Identification of AGR2 Gene-Specific Expression Patterns Associated with Epithelial-Mesenchymal Transition
Abstract
1. Introduction
2. Results
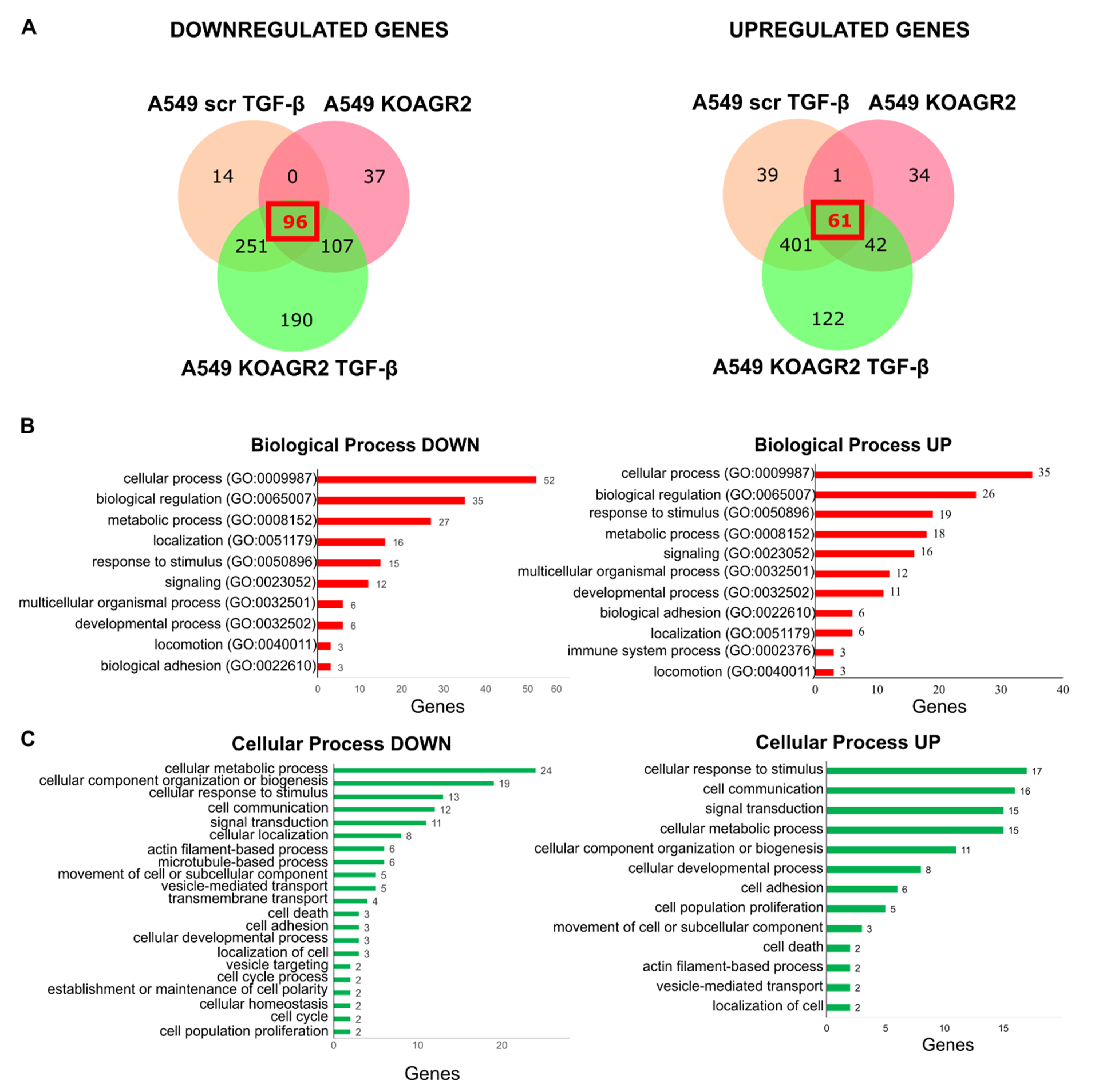
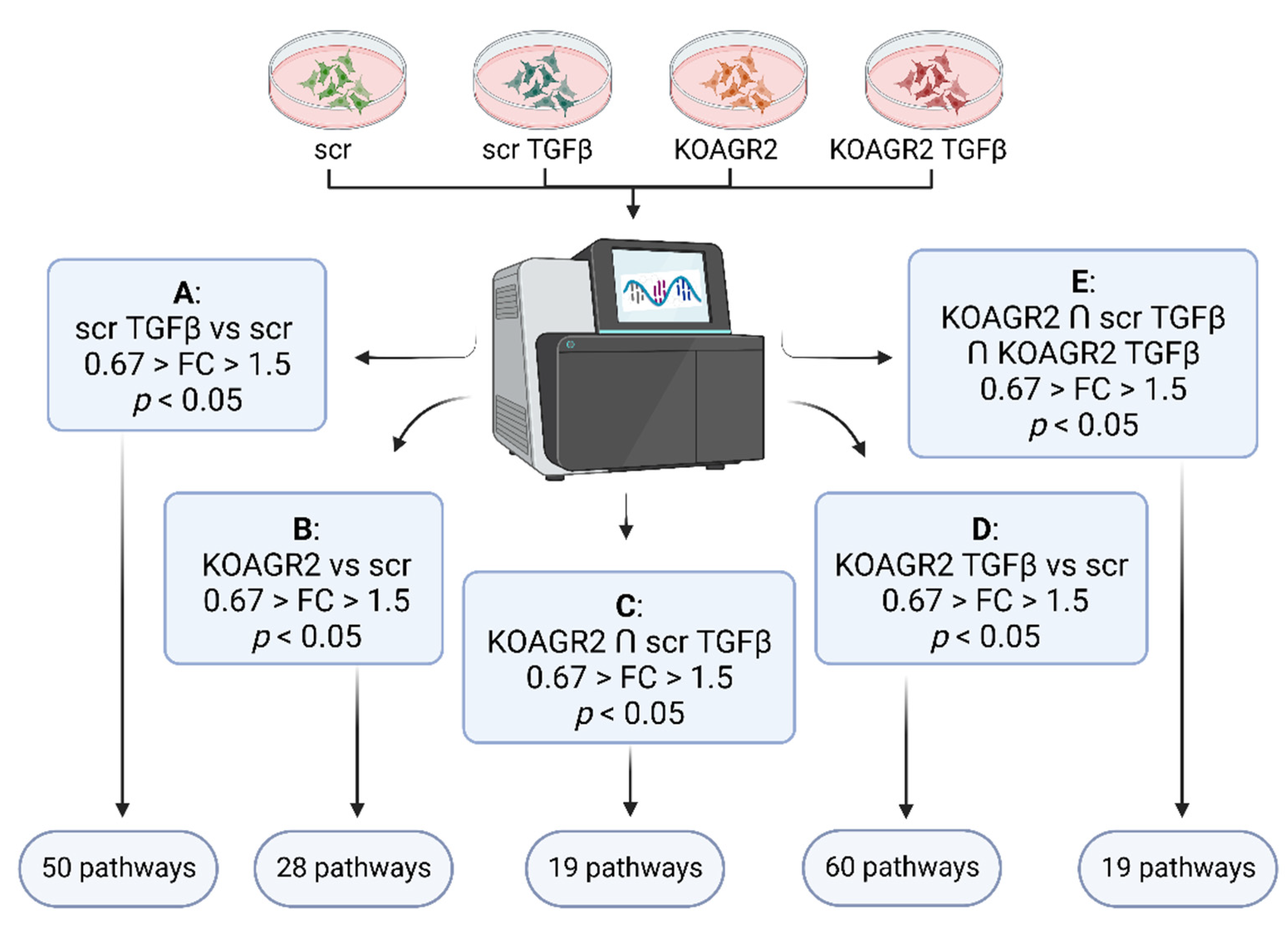
2.1. Transcriptome Analysis
2.2. Functional Annotation
| Gene | Regulation | Function |
|---|---|---|
| ROCK1 | UP | Promotes adhesion, migration, and invasion by inhibition of PTEN and activation of the PI3K/Akt signaling pathway through FAK phosphorylation. Overexpression is associated with invasive and metastatic phenotype [25]. |
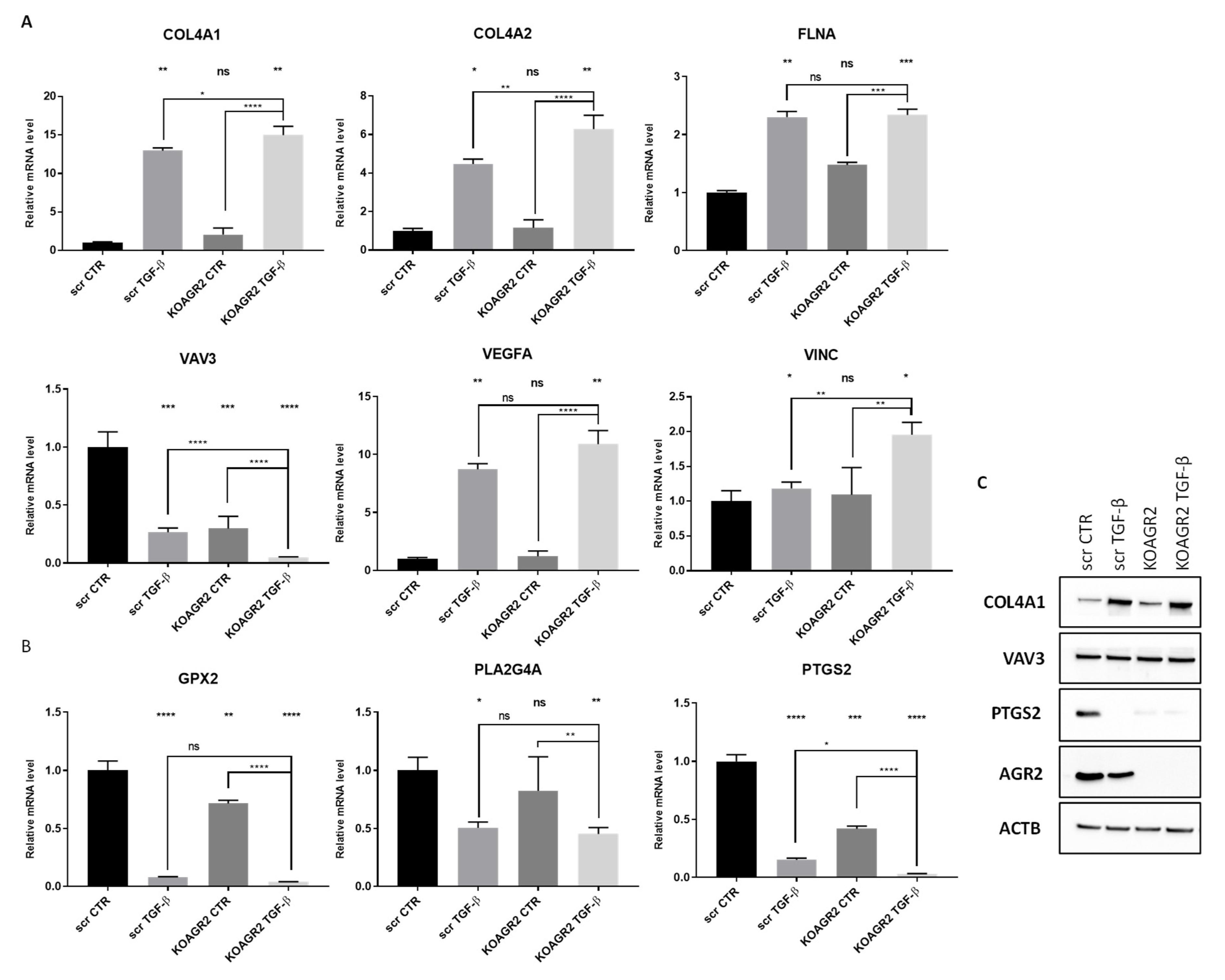
| COL4A1 | UP | Shown to be overexpressed by a number of malignancies, including hepatocellular, urothelial, or pancreatic cancer, where it contributes to migratory and invasive phenotype [26,27,28]. |
| COL4A2 | UP | Upregulated in pre-neoplastic and HCC tissue and overexpression correlated with shorter progression survival [29]. Reported to promote cell adhesion, migration, and proliferation of different cell types [30,31]. |
| FLNA | UP | Actin crosslinking protein with an important function in migration and adhesion [32]. Showed to be suppressed by miR-200c independently of ZEB1 [33]. |
| MYLK | UP | Also known as MLCK, contributes to adhesion, migration, invasion, and metastasis [34,35] and represents a miR-200c target [36]. Phosphorylates myosin light chain, which facilitates the association of myosin with F-actin and hence generates contractile forces in HCC [37]. |
| PAK3 | UP | SMAD4 effector mediating metastatic signals through the PAK3-JNK-Jun pathway in NSCLC [38]. |
| TNC | UP | An ECM glycoprotein inducing EMT and activating Src and FAK [39,40]. In Pancreatic cancer, it activates JNK/C-Jun pathway leading to production of MMP-9 and induced paxillin phosphorylation and FA formation [41]. |
| VEGFA | UP | Well-known mediator of angiogenesis and a regulator of proliferation, survival, adhesion, migration, and invasion [42]. |
| VCL | UP | Regulates polarized migration, controls integrin activity through interaction with talin, regulates recruitment and diffusion of core FA proteins in a force-dependent manner, and is required for an efficient cellular adhesion and migration [43]. |
| ZYX | UP | One of the key FA proteins contributing to migration, invasion, adhesion, and proliferation [44,45]. Induced in A549 by TGF-β through Smad3 [46]. |
| COL4A4 | DOWN | Component of basement membrane and a component of focal adhesion (according to WikiPathways and KEGG pathways). |
| FN1 | DOWN | Seemingly conflicting role in cancer and metastasis, which is well discussed in a review by Lin and colleagues [47]. |
| ITGB4 | DOWN | Transcriptionally repressed by ZEB1 [48]. |
| ITGB5 | DOWN | Plays a role in TGF-β induced EMT [49] and facilitation of migration in HCC [50]. |
| ITGB8 | DOWN | Exclusive heterodimerization with αv subunit—αvβ8 serves as a receptor for latent TGF-β and activates it in protease dependent manner [51]. αvβ8-TGF-β axis mediates cell-cell communication and its dysregulation leads to aberrant adhesion and signaling [52]. Interacts with FAK, which activates VAV and RAC1 in endometrial epithelial cells [53]. |
| LAMA3 | DOWN | Part of laminins, which are further composed of α and γ subunits. LAMA3 encodes 2 different transcripts, LAMA3A and 3B. Laminin 332 containing laminin α3A was described as a regulator of cell migration, and in focal adhesion α3A interacts with integrin α3β1, which associates with signaling molecules and connects to the actin cytoskeleton through linker molecules. Further details are available in the review by Hamill et al. [54]. |
