Mitogen-Activated Protein Kinase 4-Regulated Metabolic Networks
Abstract
1. Introduction
2. Results
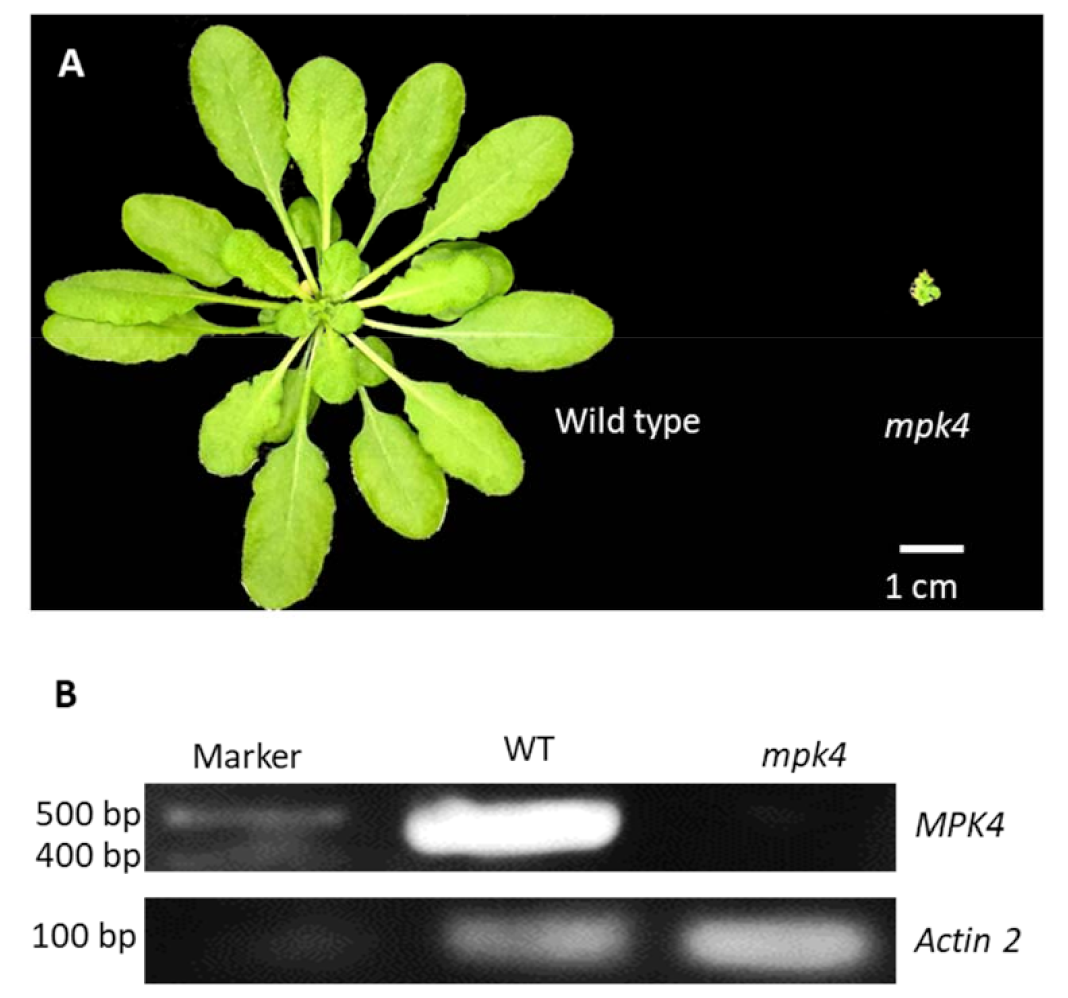
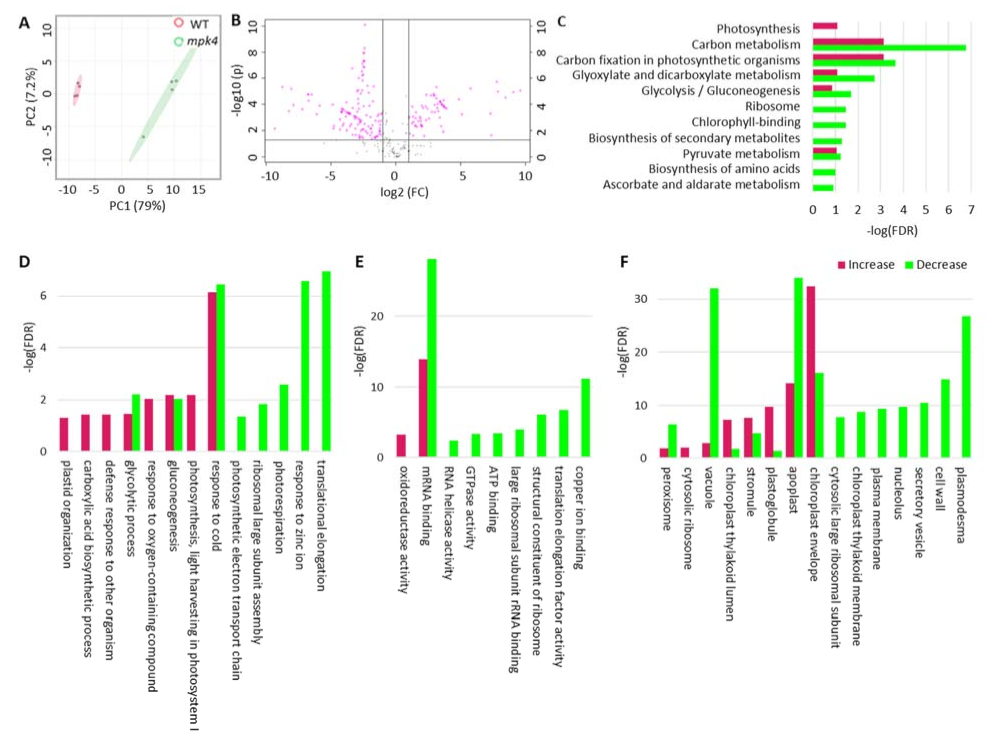
2.1. Mpk4 Mutant Phenotype and Transcription Analysis
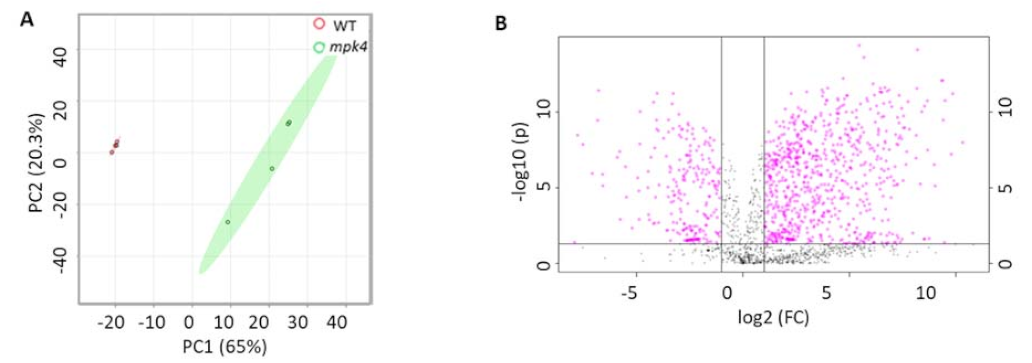
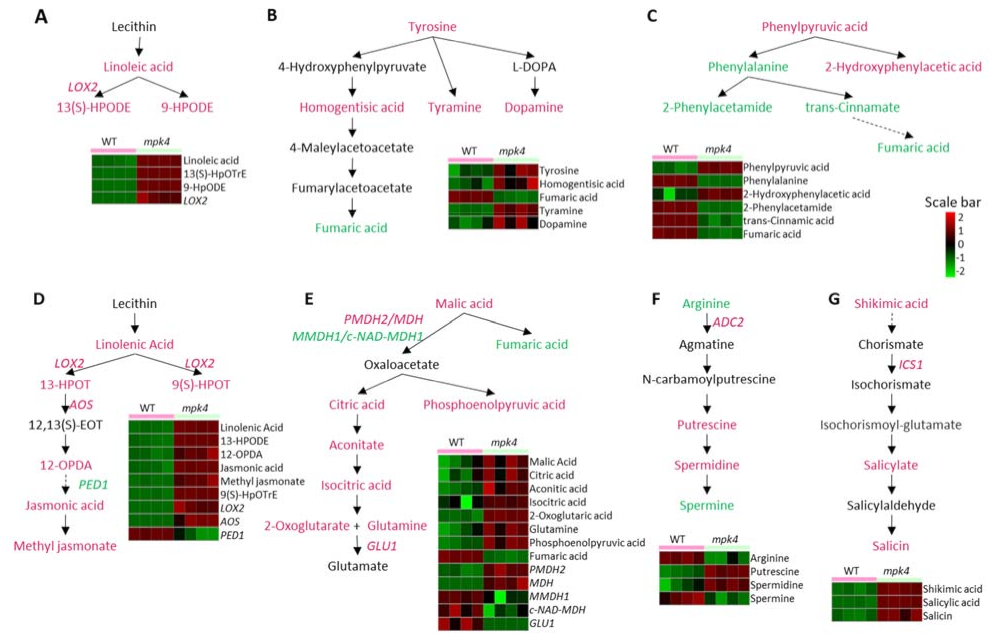
2.2. Metabolomics of WT and the Mpk4 Mutant Leaves
2.3. Proteomics of WT and the Mpk4 Mutant Leaves
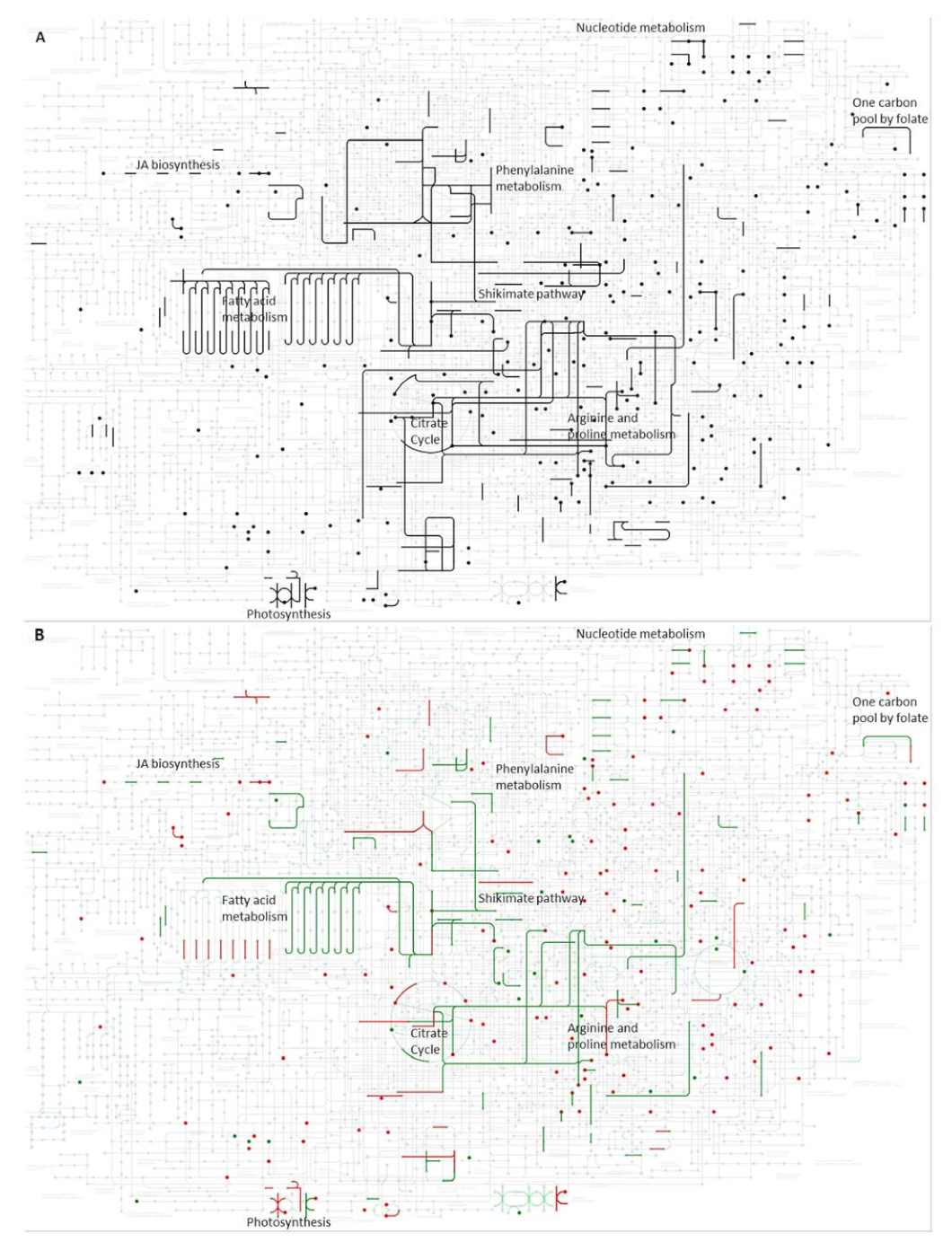
2.4. Integration of Metabolomics and Proteomics
3. Discussion
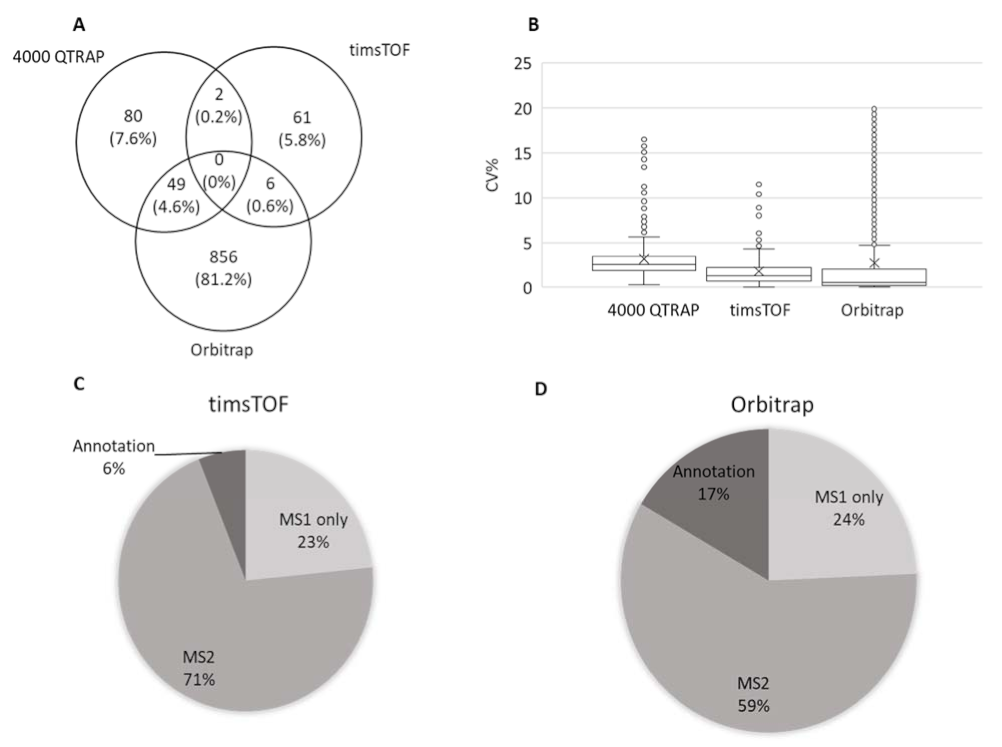
3.1. Comparison between Different Metabolomic Platforms
3.2. MPK4 Is Involved in Arginine Metabolism, JA and SA Changes
3.3. MPK4 Is Related with Chloroplast Integrity and Photosynthesis
4. Materials and Methods
4.1. Plant Materials
4.2. Semi-qPCR Analysis of MPK4 Transcript
4.3. Metabolite Extraction from WT and Mpk4 Leaves
4.4. Protein Extraction from WT and Mpk4 Leaves and Trypsin Digestion
4.5. Liquid Chromatography Tandem Mass Spectrometry for Metabolomics and Proteomics
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis MAP Kinase 4 Negatively Regulates Systemic Acquired Resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef]
- Kosetsu, K.; Matsunaga, S.; Nakagami, H.; Colcombet, J.; Sasabe, M.; Soyano, T.; Takahashi, Y.; Hirt, H.; Machida, Y. The MAP Kinase MPK4 Is Required for Cytokinesis in Arabidopsis thaliana. Plant Cell 2010, 22, 3778–3790. [Google Scholar] [CrossRef]
- Beck, M.; Komis, G.; Muller, J.; Menzel, D.; Samaj, J. Arabidopsis Homologs of Nucleus- and Phragmoplast-Localized Kinase 2 and 3 and Mitogen-Activated Protein Kinase 4 Are Essential for Microtubule Organization. Plant Cell 2010, 22, 755–771. [Google Scholar] [CrossRef]
