–465. [Google Scholar] [CrossRef]
- Saliba, S.W.; Marcotegui, A.R.; Fortwängler, E.; Ditrich, J.; Perazzo, J.C.; Muñoz, E.; de Oliveira, A.C.P.; Fiebich, B.L. AM404, paracetamol metabolite, prevents prostaglandin synthesis in activated microglia by inhibiting COX activity. J. Neuroinflamm. 2017, 14, 246. [Google Scholar] [CrossRef] [Green Version]
- Gebicke-Haerter, P.; Bauer, J.; Schobert, A.; Northoff, H. Lipopolysaccharide-free conditions in primary astrocyte cultures allow growth and isolation of microglial cells. J. Neurosci. 1989, 9, 183–194. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, A.C.P.; Candelario-Jalil, E.; Bhatia, H.S.; Lieb, K.; Hüll, M.; Fiebich, B.L. Regulation of prostaglandin E2 synthase expression in activated primary rat microglia: Evidence for uncoupled regulation of mPGES-1 and COX-2. Glia 2008, 56, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Henstridge, C.M.; Balenga, N.A.B.; Ford, L.A.; Ross, R.A.; Waldhoer, M.; Irving, A.J. The GPR55 ligand L-α-lysophosphatidylinositol promotes RhoA-dependent Ca 2+ signaling and NFAT activation. FASEB J. 2009, 23, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Fiebich, B.L.; Chrubasik, S. Effects of an ethanolic Salix extract on the release of selected inflammatory mediators in vitro. Phytomedicine 2004, 11, 135–138. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).