Identifying Soybean Pod Borer (Leguminivora glycinivorella) Resistance QTLs and the Mechanism of Induced Defense Using Linkage Mapping and RNA-Seq Analysis
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variations in Eating Seed Percentage (ESP) for SPB
2.2. QTL Mapping Identified Four QTLs Controlling ESP
2.3. Common Response Genes to SPB Feeding between Two Parents Involving Signaling, Defense, and Immune Biological Processes
2.4. Flavonoid and Isoflavonoid Anabolism Are Responsible for the Variation in Parental Resistance to SPB
2.5. Candidate Gene Prediction of ESP-Related QTLs Using Four Soybean Accessions
3. Discussion
3.1. QTL Mapping Based on SNP Markers Combined with RNA-Seq for Efficient and Rapid Identification of QTLs and Candidate Genes Related to SPB Resistance
3.2. SPB Chewing-Responsive DEGs in Soybean Pod Shells Associated with Signal Transduction, Function Proteins, Secondary Metabolite Pathways, and TFs
3.3. Phytohormone Cross-Talk and Flavonoid Metabolism May Mediate Soybean Response to SPB and Resistance Mechanisms
4. Materials and Methods
4.1. Soybean Plant Materials and Growing Environment
4.2. Determination of Eating Seed Percentage for SPB
4.3. SNP Identification by Sequencing
4.4. Construction of Linkage Map and QTL Localization
4.5. RNA Isolation and RNA-Seq Data Analysis
4.6. Bioinformatic Analysis
4.7. Quantitative Reverse Transcription PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, G.; Wang, J.; Han, Y.; Teng, W.; Sun, G.; Li, W. Identification of QTL underlying the resistance of soybean to pod borer, Leguminivora glycinivorella (Mats.) obraztsov, and correlations with plant, pod and seed traits. Euphytica 2008, 164, 275. [Google Scholar] [CrossRef]
- Zhao, G.; Jiang, Z.; Li, D.; Han, Y.; Hu, H.; Wu, L.; Wang, Y.; Gao, Y.; Teng, W.; Li, Y.; et al. Molecular loci associated with seed isoflavone content may underlie resistance to soybean pod borer (Leguminivora glycinivorella). Plant Breed. 2015, 134, 78–84. [Google Scholar] [CrossRef]
- Meng, F.; Li, Y.; Zang, Z.; Li, N.; Ran, R.; Cao, Y.; Li, T.; Zhou, Q.; Li, W. Expression of the double-stranded RNA of the soybean pod borer Leguminivora glycinivorella (Lepidoptera: Tortricidae) ribosomal protein P0 gene enhances the resistance of transgenic soybean plants. Pest Manag. Sci. 2017, 73, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yao, D.; Ma, J.; Fu, Y.P.; Qu, J.; Wang, P.W. Genetic analysis of the major gene plus polygene model in soybean resistance to Leguminivora glycinivorella. Genet. Mol. Res. 2014, 13, 4983–4989. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, T.; Ni, H.; Wang, G.; Liu, X.; Cao, Y.; Li, W.; Meng, F. Transgenic soybean plants expressing Spb18S dsRNA exhibit enhanced resistance to the soybean pod borer Leguminivora glycinivorella (Lepidoptera: Olethreutidae). Arch. Insect Biochem. 2018, 98, e21461. [Google Scholar] [CrossRef] [PubMed]
