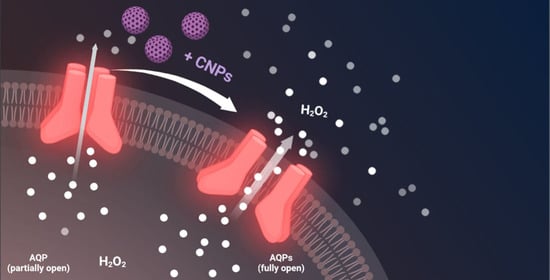
Cerium Oxide Nanoparticles Regulate Oxidative Stress in HeLa Cells by Increasing the Aquaporin-Mediated Hydrogen Peroxide Permeability
Abstract
1. Introduction
2. Results
2.1. Aquaporins mRNA and Protein Expression in HeLa Cells
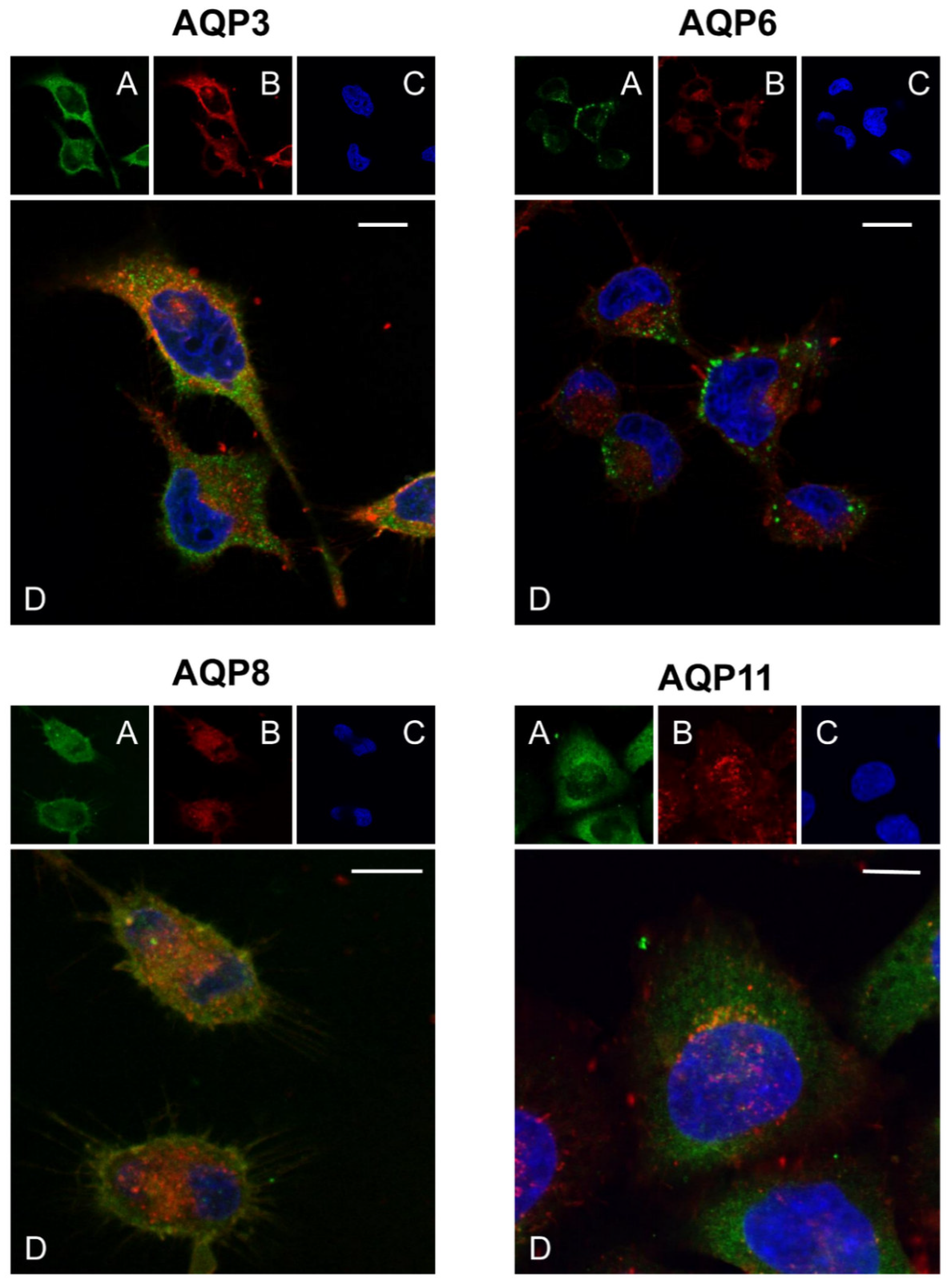
2.2. CNPs and Aquaporin-3, -6, -8, and -11 Localization in HeLa Cells