- Beck, M.; Komis, G.; Ziemann, A.; Menzel, D.; Samaj, J. Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 2011, 189, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Matsushima, C.; Nishimura, S.; Higashiyama, T.; Sasabe, M.; Machida, Y. Identification of Phosphoinositide-Binding Protein PATELLIN2 as a Substrate of Arabidopsis MPK4 MAP Kinase during Septum Formation in Cytokinesis. Plant Cell Physiol. 2016, 57, 1744–1755. [Google Scholar] [CrossRef]
- Zeng, Q.; Chen, J.; Ellis, B.E.; Laboratories, M.S. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011, 67, 895–906. [Google Scholar] [CrossRef]
- Liu, J.-Z.; Horstman, H.D.; Braun, E.; Graham, M.A.; Zhang, C.; Navarre, D.; Qiu, W.-L.; Lee, Y.; Nettleton, D.; Hill, J.H.; et al. Soybean Homologs of MPK4 Negatively Regulate Defense Responses and Positively Regulate Growth and Development. Plant Physiol. 2011, 157, 1363–1378. [Google Scholar] [CrossRef] [PubMed]
- Gawroński, P.; Witoń, D.; Vashutina, K.; Bederska, M.; Betliński, B.; Rusaczonek, A.; Karpińsk, S. Mitogen-Activated Protein Kinase 4 Is a Salicylic Acid-Independent Regulator of Growth But Not of Photosynthesis in Arabidopsis. Mol. Plant 2014, 7, 1151–1166. [Google Scholar] [CrossRef]
- Montesinos, J.C.; Abuzeineh, A.; Kopf, A.; Juanes-Garcia, A.; Ötvös, K.; Petrášek, J.; Sixt, M.; Benková, E. Phytohormone cytokinin guides microtubule dynamics during cell progression from proliferative to differentiated stage. EMBO J. 2020, 39, e104238. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Marhavý, P.; Bielach, A.; Abas, L.; Abuzeineh, A.; Duclercq, J.; Tanaka, H.; Pařezová, M.; Petrášek, J.; Friml, J.; Kleine-Vehn, J.; et al. Cytokinin Modulates Endocytic Trafficking of PIN1 Auxin Efflux Carrier to Control Plant Organogenesis. Dev. Cell 2011, 21, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Galston, A.W.; Kaur-Sawhney, R.; Altabella, T.; Tiburcio, A.F. Plant polyamines in reproductive activity and response to abiotic stress. Bot. Acta 1997, 110, 197–207. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Berriri, S.; Garcia, A.V.; dit Frey, N.F.; Rozhon, W.; Pateyron, S.; Leonhardt, N.; Montillet, J.-L.; Leung, J.; Hirt, H.; Colcombet, J. Constitutively Active Mitogen-Activated Protein Kinase Versions Reveal Functions of Arabidopsis MPK4 in Pathogen Defense Signaling. Plant Cell 2012, 24, 4281–4293. [Google Scholar] [CrossRef]
- Gomi, K.; Ogawa, D.; Katou, S.; Kamada, H.; Nakajima, N.; Saji, H.; Soyano, T.; Sasabe, M.; Machida, Y.; Mitsuhara, I.; et al. A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol. 2005, 46, 1902–1914. [Google Scholar] [CrossRef]
- Brodersen, P.; Petersen, M.; Nielsen, H.B.; Zhu, S.; Newman, M.A.; Shokat, K.M.; Rietz, S.; Parker, J.; Mundy, J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006, 47, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Korbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic Acid Suppresses Jasmonic Acid Signaling Downstream of SCFCOI1-JAZ by Targeting GCC Promoter Motifs via Transcription Factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef]
