Time-Resolved Gene Expression Analysis Monitors the Regulation of Inflammatory Mediators and Attenuation of Adaptive Immune Response by Vitamin D
Abstract
:1. Introduction
2. Results
2.1. Time-Resolved Transcriptome Changes of PBMCs in Response to 1,25(OH)2D3
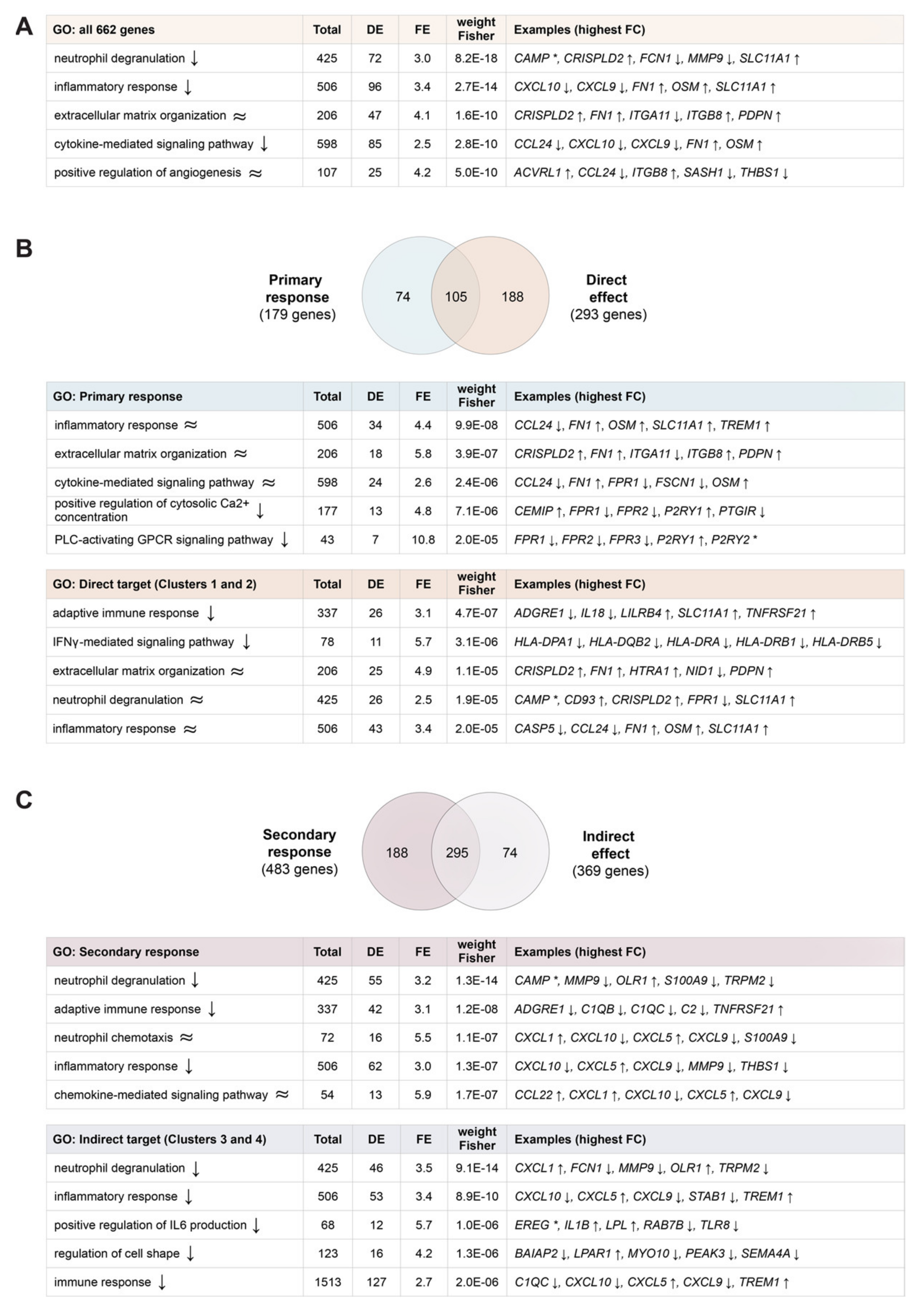
2.2. Distinction between Stimulating and Stabilizing Effects of 1,25(OH)2D3
2.3. Functional Impact of Vitamin D Target Genes
2.4. Colocalization of Vitamin D Target Genes with VDR Binding Sites
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. PBMC Isolation and Stimulation
4.3. RNA-Seq Data Generation and Processing
4.4. Transcriptome Data Analysis
4.5. Clustering and Functional Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holick, M.F. Photobiology of vitamin D. In Vitamin D, 3rd ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 45–55. [Google Scholar] [CrossRef]
- Bendik, I.; Friedel, A.; Roos, F.F.; Weber, P.; Eggersdorfer, M. Vitamin D: A critical and essential micronutrient for human health. Front. Physiol. 2014, 5, 248. [Google Scholar] [CrossRef] [PubMed]
- Chirumbolo, S.; Bjorklund, G.; Sboarina, A.; Vella, A. The role of vitamin D in the immune system as a pro-survival molecule. Clin. Ther. 2017, 39, 894–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Peppel, J.; van Leeuwen, J.P. Vitamin D and gene networks in human osteoblasts. Front. Physiol. 2014, 5, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baeke, F.; Takiishi, T.; Korf, H.; Gysemans, C.; Mathieu, C. Vitamin D: Modulator of the immune system. Curr. Opin. Pharmacol. 2010, 10, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Sita-Lumsden, A.; Lapthorn, G.; Swaminathan, R.; Milburn, H.J. Reactivation of tuberculosis and vitamin D deficiency: The contribution of diet and exposure to sunlight. Thorax 2007, 62, 1003–1007. [Google Scholar] [CrossRef] [Green Version]
- Bilezikian, J.P.; Bikle, D.; Hewison, M.; Lazaretti-Castro, M.; Formenti, A.M.; Gupta, A.; Madhavan, M.V.; Nair, N.; Babalyan, V.; Hutchings, N.; et al. Vitamin D and COVID-19. Eur. J. Endocrinol. 2020, 183, R133–R147. [Google Scholar] [CrossRef]
- Munger, K.L.; Levin, L.I.; Hollis, B.W.; Howard, N.S.; Ascherio, A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA 2006, 296, 2832–2838. [Google Scholar] [CrossRef] [Green Version]
- Sintzel, M.B.; Rametta, M.; Reder, A.T. Vitamin D and multiple sclerosis: A comprehensive review. Neurol. Ther. 2018, 7, 59–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weir, E.K.; Thenappan, T.; Bhargava, M.; Chen, Y. Does vitamin D deficiency increase the severity of COVID-19? Clin. Med. (Lond) 2020, 20, e107–e108. [Google Scholar] [CrossRef]
- Carlberg, C. Nutrigenomics of vitamin D. Nutrients 2019, 11, 676. [Google Scholar] [CrossRef] [Green Version]