| VAV3 | DOWN | Acts as an EMT promoter together with ZEB1 [55]. |
2.3. RNAseq Data Validation
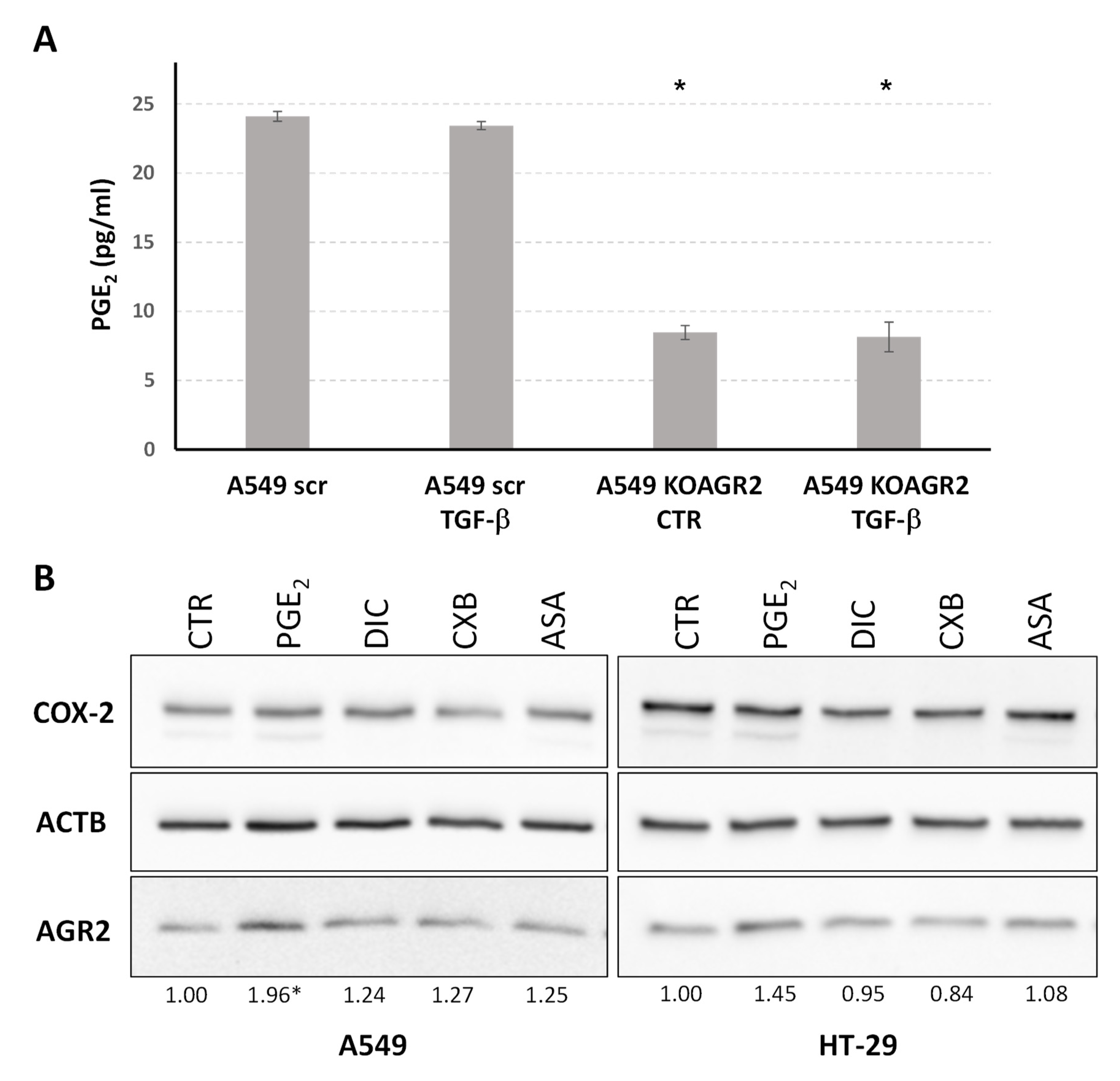
2.4. The Role of AGR2 in Relation to Arachidonic Acid Metabolism and Prostaglandin E2 Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Reagents
4.2. RNA Purification and Sequencing
4.3. Data Analysis, Statistical Analysis and Data Mining
4.4. Gene Expression
4.5. Western Blot Analysis
4.6. Immunofluorescent Staining
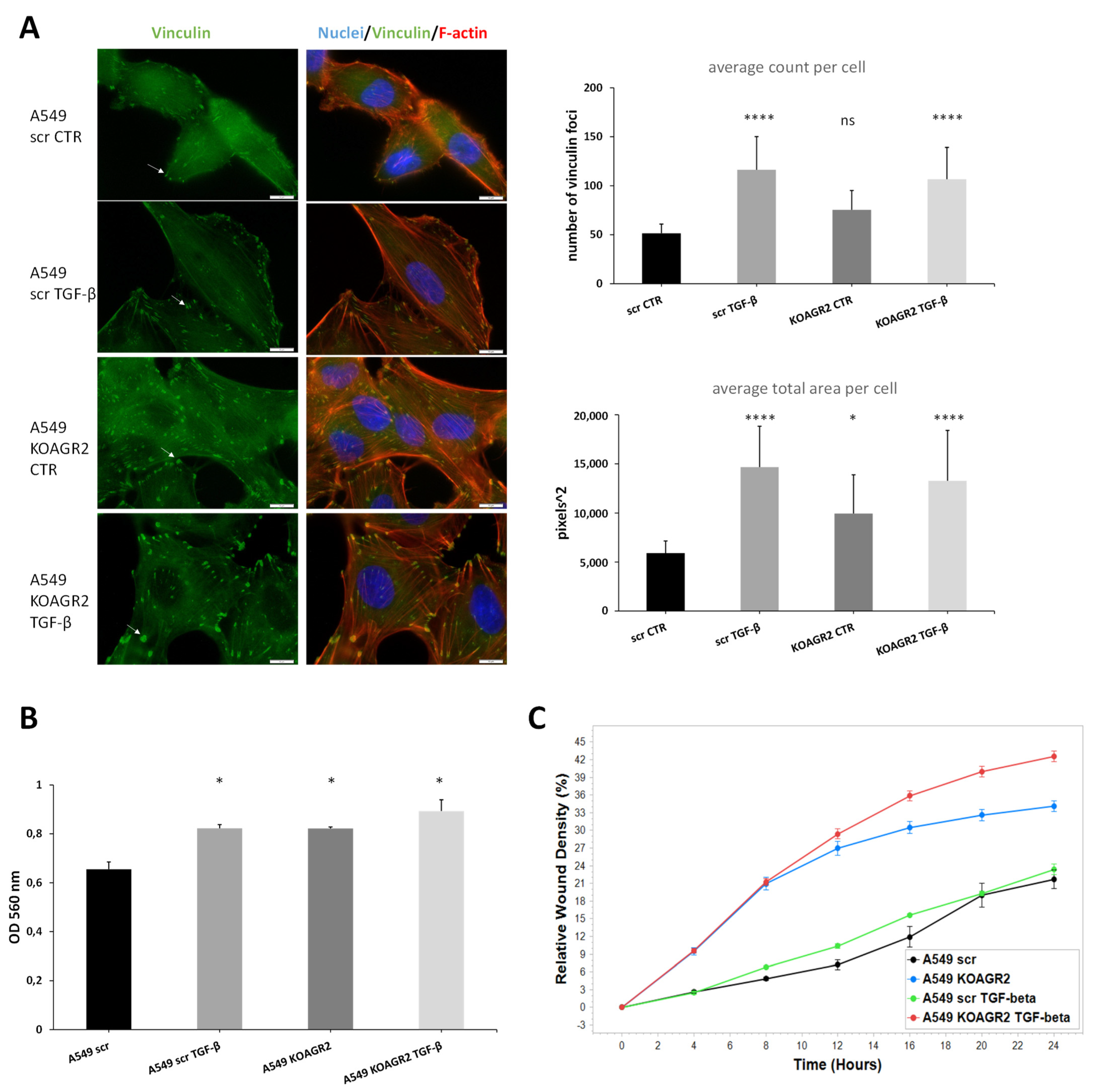
4.7. Invasion and Migration Analysis
4.8. PGE2 ELISA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Jung, A.; Reu, S.; Porzner, M.; Hlubek, F.; Kunz-Schughart, L.A.; Knuechel, R.; Kirchner, T. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA 2001, 98, 10356–10361. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Chevet, E.; Fessart, D.; Delom, F.; Mulot, A.; Vojtesek, B.; Hrstka, R.; Murray, E.; Gray, T.; Hupp, T. Emerging roles for the pro-oncogenic anterior gradient-2 in cancer development. Oncogene 2013, 32, 2499–2509. [Google Scholar] [CrossRef]
- Jach, D.; Cheng, Y.; Prica, F.; Dumartin, L.; Crnogorac-Jurcevic, T. From development to cancer—An ever-increasing role of AGR2. Am. J. Cancer Res. 2021, 11, 5249–5262. [Google Scholar]
- Hrstka, R.; Bouchalova, P.; Michalova, E.; Matoulkova, E.; Muller, P.; Coates, P.J.; Vojtesek, B. AGR2 oncoprotein inhibits p38 MAPK and p53 activation through a DUSP10-mediated regulatory pathway. Mol. Oncol. 2016, 10, 652–662. [Google Scholar] [CrossRef]
- Ramachandran, V.; Arumugam, T.; Wang, H.; Logsdon, C.D. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res. 2008, 68, 7811–7818. [Google Scholar] [CrossRef]
- Tsuji, T.; Satoyoshi, R.; Aiba, N.; Kubo, T.; Yanagihara, K.; Maeda, D.; Goto, A.; Ishikawa, K.; Yashiro, M.; Tanaka, M. Agr2 mediates paracrine effects on stromal fibroblasts that promote invasion by gastric signet-ring carcinoma cells. Cancer Res. 2015, 75, 356–366. [Google Scholar] [CrossRef]
- Wang, Z.; Hao, Y.; Lowe, A.W. The adenocarcinoma-associated antigen, AGR2, promotes tumor growth, cell migration, and cellular transformation. Cancer Res. 2008, 68, 492–497. [Google Scholar] [CrossRef]
- Hrstka, R.; Brychtova, V.; Fabian, P.; Vojtesek, B.; Svoboda, M. AGR2 predicts tamoxifen resistance in postmenopausal breast cancer patients. Dis. Markers 2013, 35, 207–212. [Google Scholar] [CrossRef]
- Mizuuchi, Y.; Aishima, S.; Ohuchida, K.; Shindo, K.; Fujino, M.; Hattori, M.; Miyazaki, T.; Mizumoto, K.; Tanaka, M.; Oda, Y. Anterior gradient 2 downregulation in a subset of pancreatic ductal adenocarcinoma is a prognostic factor indicative of epithelial-mesenchymal transition. Lab. Invest. 2015, 95, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Eitoku, M.; Moriya, S.; Kondo, S.; Kiyosawa, H.; Watanabe, T.; Suganuma, N. Regulation of Gene Expression by Sodium Valproate in Epithelial-to-Mesenchymal Transition. Lung 2015, 193, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.M.; Gore, A.; Balboni, A.; Young, A.; Longnecker, D.S.; Korc, M. AGR2 is a SMAD4-suppressible gene that modulates MUC1 levels and promotes the initiation and progression of pancreatic intraepithelial neoplasia. Oncogene 2013, 32, 3867–3876. [Google Scholar] [CrossRef] [PubMed]