- Schweiger, R.; Heise, A.M.; Persicke, M.; Müller, C. Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant Cell Environ. 2014, 37, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Schneider, J.D.; Lin, C.; Geng, S.; Ma, T.; Lawrence, S.R.; Dufresne, C.P.; Harmon, A.C.; Chen, S. MPK4 Phosphorylation Dynamics and Interacting Proteins in Plant Immunity. J. Proteome Res. 2019, 18, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Alseekh, S.; Fernie, A.R.; Luo, J. The Structure and Function of Major Plant Metabolite Modifications. Mol. Plant 2019, 12, 899–919. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Fernie, A.R.; Luo, J. Exploring the Diversity of Plant Metabolism. Trends Plant Sci. 2019, 24, 83–98. [Google Scholar] [CrossRef]
- Misra, B.B.; Assmann, S.M.; Chen, S. Plant single-cell and single-cell-type metabolomics. Trends Plant Sci. 2014, 19, 637–646. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef]
- Sawada, Y.; Akiyama, K.; Sakata, A.; Kuwahara, A.; Otsuki, H.; Sakurai, T.; Saito, K.; Hirai, M.Y. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 2009, 50, 37–47. [Google Scholar] [CrossRef]
- Schroeder, M.; Meyer, S.W.; Heyman, H.M.; Barsch, A.; Sumner, L.W. Generation of a collision cross section library for multi-dimensional plant metabolomics using UHPLC-trapped ion mobility-MS/MS. Metabolites 2020, 10, 13. [Google Scholar] [CrossRef]
- Qiu, J.; Fiil, B.K.; Petersen, K.; Nielsen, H.B.; Botanga, C.J.; Palma, K.; Suarez-rodriguez, M.C.; Sandbech-clausen, S.; Lichota, J.; Brodersen, P.; et al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008, 27, 2214–2221. [Google Scholar] [CrossRef]