- Badji, A.; Otim, M.; Machida, L.; Odong, T.; Kwemoi, D.B.; Okii, D.; Agbahoungba, S.; Mwila, N.; Kumi, F.; Ibanda, A.; et al. Maize Combined Insect Resistance Genomic Regions and Their Co-localization With Cell Wall Constituents Revealed by Tissue-Specific QTL Meta-Analyses. Front. Plant Sci. 2018, 9, 895. [Google Scholar] [CrossRef] [PubMed]
- García-Lara, S.; Burt, A.J.; Arnason, J.T.; Bergvinson, D.J. QTL Mapping of Tropical Maize Grain Components Associated with Maize Weevil Resistance. Crop Sci. 2010, 50, 815–825. [Google Scholar] [CrossRef]
- Ran, R.; Li, T.; Liu, X.; Ni, H.; Li, W.; Meng, F. RNA interference-mediated silencing of genes involved in the immune responses of the soybean pod borer Leguminivora glycinivorella (Lepidoptera: Olethreutidae). PeerJ 2018, 6, e4931. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Ran, R.X.; Li, Y.; Li, N.; Li, H.Z.; Wang, Z.K.; Li, W.B. RNA interference mediated serine protease gene (Spbtry1) knockdown affects growth and mortality in the soybean pod borer (Lepidoptera: Olethreutidae). Fla. Entomol. 2017, 100, 607–615. [Google Scholar] [CrossRef]
- Yang, C.; Yan, J.; Jiang, S.; Li, X.; Min, H.; Wang, X.; Hao, D. Resequencing 250 Soybean Accessions: New Insights into Genes Associated with Agronomic Traits and Genetic Networks. Genom. Proteom. Bioinform. 2021, in press. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Jamann, T.M.; Balint-Kurti, P.J.; Holland, J.B. QTL mapping using high-throughput sequencing. Plant Funct. Genom. 2015, 1284, 257–285. [Google Scholar]
- Hina, A.; Cao, Y.; Song, S.; Li, S.; Sharmin, R.A.; Elattar, M.A.; Bhat, J.A.; Zhao, T. High-Resolution Mapping in Two RIL Populations Refines Major “QTL Hotspot” Regions for Seed Size and Shape in Soybean (Glycine max L.). Int. J. Mol. Sci. 2020, 21, 1040. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zheng, H.; Bi, Y.; Yang, L.; Liu, H.; Wang, J.; Sun, J.; Zhao, H.; Li, X.; Li, J.; et al. Identification of a Major QTL and Candidate Gene Analysis of Salt Tolerance at the Bud Burst Stage in Rice (Oryza sativa L.) Using QTL-Seq and RNA-Seq. Rice 2020, 13, 55. [Google Scholar] [CrossRef]
- Wang, L.; Conteh, B.; Fang, L.; Xia, Q.; Nian, H. QTL mapping for soybean (Glycine max L.) leaf chlorophyll-content traits in a genotyped RIL population by using RAD-seq based high-density linkage map. BMC Genom. 2020, 21, 739. [Google Scholar] [CrossRef]
- Guo, T.; Yang, J.; Li, D.; Sun, K.; Luo, L.; Xiao, W.; Wang, J.; Liu, Y.; Wang, S.; Wang, H.; et al. Integrating GWAS, QTL, mapping and RNA-seq to identify candidate genes for seed vigor in rice (Oryza sativa L.). Mol. Breed. 2019, 39, 87. [Google Scholar] [CrossRef]
- Huang, J.; Ma, Q.; Cai, Z.; Xia, Q.; Li, S.; Jia, J.; Chu, L.; Lian, T.; Nian, H.; Cheng, Y. Identification and mapping of stable QTLs for seed oil and protein content in soybean [Glycine max (L.) Merr.]. J. Agric. Food Chem. 2020, 68, 6448–6460. [Google Scholar] [CrossRef]