- Sommerova, L.; Ondrouskova, E.; Vojtesek, B.; Hrstka, R. Suppression of AGR2 in a TGF-beta-induced Smad regulatory pathway mediates epithelial-mesenchymal transition. BMC Cancer 2017, 17, 546. [Google Scholar] [CrossRef] [PubMed]
- Sommerova, L.; Ondrouskova, E.; Martisova, A.; Zoumpourlis, V.; Galtsidis, S.; Hrstka, R. ZEB1/miR-200c/AGR2: A New Regulatory Loop Modulating the Epithelial-Mesenchymal Transition in Lung Adenocarcinomas. Cancers 2020, 12, 1614. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.R.; Mao, L.; Deng, W.W.; Li, Y.C.; Bu, L.L.; Yu, G.T.; Zhang, W.F.; Sun, Z.J. AGR2 promotes the proliferation, migration and regulates epithelial-mesenchymal transition in salivary adenoid cystic carcinoma. Am. J. Transl. Res. 2017, 9, 507–519. [Google Scholar]
- Matsuda, Y.; Miura, K.; Yamane, J.; Shima, H.; Fujibuchi, W.; Ishida, K.; Fujishima, F.; Ohnuma, S.; Sasaki, H.; Nagao, M.; et al. SERPINI1 regulates epithelial-mesenchymal transition in an orthotopic implantation model of colorectal cancer. Cancer Sci. 2016, 107, 619–628. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- The Gene Ontology, C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lehtimaki, J.I.; Rajakyla, E.K.; Tojkander, S.; Lappalainen, P. Generation of stress fibers through myosin-driven reorganization of the actin cortex. Elife 2021, 10, e60710. [Google Scholar] [CrossRef]
- Jannie, K.M.; Ellerbroek, S.M.; Zhou, D.W.; Chen, S.; Crompton, D.J.; Garcia, A.J.; DeMali, K.A. Vinculin-dependent actin bundling regulates cell migration and traction forces. Biochem. J. 2015, 465, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhou, H.; Liu, Y.; Huang, J.; Liu, W.; Zhang, Q.; Tang, Q.; Sheng, F.; Li, G.; Zhang, R. ROCK1 promotes migration and invasion of nonsmallcell lung cancer cells through the PTEN/PI3K/FAK pathway. Int J. Oncol. 2019, 55, 833–844. [Google Scholar] [CrossRef]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Toritsuka, M.; Ohnishi, S.; Shimada, K.; Furuya, H.; Khadka, V.S.; Deng, Y.; et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017, 8, 36099–36114. [Google Scholar] [CrossRef]
- Ohlund, D.; Franklin, O.; Lundberg, E.; Lundin, C.; Sund, M. Type IV collagen stimulates pancreatic cancer cell proliferation, migration, and inhibits apoptosis through an autocrine loop. BMC Cancer 2013, 13, 154. [Google Scholar] [CrossRef]
- Wang, T.; Jin, H.; Hu, J.; Li, X.; Ruan, H.; Xu, H.; Wei, L.; Dong, W.; Teng, F.; Gu, J.; et al. COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK-Src signaling. J. Exp. Clin. Cancer Res. 2020, 39, 148. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Chen, Y.; Sohel, H.; Ke, X.; Chen, J.; Li, Y.X. The correlation and role analysis of COL4A1 and COL4A2 in hepatocarcinogenesis. Aging 2020, 12, 204–223. [Google Scholar] [CrossRef]
- Brown, C.W.; Brodsky, A.S.; Freiman, R.N. Notch3 overexpression promotes anoikis resistance in epithelial ovarian cancer via upregulation of COL4A2. Mol. Cancer Res. 2015, 13, 78–85. [Google Scholar] [CrossRef]
- Ohlund, D.; Lundin, C.; Ardnor, B.; Oman, M.; Naredi, P.; Sund, M. Type IV collagen is a tumour stroma-derived biomarker for pancreas cancer. Br. J. Cancer 2009, 101, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kang, X.; An, H.; Lv, Y.; Liu, X. The function and pathogenic mechanism of filamin A. Gene 2021, 784, 145575. [Google Scholar] [CrossRef] [PubMed]