- Tiburcio, A.F. Polyamines. In Methods in Molecular Biology; Alcázar, R., Tiburcio, A.F., Eds.; Springer: New York, NY, USA, 2018; Volume 1694, ISBN 978-1-4939-7397-2. [Google Scholar]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Djamei, A.; Bitton, F.; Hirt, H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2009, 2, 120–137. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chen, S. New functions of an old kinase MPK4 in guard cells. Plant Signal. Behav. 2018, 13, e1477908. [Google Scholar] [CrossRef]
- Imai, A.; Matsuyama, T.; Hanzawa, Y.; Akiyama, T.; Tamaoki, M.; Saji, H.; Shirano, Y.; Kato, T.; Hayashi, H.; Shibata, D.; et al. Spermidine Synthase Genes Are Essential for Survival of Arabidopsis. Plant Physiol. 2004, 135, 1565–1573. [Google Scholar] [CrossRef]
- Mollá-Morales, A.; Sarmiento-Mañús, R.; Robles, P.; Quesada, V.; Pérez-Pérez, J.M.; González-Bayón, R.; Hannah, M.A.; Willmitzer, L.; Ponce, M.R.; Micol, J.L. Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J. 2011, 65, 335–345. [Google Scholar] [CrossRef]
- Yamakawa, H.; Kamada, H.; Satoh, M.; Ohashi, Y. Spermine Is a Salicylate-Independent Endogenous Inducer for Both Tobacco Acidic Pathogenesis-Related Proteins and Resistance against Tobacco Mosaic Virus Infection. Plant Physiol. 1998, 118, 1213–1222. [Google Scholar] [CrossRef]
- Perez-Amador, M.A. Induction of the Arginine Decarboxylase ADC2 Gene Provides Evidence for the Involvement of Polyamines in the Wound Response in Arabidopsis. Plant Physiol. 2002, 130, 1454–1463. [Google Scholar] [CrossRef]
- Urano, K.; Yoshiba, Y.; Nanjo, T.; Ito, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Arabidopsis stress-inducible gene for arginine decarboxylase AtADC2 is required for accumulation of putrescine in salt tolerance. Biochem. Biophys. Res. Commun. 2004, 313, 369–375. [Google Scholar] [CrossRef]