- Ye, J.; Yang, Y.; Chen, B.; Shi, J.; Luo, M.; Zhan, J.; Wang, X.; Liu, G.; Wang, H. An integrated analysis of QTL mapping and RNA sequencing provides further insights and promising candidates for pod number variation in rapeseed (Brassica napus L.). BMC Genom. 2017, 18, 71. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, C.; Zhu, D.; He, H.; Wei, Z.; Yuan, Q.; Li, X.; Gao, X.; Zhang, B.; Gao, H. Identifying candidate genes and patterns of heat-stress response in rice using a genome-wide association study and transcriptome analyses. Crop J. 2022, in press. [Google Scholar] [CrossRef]
- Lyu, D.; Xu, W.; Lu, H.; Shi, S. Structure of soybean pod of 160 spring soybean varieties and analysis of resistance to Leguminivora glycinivorella. Chin. J. Oil Crop Sci. 2018, 40, 413–419. (In Chinese) [Google Scholar]
- Lyu, D.; Xu, W.; Shi, S. The Structure of Soybean Pod of 45 Spring Soybean Varieties and Evaluation of Resistance to Leguminivora glycinivorell. Soybean Sci. 2018, 37, 275–283. (In Chinese) [Google Scholar]
- Fu, X.; Xu, W.; Lu, A.; Bi, J.; Su, X.; Shi, S. Relationship Between Components Change of Soybean Volatile and Leguminivora glycinivorella Infestation. J. Jilin Agric. Univ. 2014, 36, 389–394. (In Chinese) [Google Scholar] [CrossRef]
- Wari, D.; Aboshi, T.; Shinya, T.; Galis, I. Integrated view of plant metabolic defense with particular focus on chewing herbivores. J. Integr. Plant Biol. 2022, 64, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Browse, J. Jasmonate: An Oxylipin Signal with Many Roles in Plants. In Vitamins & Hormones; Litwack, G., Ed.; Academic Press: Cambridge, MA, USA, 2005; Volume 72, pp. 431–456. ISBN 0083-6729. [Google Scholar]
- Schmelz, E.A.; Engelberth, J.; Alborn, H.T.; Tumlinson, J.H.; Teal, P.E.A. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc. Natl. Acad. Sci. USA 2009, 106, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, M.; Molla, K.A.; Karmakar, S.; Datta, K.; Datta, S.K. Development of pod borer-resistant transgenic chickpea using a pod-specific and a constitutive promoter-driven fused cry1Ab/Ac gene. Theor. Appl. Genet. 2014, 127, 2555–2565. [Google Scholar] [CrossRef]
- Dillon, F.M.; Chludil, H.D.; Mithöfer, A.; Zavala, J.A. Solar UVB-inducible ethylene alone induced isoflavonoids in pods of field-grown soybean, an important defense against stink bugs. Environ. Exp. Bot. 2020, 178, 104167. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Shan, Z.; Hao, Q.; Zhang, C.; Yang, Z.; Zhang, X.; Yuan, S.; Qiu, D.; Chen, S.; et al. Herbivore defense responses and associated herbivore defense mechanism as revealed by comparing a resistant wild soybean with a susceptible cultivar. Crop J. 2015, 3, 451–467. [Google Scholar] [CrossRef]
- Kirsch, R.; Vurmaz, E.; Schaefer, C.; Eberl, F.; Sporer, T.; Haeger, W.; Pauchet, Y. Plants use identical inhibitors to protect their cell wall pectin against microbes and insects. Ecol. Evol. 2020, 10, 3814–3824. [Google Scholar] [CrossRef]