- Ljepoja, B.; Schreiber, C.; Gegenfurtner, F.A.; Garcia-Roman, J.; Kohler, B.; Zahler, S.; Radler, J.O.; Wagner, E.; Roidl, A. Inducible microRNA-200c decreases motility of breast cancer cells and reduces filamin A. PLoS ONE 2019, 14, e0224314. [Google Scholar] [CrossRef]
- Khapchaev, A.Y.; Shirinsky, V.P. Myosin Light Chain Kinase MYLK1: Anatomy, Interactions, Functions, and Regulation. Biochemistry 2016, 81, 1676–1697. [Google Scholar] [CrossRef]
- Park, J.; Kim, D.H.; Kim, H.N.; Wang, C.J.; Kwak, M.K.; Hur, E.; Suh, K.Y.; An, S.S.; Levchenko, A. Directed migration of cancer cells guided by the graded texture of the underlying matrix. Nat. Mater. 2016, 15, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Sundararajan, V.; Gengenbacher, N.; Stemmler, M.P.; Kleemann, J.A.; Brabletz, T.; Brabletz, S. The ZEB1/miR-200c feedback loop regulates invasion via actin interacting proteins MYLK and TKS5. Oncotarget 2015, 6, 27083–27096. [Google Scholar] [CrossRef] [PubMed]
- Stull, J.T.; Tansey, M.G.; Tang, D.C.; Word, R.A.; Kamm, K.E. Phosphorylation of myosin light chain kinase: A cellular mechanism for Ca2+ desensitization. Mol. Cell Biochem. 1993, 127–128, 229–237. [Google Scholar] [CrossRef]
- Tan, X.; Tong, L.; Li, L.; Xu, J.; Xie, S.; Ji, L.; Fu, J.; Liu, Q.; Shen, S.; Liu, Y.; et al. Loss of Smad4 promotes aggressive lung cancer metastasis by de-repression of PAK3 via miRNA regulation. Nat. Commun. 2021, 12, 4853. [Google Scholar] [CrossRef]
- Lowy, C.M.; Oskarsson, T. Tenascin C in metastasis: A view from the invasive front. Cell Adh. Migr. 2015, 9, 112–124. [Google Scholar] [CrossRef]
- Nagaharu, K.; Zhang, X.; Yoshida, T.; Katoh, D.; Hanamura, N.; Kozuka, Y.; Ogawa, T.; Shiraishi, T.; Imanaka-Yoshida, K. Tenascin C induces epithelial-mesenchymal transition-like change accompanied by SRC activation and focal adhesion kinase phosphorylation in human breast cancer cells. Am. J. Pathol. 2011, 178, 754–763. [Google Scholar] [CrossRef]
- Rodriguez de Cordoba, S.; Marshall, P.; Rubinstein, P. Twenty-six DR beta and 16 DQ beta chain IEF variants and their associated HLA-DR, HLA-DQ, and HLA-Dw specificities. Immunogenetics 1989, 29, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Applanat, M.; Di Benedetto, M. Autocrine functions of VEGF in breast tumor cells: Adhesion, survival, migration and invasion. Cell Adh. Migr. 2012, 6, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Carisey, A.; Tsang, R.; Greiner, A.M.; Nijenhuis, N.; Heath, N.; Nazgiewicz, A.; Kemkemer, R.; Derby, B.; Spatz, J.; Ballestrem, C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 2013, 23, 271–281. [Google Scholar] [CrossRef]
- Legerstee, K.; Geverts, B.; Slotman, J.A.; Houtsmuller, A.B. Dynamics and distribution of paxillin, vinculin, zyxin and VASP depend on focal adhesion location and orientation. Sci. Rep. 2019, 9, 10460. [Google Scholar] [CrossRef]
- Lemos Gomes, M.; Lopes, A.; Sobrinho, G.; Mendes Pedro, L. Restenosis of Aorto-renal Venous Grafts: Report of Two Patients Treated by Endovascular Stenting. EJVES Short Rep. 2018, 40, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Mise, N.; Savai, R.; Yu, H.; Schwarz, J.; Kaminski, N.; Eickelberg, O. Zyxin is a transforming growth factor-beta (TGF-beta)/Smad3 target gene that regulates lung cancer cell motility via integrin alpha5beta1. J. Biol. Chem. 2012, 287, 31393–31405. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Yang, C.H.; Cheng, L.H.; Chang, W.T.; Lin, Y.R.; Cheng, H.C. Fibronectin in Cancer: Friend or Foe. Cells 2019, 9, 27. [Google Scholar] [CrossRef]