- Haussler, M.R.; Haussler, C.A.; Bartik, L.; Whitfield, G.K.; Hsieh, J.C.; Slater, S.; Jurutka, P.W. Vitamin D receptor: Molecular signaling and actions of nutritional ligands in disease prevention. Nutr. Rev. 2008, 66, S98–S112. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C. Genome-wide (over)view on the actions of vitamin D. Front. Physiol. 2014, 5, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, R.M. The nuclear receptor superfamily: A rosetta stone for physiology. Mol. Endocrinol. 2005, 19, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.J. Vitamin D and the RNA transcriptome: More than mRNA regulation. Front. Physiol. 2014, 5, 181. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C. Vitamin D genomics: From in vitro to in vivo. Front. Endocrinol. (Lausanne) 2018, 9, 250. [Google Scholar] [CrossRef]
- Heikkinen, S.; Väisänen, S.; Pehkonen, P.; Seuter, S.; Benes, V.; Carlberg, C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011, 39, 9181–9193. [Google Scholar] [CrossRef] [Green Version]
- Verway, M.; Bouttier, M.; Wang, T.T.; Carrier, M.; Calderon, M.; An, B.S.; Devemy, E.; McIntosh, F.; Divangahi, M.; Behr, M.A.; et al. Vitamin D induces interleukin-1beta expression: Paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 2013, 9, e1003407. [Google Scholar] [CrossRef] [Green Version]
- Seuter, S.; Neme, A.; Carlberg, C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016, 44, 4090–4104. [Google Scholar] [CrossRef] [Green Version]
- Nurminen, V.; Neme, A.; Seuter, S.; Carlberg, C. Modulation of vitamin D signaling by the pioneer factor CEBPA. Biochim. Biophys. Acta 2019, 1862, 96–106. [Google Scholar] [CrossRef]
- Carlberg, C. Vitamin D signaling in the context of innate immunity: Focus on human monocytes. Front. Immunol. 2019, 10, 2211. [Google Scholar] [CrossRef] [Green Version]
- Neme, A.; Seuter, S.; Malinen, M.; Nurmi, T.; Tuomainen, T.P.; Virtanen, J.K.; Carlberg, C. In Vivo transcriptome changes of human white blood cells in response to vitamin D. J. Steroid Biochem. Mol. Biol 2019, 188, 71–76. [Google Scholar] [CrossRef]
- Carlberg, C. Molecular approaches for optimizing vitamin D supplementation. Vitam. Horm. 2016, 100, 255–271. [Google Scholar] [CrossRef]
- Tuoresmäki, P.; Väisänen, S.; Neme, A.; Heikkinen, S.; Carlberg, C. Patterns of genome-wide VDR locations. PLoS ONE 2014, 9, e96105. [Google Scholar] [CrossRef] [Green Version]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta 2017, 1860, 952–961. [Google Scholar] [CrossRef] [Green Version]
- Ramagopalan, S.V.; Heger, A.; Berlanga, A.J.; Maugeri, N.J.; Lincoln, M.R.; Burrell, A.; Handunnetthi, L.; Handel, A.E.; Disanto, G.; Orton, S.M.; et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010, 20, 1352–1360. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Goetsch, P.D.; Pike, J.W. VDR/RXR and TCF4/beta-catenin cistromes in colonic cells of colorectal tumor origin: Impact on c-FOS and c-MYC gene expression. Mol. Endocrinol. 2012, 26, 37–51. [Google Scholar] [CrossRef]
- Ding, N.; Yu, R.T.; Subramaniam, N.; Sherman, M.H.; Wilson, C.; Rao, R.; Leblanc, M.; Coulter, S.; He, M.; Scott, C.; et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 2013, 153, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Fleet, J.C.; Kovalenko, P.L.; Li, Y.; Smolinski, J.; Spees, C.; Yu, J.G.; Thomas-Ahner, J.M.; Cui, M.; Neme, A.; Carlberg, C.; et al. Vitamin D signaling suppresses early prostate carcinogenesis in TgAPT121 mice. Cancer Prev. Res. (Phila) 2019, 12, 343–356. [Google Scholar] [CrossRef]