- Bentivenha, J.P.F.; Canassa, V.F.; Baldin, E.L.L.; Borguini, M.G.; Lima, G.P.P.; Lourenção, A.L. Role of the Rutin and Genistein Flavonoids in Soybean Resistance to Piezodorus guildinii (Hemiptera: Pentatomidae). Arthropod-Plant Inte. 2018, 12, 311–320. [Google Scholar] [CrossRef]
- Piubelli, G.C.; Hoffmann-Campo, C.B.; De Arruda, I.C.; Franchini, J.C.; Lara, F.M. Flavonoid Increase in Soybean as a Response to Nezara viridula Injury and Its Effect on Insect-Feeding Preference. J. Chem. Ecol. 2003, 29, 1223–1233. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, W.; Chen, L.; Shen, X.; Yang, H.; Fang, Y.; Ouyang, W.; Mai, S.; Chen, H.; Chen, S.; et al. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmUGT Enhanced Soybean Resistance Against Leaf-Chewing Insects Through Flavonoids Biosynthesis. Front. Plant Sci. 2022, 13, 802716. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, H.; Tao, J.; Ren, Y.; Xu, C.; Wu, K.; Zou, C.; Zhang, J.; Xu, Y. Development of multiple SNP marker panels affordable to breeders through genotyping by target sequencing (GBTS) in maize. Mol. Breed. 2019, 39, 37. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Rajput, S.G.; Santra, D.K.; Schnable, J. Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol. Breed. 2016, 36, 37. [Google Scholar] [CrossRef]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010, 38, D843–D846. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.; Meng, Y.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397. [Google Scholar] [CrossRef]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 2013, 14, 689. [Google Scholar] [CrossRef]
- Li, R.; Jiang, H.; Zhang, Z.; Zhao, Y.; Xie, J.; Wang, Q.; Zheng, H.; Hou, L.; Xiong, X.; Xin, D.; et al. Combined Linkage Mapping and BSA to Identify QTL and Candidate Genes for Plant Height and the Number of Nodes on the Main Stem in Soybean. Int. J. Mol. Sci. 2019, 21, 42. [Google Scholar] [CrossRef]
- Cao, Y.; Li, S.; Chen, G.; Wang, Y.; Bhat, J.A.; Karikari, B.; Kong, J.; Gai, J.; Zhao, T. Deciphering the Genetic Architecture of Plant Height in Soybean Using Two RIL Populations Sharing a Common M8206 Parent. Plants 2019, 8, 373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kjemtrup-Lovelace, S.; Li, C.; Luo, Y.; Chen, L.P.; Song, B. Comparative RNA-seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja). Sci. Rep. 2017, 7, 9699. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.Z.; Ishiga, Y.; Yamanaka, N.; Ogiso-Tanaka, E.; Yamaoka, Y. Soybean leaves transcriptomic data dissects the phenylpropanoid pathway genes as a defence response against Phakopsora pachyrhizi. Plant Physiol. Bioch. 2018, 132, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, D.; Cai, L.; Wang, H.; Liu, X.; Du, H.; Yang, Z.; Zhang, H.; Hu, Z.; Huang, F.; et al. CALCIUM-DEPENDENT PROTEIN KINASE38 regulates flowering time and common cutworm resistance in soybean. Plant Physiol. 2022, 190, 480–499. [Google Scholar] [CrossRef] [PubMed]