- Drake, J.M.; Barnes, J.M.; Madsen, J.M.; Domann, F.E.; Stipp, C.S.; Henry, M.D. ZEB1 coordinately regulates laminin-332 and {beta}4 integrin expression altering the invasive phenotype of prostate cancer cells. J. Biol. Chem 2010, 285, 33940–33948. [Google Scholar] [CrossRef]
- Bianchi, A.; Gervasi, M.E.; Bakin, A. Role of beta5-integrin in epithelial-mesenchymal transition in response to TGF-beta. Cell Cycle 2010, 9, 1647–1659. [Google Scholar] [CrossRef]
- Lin, Z.; He, R.; Luo, H.; Lu, C.; Ning, Z.; Wu, Y.; Han, C.; Tan, G.; Wang, Z. Integrin-beta5, a miR-185-targeted gene, promotes hepatocellular carcinoma tumorigenesis by regulating beta-catenin stability. J. Exp. Clin. Cancer Res. 2018, 37, 17. [Google Scholar] [CrossRef]
- Zhou, M.; Niu, J.; Wang, J.; Gao, H.; Shahbaz, M.; Niu, Z.; Li, Z.; Zou, X.; Liang, B. Integrin alphavbeta8 serves as a Novel Marker of Poor Prognosis in Colon Carcinoma and Regulates Cell Invasiveness through the Activation of TGF-beta1. J. Cancer 2020, 11, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- McCarty, J.H. alphavbeta8 integrin adhesion and signaling pathways in development, physiology and disease. J. Cell Sci. 2020, 133, jcs239434. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Soni, U.K.; Maurya, V.K.; Singh, K.; Jha, R.K. Integrin beta8 (ITGB8) activates VAV-RAC1 signaling via FAK in the acquisition of endometrial epithelial cell receptivity for blastocyst implantation. Sci. Rep. 2017, 7, 1885. [Google Scholar] [CrossRef]
- Hamill, K.J.; Paller, A.S.; Jones, J.C. Adhesion and migration, the diverse functions of the laminin alpha3 subunit. Dermatol. Clin. 2010, 28, 79–87. [Google Scholar] [CrossRef]
- Moncho-Amor, V.; Pintado-Berninches, L.; Ibanez de Caceres, I.; Martin-Villar, E.; Quintanilla, M.; Chakravarty, P.; Cortes-Sempere, M.; Fernandez-Varas, B.; Rodriguez-Antolin, C.; de Castro, J.; et al. Role of Dusp6 Phosphatase as a Tumor Suppressor in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2019, 20, 2036. [Google Scholar] [CrossRef] [PubMed]
- Horzum, U.; Ozdil, B.; Pesen-Okvur, D. Step-by-step quantitative analysis of focal adhesions. MethodsX 2014, 1, 56–59. [Google Scholar] [CrossRef]
- Yui, K.; Imataka, G.; Nakamura, H.; Ohara, N.; Naito, Y. Eicosanoids Derived From Arachidonic Acid and Their Family Prostaglandins and Cyclooxygenase in Psychiatric Disorders. Curr. Neuropharmacol. 2015, 13, 776–785. [Google Scholar] [CrossRef]
- Wang, T.; Fu, X.; Chen, Q.; Patra, J.K.; Wang, D.; Wang, Z.; Gai, Z. Arachidonic Acid Metabolism and Kidney Inflammation. Int. J. Mol. Sci. 2019, 20, 3683. [Google Scholar] [CrossRef]
- Kudo, I.; Murakami, M. Prostaglandin E synthase, a terminal enzyme for prostaglandin E2 biosynthesis. J. Biochem. Mol. Biol. 2005, 38, 633–638. [Google Scholar] [CrossRef]
- Komoto, J.; Yamada, T.; Watanabe, K.; Takusagawa, F. Crystal structure of human prostaglandin F synthase (AKR1C3). Biochemistry 2004, 43, 2188–2198. [Google Scholar] [CrossRef]
- Koeberle, S.C.; Gollowitzer, A.; Laoukili, J.; Kranenburg, O.; Werz, O.; Koeberle, A.; Kipp, A.P. Distinct and overlapping functions of glutathione peroxidases 1 and 2 in limiting NF-kappaB-driven inflammation through redox-active mechanisms. Redox Biol. 2020, 28, 101388. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Xu, K.P.; Tan, G.S. Cyclooxygenase-2 inhibitors in lung cancer treatment: Bench to bed. Eur. J. Pharmacol. 2015, 769, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Kawada, K.; Sakai, Y. Prostaglandin E2/EP Signaling in the Tumor Microenvironment of Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 6254. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Yarla, N.S.; Madka, V.; Rao, C.V. Clinically Relevant Anti-Inflammatory Agents for Chemoprevention of Colorectal Cancer: New Perspectives. Int. J. Mol. Sci. 2018, 19, 2332. [Google Scholar] [CrossRef]