- Carlberg, C. Molecular endocrinology of vitamin D on the epigenome level. Mol. Cell Endocrinol. 2017, 453, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C.; Neme, A. Machine learning approaches infer vitamin D signaling: Critical impact of vitamin D receptor binding within topologically associated domains. J. Steroid Biochem. Mol. Biol. 2019, 185, 103–109. [Google Scholar] [CrossRef]
- Malmberg, H.R.; Hanel, A.; Taipale, M.; Heikkinen, S.; Carlberg, C. Vitamin D treatment sequence is critical for transcriptome modulation of immune challenged primary human cells. Front. Immunol. 2021, 12, 754056. [Google Scholar] [CrossRef] [PubMed]
- Hanel, A.; Bendik, I.; Carlberg, C. Transcriptome-wide profile of 25-hydroxyvitamin D3 in pimary immune cells from human peripheral blood. Nutrients 2021, 13, 4100. [Google Scholar] [CrossRef] [PubMed]
- Nurminen, V.; Seuter, S.; Carlberg, C. Primary vitamin D target genes of human monocytes. Front. Physiol. 2019, 10, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanel, A.; Malmberg, H.R.; Carlberg, C. Genome-wide effects of chromatin on vitamin D signaling. J. Mol. Endocrinol. 2020, 64, R45–R56. [Google Scholar] [CrossRef] [PubMed]
- Zierold, C.; Darwish, H.M.; DeLuca, H.F. Identification of a vitamin D-response element in the rat calcidiol (25-hydroxyvitamin D3) 24-hydroxylase gene. Proc. Natl. Acad. Sci. USA 1994, 91, 900–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seuter, S.; Pehkonen, P.; Heikkinen, S.; Carlberg, C. Dynamics of 1α,25-dihydroxyvitamin D-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim. Biophys. Acta 2013, 1829, 1266–1275. [Google Scholar] [CrossRef] [PubMed]
- Skendros, P.; Mitroulis, I.; Ritis, K. Autophagy in neutrophils: From granulopoiesis to neutrophil extracellular traps. Front. Cell Dev. Biol. 2018, 6, 109. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. The chemokine superfamily revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef] [Green Version]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef]
- Quesada-Gomez, J.M.; Entrenas-Castillo, M.; Bouillon, R. Vitamin D receptor stimulation to reduce acute respiratory distress syndrome (ARDS) in patients with coronavirus SARS-CoV-2 infections. J. Steroid Biochem. Mol. Biol. 2020, 202, 105719. [Google Scholar] [CrossRef]
- Vogl, T.; Eisenblatter, M.; Voller, T.; Zenker, S.; Hermann, S.; van Lent, P.; Faust, A.; Geyer, C.; Petersen, B.; Roebrock, K.; et al. Alarmin S100A8/S100A9 as a biomarker for molecular imaging of local inflammatory activity. Nat. Commun. 2014, 5, 4593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzinger, D.; Tenbrock, K.; Roth, J. Alarmins of the S100-family in juvenile autoimmune and auto-inflammatory diseases. Front. Immunol. 2019, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Trowsdale, J.; Knight, J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet. 2013, 14, 301–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wosen, J.E.; Mukhopadhyay, D.; Macaubas, C.; Mellins, E.D. Epithelial MHC Class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front. Immunol. 2018, 9, 2144. [Google Scholar] [CrossRef]