- Hettenhausen, C.; Sun, G.; He, Y.; Zhuang, H.; Sun, T.; Qi, J.; Wu, J. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 2016, 6, 18973. [Google Scholar] [CrossRef] [PubMed]
- Jun, T.; Rouf Mian, M.A.; Michel, A.P. Genetic mapping revealed two loci for soybean aphid resistance in PI 567301B. Theor. Appl. Genet. 2012, 124, 13–22. [Google Scholar] [CrossRef]
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef]
- Schilling, S.; Wasternack, C.; Demuth, H. Glutaminyl cyclases from animals and plants: A case of functionally convergent protein evolution. Biol. Chem. 2008, 389, 983–991. [Google Scholar] [CrossRef]
- Rajaraman, J.; Douchkov, D.; Hensel, G.; Stefanato, F.L.; Gordon, A.; Ereful, N.; Caldararu, O.F.; Petrescu, A.; Kumlehn, J.; Boyd, L.A.; et al. An LRR/Malectin Receptor-Like Kinase Mediates Resistance to Non-adapted and Adapted Powdery Mildew Fungi in Barley and Wheat. Front. Plant Sci. 2016, 7, 1836. [Google Scholar] [CrossRef]
- Shao, Z.; Xue, J.; Wang, Q.; Wang, B.; Chen, J. Revisiting the Origin of Plant NBS-LRR Genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S.; Hashem, A.; Abd Allah, E.F.; Ansari, M.I. Plant Defense Responses to Biotic Stress and Its Interplay With Fluctuating Dark/Light Conditions. Front. Plant Sci. 2021, 12, 631810. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.W.; Abeysinghe, J.K.; Kamali, M. Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [PubMed]
- de Bobadilla, M.F.; Vitiello, A.; Erb, M.; Poelman, E.H. Plant defense strategies against attack by multiple herbivores. Trends Plant Sci. 2022, 27, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Kariola, T.; Brader, G.; Li, J.; Palva, E.T. Chlorophyllase 1, a Damage Control Enzyme, Affects the Balance between Defense Pathways in Plants. Plant Cell 2005, 17, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Alves, P.C.; Ahmad, I.; Gaffoor, I.; Acevedo, F.E.; Peiffer, M.; Jin, S.; Han, Y.; Shakeel, S.; Felton, G.W. Turnabout is fair play: Herbivory-induced plant chitinases excreted in fall armyworm frass suppress herbivore defenses in maize. Plant Physiol. 2016, 171, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, H.; Long, Y.; Shu, Y.; Zhai, J. Plant Public RNA-seq Database: A comprehensive online database for expression analysis of ~45,000 plant public RNA-Seq libraries. Plant Biotechnol. J. 2022, 20, 806–808. [Google Scholar] [CrossRef]
- Whiteman, N.K.; Groen, S.C.; Chevasco, D.; Bear, A.; Beckwith, N.; Gregory, T.R.; Denoux, C.; Mammarella, N.; Ausubel, F.M.; Pierce, N.E. Mining the plant–herbivore interface with a leafmining Drosophila of Arabidopsis. Mol. Ecol. 2011, 20, 995–1014. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Lin, H. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Heinzel, N.; Schöttner, M.; Baldwin, I.T.; Gális, I. R2R3-NaMYB8 Regulates the Accumulation of Phenylpropanoid-Polyamine Conjugates, Which Are Essential for Local and Systemic Defense against Insect Herbivores in Nicotiana attenuata. Plant Physiol. 2010, 152, 1731–1747. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.M.; Marchi-Werle, L.; Hunt, T.E.; Heng-Moss, T.M.; Louis, J. Abscisic and Jasmonic Acids Contribute to Soybean Tolerance to the Soybean Aphid (Aphis glycines Matsumura). Sci. Rep. 2018, 8, 15148. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Ma, Y.; Yang, W.; Yang, Q.; Yu, D. Identification of soybean herbivory-regulated genes and a transgenic investigation of their potential in insect resistance. Plant Cell Tissue Organ Cult. 2015, 123, 321–340. [Google Scholar] [CrossRef]
- Selig, P.; Keough, S.; Nalam, V.J.; Nachappa, P. Jasmonate-dependent plant defenses mediate soybean thrips and soybean aphid performance on soybean. Arthropod-Plant Inte. 2016, 10, 273–282. [Google Scholar] [CrossRef]
- Dong, H.; Peng, J.; Bao, Z.; Meng, X.; Bonasera, J.M.; Chen, G.; Beer, S.V.; Dong, H. Downstream Divergence of the Ethylene Signaling Pathway for Harpin-Stimulated Arabidopsis Growth and Insect Defense. Plant Physiol. 2004, 136, 3628–3638. [Google Scholar] [CrossRef] [PubMed]
- Harfouche, A.L.; Shivaji, R.; Stocker, R.; Williams, P.W.; Luthe, D.S. Ethylene Signaling Mediates a Maize Defense Response to Insect Herbivory. Mol. Plant Microbe Interact. 2006, 19, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Dezar, C.A.; Giacomelli, J.I.; Manavella, P.A.; Ré, D.A.; Alves-Ferreira, M.; Baldwin, I.T.; Bonaventure, G.; Chan, R.L. HAHB10, a sunflower HD-Zip II transcription factor, participates in the induction of flowering and in the control of phytohormone-mediated responses to biotic stress. J. Exp. Bot. 2011, 62, 1061–1076. [Google Scholar] [CrossRef]
- Shani, E.; Salehin, M.; Zhang, Y.; Sanchez, S.E.; Doherty, C.; Wang, R.; Mangado, C.C.; Song, L.; Tal, I.; Pisanty, O.; et al. Plant Stress Tolerance Requires Auxin-Sensitive Aux/IAA Transcriptional Repressors. Curr. Biol. 2017, 27, 437–444. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Wild, M.; Davière, J.; Cheminant, S.; Regnault, T.; Baumberger, N.; Heintz, D.; Baltz, R.; Genschik, P.; Achard, P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012, 24, 3307–3319. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Fan, Y.; Cao, H.; Song, Z.; Dong, B.; Liu, T.; Yang, W.; Wang, M.; Niu, L.; Yang, Q.; et al. Transcriptome analysis revealed key genes involved in flavonoid metabolism in response to jasmonic acid in pigeon pea (Cajanus cajan (L.) Millsp.). Plant Physiol. Bioch. 2021, 168, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Shen, G.; Di, S.; Fan, C.; Chang, Z.; Pang, Y. Genome-Wide Identification and Functional Characterization of UDP-Glucosyltransferase Genes Involved in Flavonoid Biosynthesis in Glycine max. Plant Cell Physiol. 2017, 58, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S. Mapping Quantitative Trait Loci in F2 Incorporating Phenotypes of F3 Progeny. Genetics 2004, 166, 1981–1993. [Google Scholar] [CrossRef]