- Nanda, N.; Dhawan, D.K. Role of Cyclooxygenase-2 in colorectal cancer patients. Front. Biosci. 2021, 26, 706–716. [Google Scholar] [CrossRef]
- Williams, C.S.; Mann, M.; DuBois, R.N. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 1999, 18, 7908–7916. [Google Scholar] [CrossRef]
- Fijneman, R.J.; Cormier, R.T. The roles of sPLA2-IIA (Pla2g2a) in cancer of the small and large intestine. Front. Biosci. 2008, 13, 4144–4174. [Google Scholar] [CrossRef]
- Adler, D.H.; Cogan, J.D.; Phillips, J.A., 3rd; Schnetz-Boutaud, N.; Milne, G.L.; Iverson, T.; Stein, J.A.; Brenner, D.A.; Morrow, J.D.; Boutaud, O.; et al. Inherited human cPLA(2alpha) deficiency is associated with impaired eicosanoid biosynthesis, small intestinal ulceration, and platelet dysfunction. J. Clin. Investig. 2008, 118, 2121–2131. [Google Scholar] [CrossRef]
- Zhan, Y.; Zheng, L.; Liu, J.; Hu, D.; Wang, J.; Liu, K.; Guo, J.; Zhang, T.; Kong, D. PLA2G4A promotes right-sided colorectal cancer progression by inducing CD39+gammadelta Treg polarization. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, X.; Zhang, L.; Geng, D.; Wang, Y.; Sun, D.; Sui, P.; Zhao, X.; Xin, C.; Jiang, J.; et al. Inhibition of PLA2G4A Reduces the Expression of Lung Cancer-Related Cytokines. DNA Cell Biol. 2018, 37, 1076–1081. [Google Scholar] [CrossRef]
- Mounier, C.M.; Wendum, D.; Greenspan, E.; Flejou, J.F.; Rosenberg, D.W.; Lambeau, G. Distinct expression pattern of the full set of secreted phospholipases A2 in human colorectal adenocarcinomas: sPLA2-III as a biomarker candidate. Br. J. Cancer 2008, 98, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Brychtova, V.; Vojtesek, B.; Hrstka, R. Anterior gradient 2: A novel player in tumor cell biology. Cancer Lett. 2011, 304, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Guo, Y.; Zhu, D.; Zhang, N.; Li, L.; Jiang, J.; Dong, Y.; Xu, Q.; Zhang, X.; Wang, M.; et al. Pro-metastatic activity of AGR2 interrupts angiogenesis target bevacizumab efficiency via direct interaction with VEGFA and activation of NF-kappaB pathway. Biochim Biophys Acta Mol. Basis Dis. 2018, 1864, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Ondrouskova, E.; Sommerova, L.; Nenutil, R.; Coufal, O.; Bouchal, P.; Vojtesek, B.; Hrstka, R. AGR2 associates with HER2 expression predicting poor outcome in subset of estrogen receptor negative breast cancer patients. Exp. Mol. Pathol. 2017, 102, 280–283. [Google Scholar] [CrossRef]
- Tian, S.B.; Tao, K.X.; Hu, J.; Liu, Z.B.; Ding, X.L.; Chu, Y.N.; Cui, J.Y.; Shuai, X.M.; Gao, J.B.; Cai, K.L.; et al. The prognostic value of AGR2 expression in solid tumours: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 15500. [Google Scholar] [CrossRef]
- Bouchard, V.; Demers, M.J.; Thibodeau, S.; Laquerre, V.; Fujita, N.; Tsuruo, T.; Beaulieu, J.F.; Gauthier, R.; Vezina, A.; Villeneuve, L.; et al. Fak/Src signaling in human intestinal epithelial cell survival and anoikis: Differentiation state-specific uncoupling with the PI3-K/Akt-1 and MEK/Erk pathways. J. Cell Physiol. 2007, 212, 717–728. [Google Scholar] [CrossRef]