- Gonzalez, M.E.; Marco, F.; Minguet, E.G.; Carrasco-Sorli, P.; Blázquez, M.A.; Carbonell, J.; Ruiz, O.A.; Pieckenstain, F.L. Perturbation of spermine synthase Gene Expression and Transcript Profiling Provide New Insights on the Role of the Tetraamine Spermine in Arabidopsis Defense against Pseudomonas viridiflava. Plant Physiol. 2011, 156, 2266–2277. [Google Scholar] [CrossRef]
- Marco, F.; Busó, E.; Carrasco, P. Overexpression of SAMDC1 gene in Arabidopsis thaliana increases expression of defense-related genes as well as resistance to Pseudomonas syringae and Hyaloperonospora arabidopsidis. Front. Plant Sci. 2014, 5, 115. [Google Scholar] [CrossRef]
- Frei dit Frey, N.; Garcia, A.V.; Bigeard, J.; Zaag, R.; Bueso, E.; Garmier, M.; Pateyron, S.; De Tauzia-Moreau, M.L.; Brunaud, V.; Balzergue, S.; et al. Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 2014, 15, R87. [Google Scholar] [CrossRef]
- Glauser, G.; Grata, E.; Dubugnon, L.; Rudaz, S.; Farmer, E.E.; Wolfender, J.-L. Spatial and Temporal Dynamics of Jasmonate Synthesis and Accumulation in Arabidopsis in Response to Wounding. J. Biol. Chem. 2008, 283, 16400–16407. [Google Scholar] [CrossRef]
- Reymond, P.; Weber, H.; Damond, M.; Farmer, E.E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 2000, 12, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic Acid and Jasmonic Acid Pathways are Activated in Spatially Different Domains around the Infection Site during Effector-Triggered Immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Su, S.