- Tang, Q.; Zhang, C. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Fralick, D.; Zheng, J.Z.; Wang, B.; Changyong, F. The differences and similarities between two-sample t-test and paired t-test. Shanghai Arch. Psychiatry 2017, 29, 184. [Google Scholar] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, X.; Pan, Q.; Li, P.; Liu, Y.; Lu, X.; Zhong, W.; Li, M.; Han, L.; Li, J.; et al. QTG-Seq Accelerates QTL Fine Mapping through QTL Partitioning and Whole-Genome Sequencing of Bulked Segregant Samples. Mol. Plant 2019, 12, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
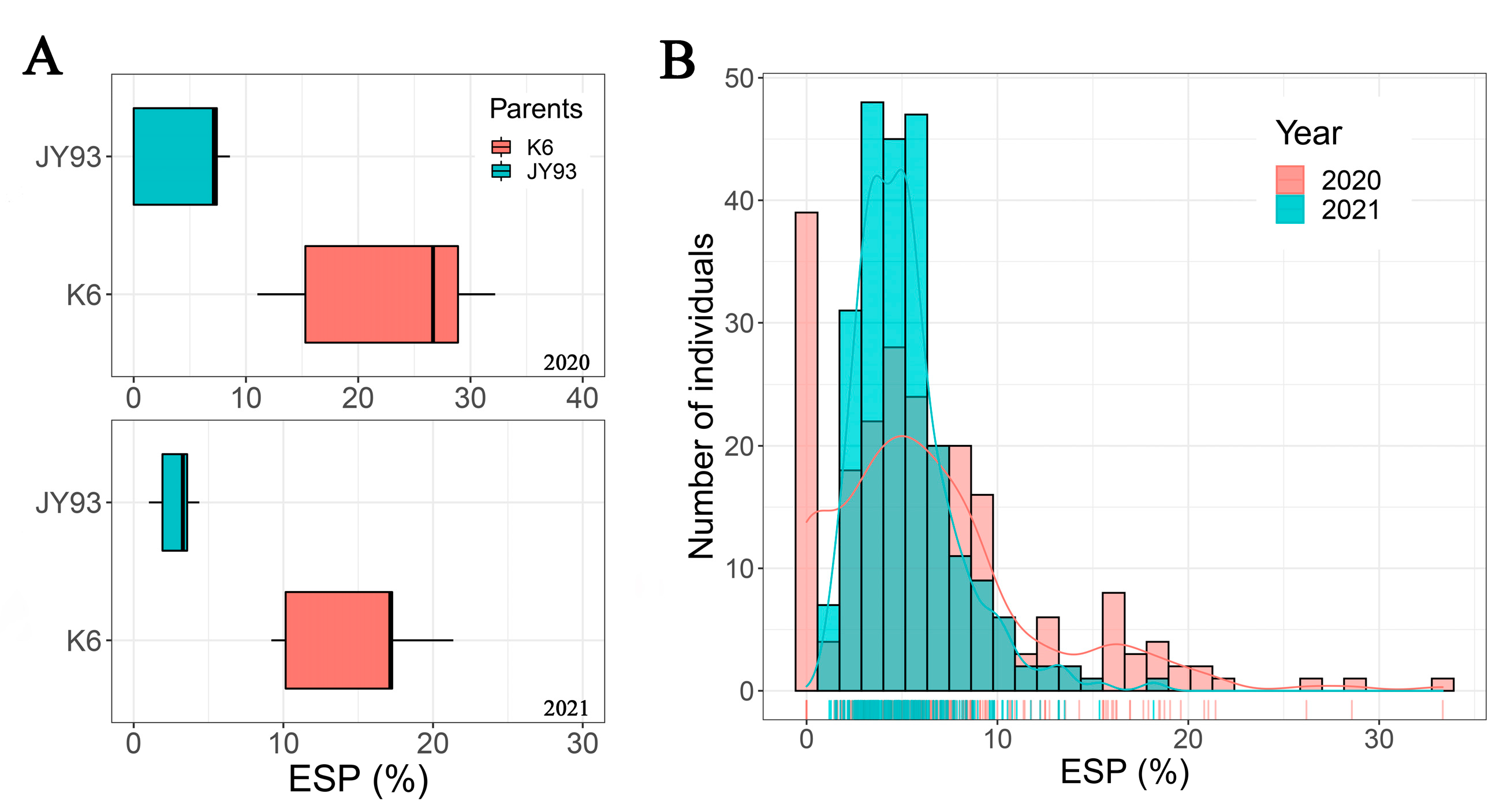
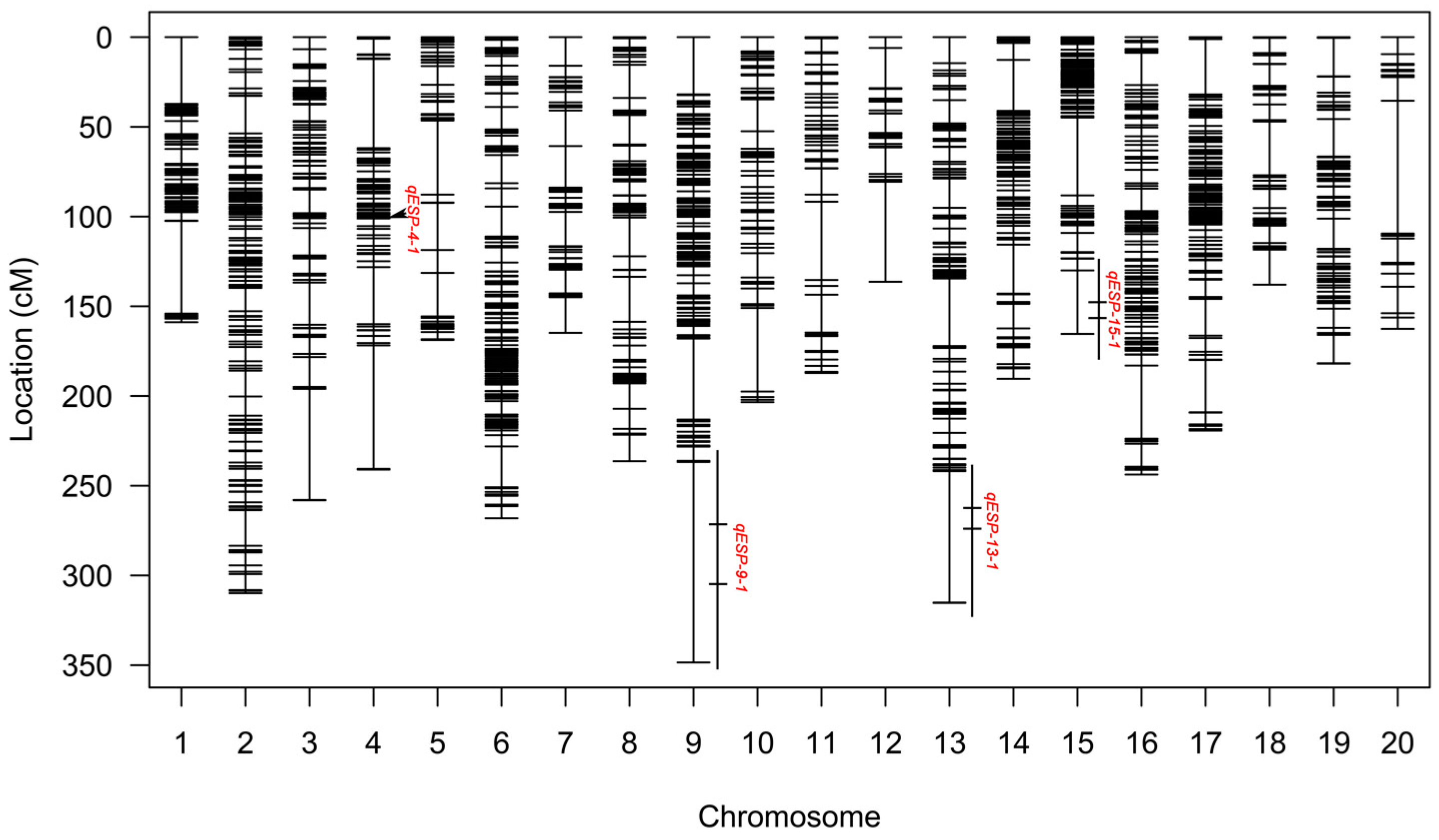


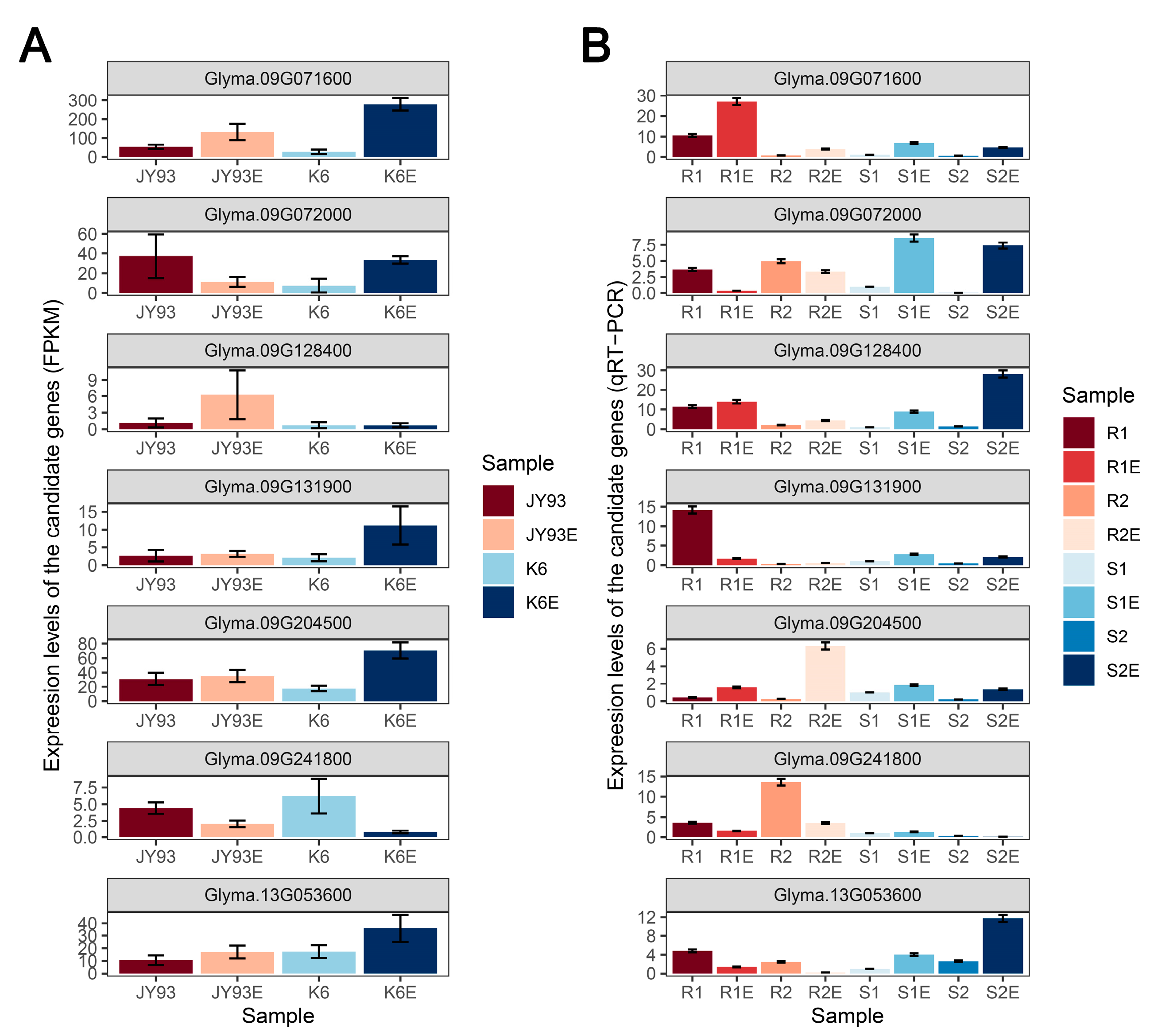
| Year | Parents | Offspring of JY93 × K6 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| JY93 + | K6 + | Min | Max | Mean | SD | CV (%) | Skewness | Kurtosis | |
| 2020 | 4.60 A,a | 22.81 B,a | 0.00 | 33.33 | 6.44 b | 5.63 | 87.39 | 1.48 | 3.17 |
| 2021 | 2.84 A,a | 15.02 B,a | 1.19 | 18.18 | 5.19 a | 2.65 | 51.09 | 1.48 | 3.38 |
| Chromosome | Linkage Group | Number of SNPs | Linkage Distance (cM) | Average Distance between Markers (cM) |
|---|---|---|---|---|
| Chr01 | D1a | 69 | 158.87 | 2.30 |
| Chr02 | D1b | 155 | 309.88 | 2.00 |
| Chr03 | N | 78 | 258.15 | 3.31 |
| Chr04 | C1 | 68 | 241.04 | 3.54 |
| Chr05 | A1 | 45 | 168.80 | 3.75 |
| Chr06 | C2 | 147 | 268.14 | 1.82 |
| Chr07 | M | 58 | 164.83 | 2.84 |
| Chr08 | A2 | 82 | 236.34 | 2.88 |
| Chr09 | K | 135 | 348.46 | 2.58 |
| Chr10 | O | 60 | 203.45 | 3.39 |
| Chr11 | B1 | 51 | 187.19 | 3.67 |
| Chr12 | H | 28 | 136.37 | 4.87 |
| Chr13 | F | 90 | 315.31 | 3.50 |
| Chr14 | B2 | 95 | 190.43 | 2.00 |
| Chr15 | E | 87 | 165.41 | 1.90 |
| Chr16 | J | 120 | 243.79 | 2.03 |
| Chr17 | D2 | 122 | 219.30 | 1.80 |
| Chr18 | G | 43 | 137.98 | 3.21 |
| Chr19 | L | 77 | 181.97 | 2.36 |
| Chr20 | I | 24 | 162.59 | 6.77 |
| Total | − | 1634 | 4298.30 | 2.63 |
| QTL Name | Year | Chr. a | Position (cM) | LeftCI | RightCI | LOD | Interval Region (bp) c | PVE | Additive Effect |
|---|---|---|---|---|---|---|---|---|---|
| (cM) b | (cM) b | (%) | |||||||
| qESP-4-1 | 2021 | 4 | 105 | 100.5 | 107.5 | 3.24 | 8,394,899–9,376,933 | 0.8439 | 0.0059 |
| qESP-9-1 | 2020 | 9 | 284 | 271.5 | 293.5 | 8.05 | 2,021,437–49,893,816 | 4.3313 | −0.0632 |
| qESP-9-1 | 2021 | 9 | 292 | 276.5 | 309.5 | 2.64 | 2,021,437–49,893,816 | 6.0935 | 0.0231 |
| qESP-13-1 | 2021 | 13 | 273 | 268.5 | 275.5 | 3.16 | 14,613,981–26,717,362 | 5.7935 | −0.0219 |
| qESP-15-1 | 2021 | 15 | 165 | 149.5 | 165 | 2.82 | 583,964–4,836,857 | 0.7931 | −0.0013 |
| Gene Locus | QTL Name | A. thaliana Homolog | Response Pattern a | Description |
|---|---|---|---|---|
| Glyma.04G098000 | qESP-4-1 | AT4G40080 | Reverse | ENTH/ANTH/VHS superfamily protein |