- Paul, R.; Luo, M.; Mo, X.; Lu, J.; Yeo, S.K.; Guan, J.L. FAK activates AKT-mTOR signaling to promote the growth and progression of MMTV-Wnt1-driven basal-like mammary tumors. Breast Cancer Res. 2020, 22, 59. [Google Scholar] [CrossRef]
- Webb, D.J.; Donais, K.; Whitmore, L.A.; Thomas, S.M.; Turner, C.E.; Parsons, J.T.; Horwitz, A.F. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004, 6, 154–161. [Google Scholar] [CrossRef]
- Fessart, D.; Villamor, I.; Chevet, E.; Delom, F.; Robert, J. Integrative analysis of genomic and transcriptomic alterations of AGR2 and AGR3 in cancer. Open Biol. 2022, 12, 220068. [Google Scholar] [CrossRef]
- Hanna, V.S.; Hafez, E.A.A. Synopsis of arachidonic acid metabolism: A review. J. Adv. Res. 2018, 11, 23–32. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene 2010, 29, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; DuBois, R.N. An inflammatory mediator, prostaglandin E2, in colorectal cancer. Cancer J. 2013, 19, 502–510. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. The role of anti-inflammatory drugs in colorectal cancer. Annu. Rev. Med. 2013, 64, 131–144. [Google Scholar] [CrossRef]
- Zhang, H.; Chi, J.; Hu, J.; Ji, T.; Luo, Z.; Zhou, C.; Huang, L.; Dai, Z.; Li, J.; Wang, G.; et al. Intracellular AGR2 transduces PGE2 stimuli to promote epithelial-mesenchymal transition and metastasis of colorectal cancer. Cancer Lett. 2021, 518, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Rosenberg, D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin. Immunopathol. 2013, 35, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; DuBois, R.N. Role of prostanoids in gastrointestinal cancer. J. Clin. Investig. 2018, 128, 2732–2742. [Google Scholar] [CrossRef]
- Menter, D.G.; Dubois, R.N. Prostaglandins in cancer cell adhesion, migration, and invasion. Int. J. Cell Biol. 2012, 2012, 723419. [Google Scholar] [CrossRef]
- Wang, Q.; Morris, R.J.; Bode, A.M.; Zhang, T. Prostaglandin Pathways: Opportunities for Cancer Prevention and Therapy. Cancer Res. 2022, 82, 949–965. [Google Scholar] [CrossRef]
- Alsereihi, R.; Schulten, H.J.; Bakhashab, S.; Saini, K.; Al-Hejin, A.M.; Hussein, D. Leveraging the Role of the Metastatic Associated Protein Anterior Gradient Homologue 2 in Unfolded Protein Degradation: A Novel Therapeutic Biomarker for Cancer. Cancers 2019, 11, 890. [Google Scholar] [CrossRef]
- Gupta, A.; Wodziak, D.; Tun, M.; Bouley, D.M.; Lowe, A.W. Loss of anterior gradient 2 (Agr2) expression results in hyperplasia and defective lineage maturation in the murine stomach. J. Biol. Chem. 2013, 288, 4321–4333. [Google Scholar] [CrossRef]
- Bouchalova, P.; Sommerova, L.; Potesil, D.; Martisova, A.; Lapcik, P.; Koci, V.; Scherl, A.; Vonka, P.; Planas-Iglesias, J.; Chevet, E.; et al. Characterization of the AGR2 Interactome Uncovers New Players of Protein Disulfide Isomerase Network in Cancer Cells. Mol. Cell Proteom. 2022, 21, 100188. [Google Scholar] [CrossRef] [PubMed]
- Moidu, N.A.; NS, A.R.; Syafruddin, S.E.; Low, T.Y.; Mohtar, M.A. Secretion of pro-oncogenic AGR2 protein in cancer. Heliyon 2020, 6, e05000. [Google Scholar] [CrossRef] [PubMed]