-H.; Bush, S.M.; Zaman, N.; Stecker, K.; Sussman, M.R.; Krysan, P. Deletion of a Tandem Gene Family in Arabidopsis: Increased MEKK2 Abundance Triggers Autoimmunity when the MEKK1-MKK1/2-MPK4 Signaling Cascade Is Disrupted. Plant Cell 2013, 25, 1895–1910. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Wijk, K.J.; Kessler, F. Plastoglobuli: Plastid Microcompartments with Integrated Functions in Metabolism, Plastid Developmental Transitions, and Environmental Adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef]
- Rudella, A.; Friso, G.; Alonso, J.M.; Ecker, J.R.; van Wijk, K.J. Downregulation of ClpR2 Leads to Reduced Accumulation of the ClpPRS Protease Complex and Defects in Chloroplast Biogenesis in Arabidopsis. Plant Cell 2006, 18, 1704–1721. [Google Scholar] [CrossRef]
- Youssef, A.; Laizet, Y.; Block, M.A.; Maréchal, E.; Alcaraz, J.-P.; Larson, T.R.; Pontier, D.; Gaffé, J.; Kuntz, M. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 2010, 61, 436–445. [Google Scholar] [CrossRef]
- Singh, D.K.; Maximova, S.N.; Jensen, P.J.; Lehman, B.L.; Ngugi, H.K.; McNellis, T.W. FIBRILLIN4 Is Required for Plastoglobule Development and Stress Resistance in Apple and Arabidopsis. Plant Physiol. 2010, 154, 1281–1293. [Google Scholar] [CrossRef]
- Zhang, T.; Chhajed, S.; Schneider, J.D.; Feng, G.; Song, W.Y.; Chen, S. Proteomic characterization of MPK4 signaling network and putative substrates. Plant Mol. Biol. 2019, 101, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Rayapuram, N.; Bigeard, J.; Alhoraibi, H.; Bonhomme, L.; Hesse, A.-M.; Vinh, J.; Hirt, H.; Pflieger, D. Quantitative Phosphoproteomic Analysis Reveals Shared and Specific Targets of Arabidopsis Mitogen-Activated Protein Kinases (MAPKs) MPK3, MPK4, and MPK6. Mol. Cell. Proteom. 2018, 17, 61–80. [Google Scholar] [CrossRef]