- Adorini, L.; Penna, G. Control of autoimmune diseases by the vitamin D endocrine system. Nat. Clin. Pract. Rheumatol. 2008, 4, 404–412. [Google Scholar] [CrossRef]
- Kular, L.; Liu, Y.; Ruhrmann, S.; Zheleznyakova, G.; Marabita, F.; Gomez-Cabrero, D.; James, T.; Ewing, E.; Linden, M.; Gornikiewicz, B.; et al. DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun. 2018, 9, 2397. [Google Scholar] [CrossRef]
- Handunnetthi, L.; Ramagopalan, S.V.; Ebers, G.C.; Knight, J.C. Regulation of major histocompatibility complex class II gene expression, genetic variation and disease. Genes Immun. 2010, 11, 99–112. [Google Scholar] [CrossRef]
- Monaco, G.; Lee, B.; Xu, W.; Mustafah, S.; Hwang, Y.Y.; Carre, C.; Burdin, N.; Visan, L.; Ceccarelli, M.; Poidinger, M.; et al. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019, 26, 1627–1640. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Hanel, A.; Neme, A.; Malinen, M.; Hamalainen, E.; Malmberg, H.R.; Etheve, S.; Tuomainen, T.P.; Virtanen, J.K.; Bendik, I.; Carlberg, C. Common and personal target genes of the micronutrient vitamin D in primary immune cells from human peripheral blood. Sci. Rep. 2020, 10, 21051. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 2016, 5, 1438. [Google Scholar] [CrossRef] [Green Version]
- Spies, D.; Renz, P.F.; Beyer, T.A.; Ciaudo, C. Comparative analysis of differential gene expression tools for RNA sequencing time course data. Brief. Bioinform. 2019, 20, 288–298. [Google Scholar] [CrossRef] [Green Version]
- Grant, R.A.; Morales-Nebreda, L.; Markov, N.S.; Swaminathan, S.; Querrey, M.; Guzman, E.R.; Abbott, D.A.; Donnelly, H.K.; Donayre, A.; Goldberg, I.A.; et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 2021, 590, 635–641. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanel, A.; Carlberg, C. Time-Resolved Gene Expression Analysis Monitors the Regulation of Inflammatory Mediators and Attenuation of Adaptive Immune Response by Vitamin D. Int. J. Mol. Sci. 2022, 23, 911. https://doi.org/10.3390/ijms23020911
Hanel A, Carlberg C. Time-Resolved Gene Expression Analysis Monitors the Regulation of Inflammatory Mediators and Attenuation of Adaptive Immune Response by Vitamin D. International Journal of Molecular Sciences. 2022; 23(2):911. https://doi.org/10.3390/ijms23020911
Chicago/Turabian StyleHanel, Andrea, and Carsten Carlberg. 2022. "Time-Resolved Gene Expression Analysis Monitors the Regulation of Inflammatory Mediators and Attenuation of Adaptive Immune Response by Vitamin D" International Journal of Molecular Sciences 23, no. 2: 911. https://doi.org/10.3390/ijms23020911
APA StyleHanel, A., & Carlberg, C. (2022). Time-Resolved Gene Expression Analysis Monitors the Regulation of Inflammatory Mediators and Attenuation of Adaptive Immune Response by Vitamin D. International Journal of Molecular Sciences, 23(2), 911. https://doi.org/10.3390/ijms23020911