- Maurel, M.; Obacz, J.; Avril, T.; Ding, Y.P.; Papadodima, O.; Treton, X.; Daniel, F.; Pilalis, E.; Horberg, J.; Hou, W.; et al. Control of anterior GRadient 2 (AGR2) dimerization links endoplasmic reticulum proteostasis to inflammation. EMBO Mol. Med. 2019, 11, e10120. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Forbes, S.A.; Bhamra, G.; Bamford, S.; Dawson, E.; Kok, C.; Clements, J.; Menzies, A.; Teague, J.W.; Futreal, P.A.; Stratton, M.R. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum. Genet. 2008. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Sage, D.; Neumann, F.R.; Hediger, F.; Gasser, S.M.; Unser, M. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans. Image Process. 2005, 14, 1372–1383. [Google Scholar] [CrossRef]
| Gene | Regulation | Function |
|---|---|---|
| PLA2G4A | DOWN | Hydrolysis of membrane phospholipids, which leads to release of arachidonic acid that is further metabolized into eicosanoids through one of three pathways (COX, LOX, CYP450) [58]. |
| PTGS2 | DOWN | Also known as COX-2, a rate-limiting enzyme involved in the conversion of arachidonic acid into prostaglandins [58]. |
| PTGES | DOWN | Enzyme catalyzing conversion of COX derived PDH2 into PGE2 [59]. |
| AKR1C3 | DOWN | Downstream of COX catalyzes reduction of PGH2 and PGD2 into PGF2α [60]. |
| GPX2 | DOWN | Reduces fatty acid-derived hydroperoxides and inhibits NF-κB activity [61]. |
| Gene | Lung Tumors | Large Intestine Tumors | Lung Cell Lines | Colorectal Cell Lines |
|---|---|---|---|---|
| ALOX12BA | −0.154 | 0.330 | − | −0.214 |
| ALOX15 | 0.053 | −0.324 | − | − |
| CBR3 | 0.052 | 0.391 | 0.113 | −0.137 |
| CYP2B6 | −0.053 | −0.622 | − | − |
| CYP2C18 | 0.341 | 0.427 | − | − |
| CYP2C9 | 0.355 | 0.245 | − | − |
| CYP4F2 | 0.060 | −0.576 | − | − |
| CYP4F3 | 0.363 | −0.599 | −0.046 | −0.088 |
| DHRS4 | 0.036 | 0.471 | −0.097 | 0.165 |
| EPHX2 | 0.305 | −0.082 | 0.317 | 0.066 |
| GGT1 | 0.040 | −0.445 | 0.021 | 0.072 |
| LTC4S | 0.053 | −0.395 | − | − |
| PLA2G10 | 0.277 | 0.156 | 0.315 | 0.150 |
| PLA2G12A | 0.103 | 0.253 | 0.492 | 0.334 |
| PLA2G12B | −0.022 | −0.622 | − | − |
| PLA2G2A | 0.094 | 0.462 | − | − |
| PLA2G3 | 0.093 | 0.347 | − | − |
| PLA2G4A | 0.068 | 0.569 | 0.176 | 0.264 |
| PLA2G6 | 0.226 | −0.091 | 0.255 | 0.474 |
| PTGS2 | −0.190 | 0.332 | 0.017 | −0.074 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martisova, A.; Sommerova, L.; Krejci, A.; Selingerova, I.; Kolarova, T.; Zavadil Kokas, F.; Holanek, M.; Podhorec, J.; Kazda, T.; Hrstka, R. Identification of AGR2 Gene-Specific Expression Patterns Associated with Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2022, 23, 10845. https://doi.org/10.3390/ijms231810845
Martisova A, Sommerova L, Krejci A, Selingerova I, Kolarova T, Zavadil Kokas F, Holanek M, Podhorec J, Kazda T, Hrstka R. Identification of AGR2 Gene-Specific Expression Patterns Associated with Epithelial-Mesenchymal Transition. International Journal of Molecular Sciences. 2022; 23(18):10845. https://doi.org/10.3390/ijms231810845
Chicago/Turabian StyleMartisova, Andrea, Lucia Sommerova, Adam Krejci, Iveta Selingerova, Tamara Kolarova, Filip Zavadil Kokas, Milos Holanek, Jan Podhorec, Tomas Kazda, and Roman Hrstka. 2022. "Identification of AGR2 Gene-Specific Expression Patterns Associated with Epithelial-Mesenchymal Transition" International Journal of Molecular Sciences 23, no. 18: 10845. https://doi.org/10.3390/ijms231810845
APA StyleMartisova, A., Sommerova, L., Krejci, A., Selingerova, I., Kolarova, T., Zavadil Kokas, F., Holanek, M., Podhorec, J., Kazda, T., & Hrstka, R. (2022). Identification of AGR2 Gene-Specific Expression Patterns Associated with Epithelial-Mesenchymal Transition. International Journal of Molecular Sciences, 23(18), 10845. https://doi.org/10.3390/ijms231810845