- David, L.; Kang, J.; Chen, S. Targeted Metabolomics of Plant Hormones and Redox Metabolites in Stomatal Immunity. In Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2085, pp. 79–92. ISBN 9781071601426. [Google Scholar]
- Hurkman, W.J.; Tanaka, C.K. Solubilization of Plant Membrane Proteins for Analysis by Two-Dimensional Gel Electrophoresis. Plant Physiol. 1986, 81, 802–806. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Misra, B.B.; de Armas, E.; Huhman, D.V.; Alborn, H.T.; Sumner, L.W.; Chen, S. Jasmonate-mediated stomatal closure under elevated CO2 revealed by time-resolved metabolomics. Plant J. 2016, 88, 947–962. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Lott, A.A.; Zhu, W.; Dufresne, C.P.; Chen, S. Mitogen-Activated Protein Kinase 4-Regulated Metabolic Networks. Int. J. Mol. Sci. 2022, 23, 880. https://doi.org/10.3390/ijms23020880
Lin C, Lott AA, Zhu W, Dufresne CP, Chen S. Mitogen-Activated Protein Kinase 4-Regulated Metabolic Networks. International Journal of Molecular Sciences. 2022; 23(2):880. https://doi.org/10.3390/ijms23020880
Chicago/Turabian StyleLin, Chuwei, Aneirin Alan Lott, Wei Zhu, Craig P. Dufresne, and Sixue Chen. 2022. "Mitogen-Activated Protein Kinase 4-Regulated Metabolic Networks" International Journal of Molecular Sciences 23, no. 2: 880. https://doi.org/10.3390/ijms23020880
APA StyleLin, C., Lott, A. A., Zhu, W., Dufresne, C. P., & Chen, S. (2022). Mitogen-Activated Protein Kinase 4-Regulated Metabolic Networks. International Journal of Molecular Sciences, 23(2), 880. https://doi.org/10.3390/ijms23020880