| Glyma.09G040500 | qESP-9-1 | NA | Up (JY93) | Pathogenesis-related protein |
| Glyma.09G062900 | qESP-9-1 | AT4G26140 | Up (JY93) | Beta-galactosidase |
| Glyma.09G071600 | qESP-9-1 | AT1G19180 | Co-up | Jasmonate-zim-domain protein |
| Glyma.09G072000 | qESP-9-1 | AT1G19210 | Reverse | ERF family proteins |
| Glyma.09G128400 | qESP-9-1 | AT3G16520 | Up (JY93) | UDP-glucosyl transferase |
| Glyma.09G131900 | qESP-9-1 | AT4G25720 | Up (K6) | Glutaminyl cyclase |
| Glyma.09G143800 | qESP-9-1 | AT1G63120 | Up (K6) | RHOMBOID-like protein |
| Glyma.09G149400 | qESP-9-1 | AT5G62180 | Down (JY93) | Carboxylesterase |
| Glyma.09G204500 | qESP-9-1 | AT1G32640 | Up (K6) | MYC family protein |
| Glyma.09G241800 | qESP-9-1 | AT3G60390 | Co-down | HD-ZIP family proteins |
| Glyma.13G053600 | qESP-13-1 | AT3G51550 | Up (K6) | Malectin family protein |
| Glyma.13G054200 | qESP-13-1 | AT3G51550 | Up (K6) | Malectin family protein |
| Glyma.13G054400 | qESP-13-1 | AT3G51550 | Up (K6) | Malectin family protein |
| Glyma.13G054600 | qESP-13-1 | AT4G20050 | Up (JY93) | Pectin lyase-like superfamily protein |
| Glyma.13G113800 | qESP-13-1 | AT5G20820 | Up (JY93) | SAUR-like auxin-responsive protein family |
| Glyma.15G024800 | qESP-15-1 | AT5G17680 | Down (JY93) | Disease-resistant protein |
| Glyma.15G062300 | qESP-15-1 | AT2G19990 | Up (JY93) | Pathogenesis-related protein like |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Song, B.; Yu, C.; Zhang, J.; Zhang, J.; Bi, R.; Li, X.; Ren, X.; Zhu, Y.; Yao, D.; et al. Identifying Soybean Pod Borer (Leguminivora glycinivorella) Resistance QTLs and the Mechanism of Induced Defense Using Linkage Mapping and RNA-Seq Analysis. Int. J. Mol. Sci. 2022, 23, 10910. https://doi.org/10.3390/ijms231810910
Chen L, Song B, Yu C, Zhang J, Zhang J, Bi R, Li X, Ren X, Zhu Y, Yao D, et al. Identifying Soybean Pod Borer (Leguminivora glycinivorella) Resistance QTLs and the Mechanism of Induced Defense Using Linkage Mapping and RNA-Seq Analysis. International Journal of Molecular Sciences. 2022; 23(18):10910. https://doi.org/10.3390/ijms231810910
Chicago/Turabian StyleChen, Liangyu, Baixing Song, Cheng Yu, Jun Zhang, Jian Zhang, Rui Bi, Xueying Li, Xiaobo Ren, Yanyu Zhu, Dan Yao, and et al. 2022. "Identifying Soybean Pod Borer (Leguminivora glycinivorella) Resistance QTLs and the Mechanism of Induced Defense Using Linkage Mapping and RNA-Seq Analysis" International Journal of Molecular Sciences 23, no. 18: 10910. https://doi.org/10.3390/ijms231810910
APA StyleChen, L., Song, B., Yu, C., Zhang, J., Zhang, J., Bi, R., Li, X., Ren, X., Zhu, Y., Yao, D., Song, Y., Yang, S., & Zhao, R. (2022). Identifying Soybean Pod Borer (Leguminivora glycinivorella) Resistance QTLs and the Mechanism of Induced Defense Using Linkage Mapping and RNA-Seq Analysis. International Journal of Molecular Sciences, 23(18), 10910. https://doi.org/10.3390/ijms231810910