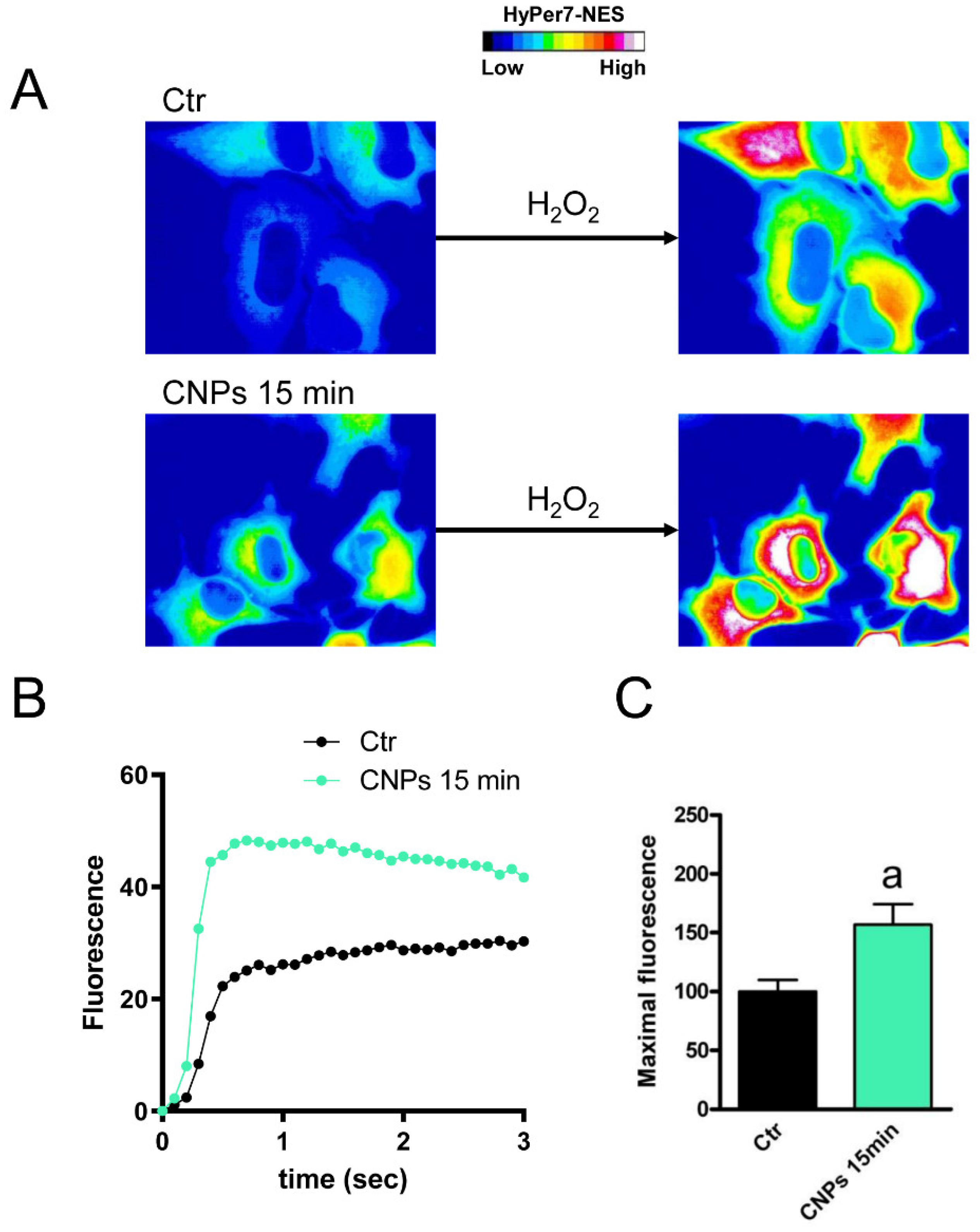
2.3. The Effect of CNPs on Water Permeability of HeLa Cells in Normal and Oxidative Stress Conditions
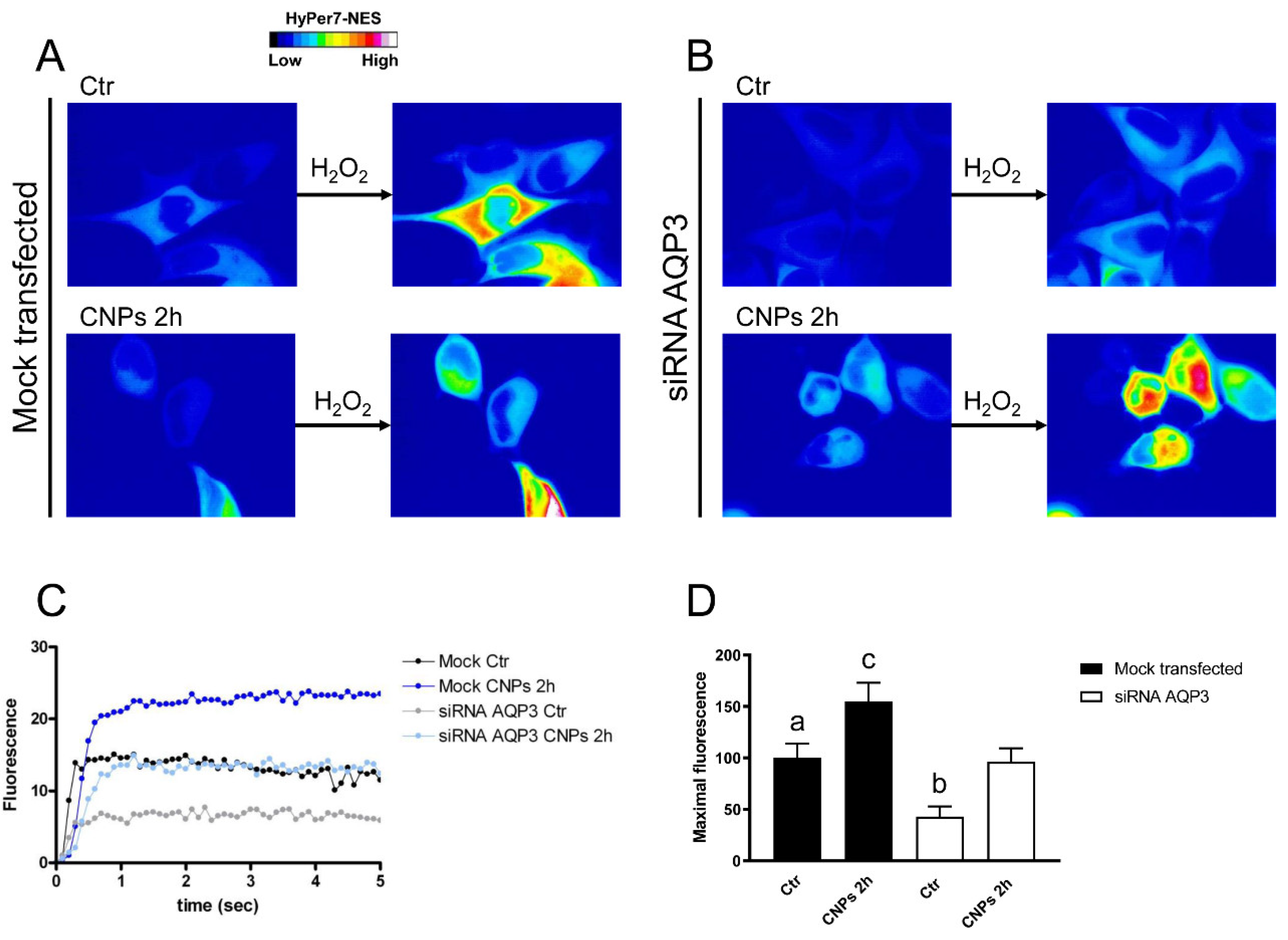
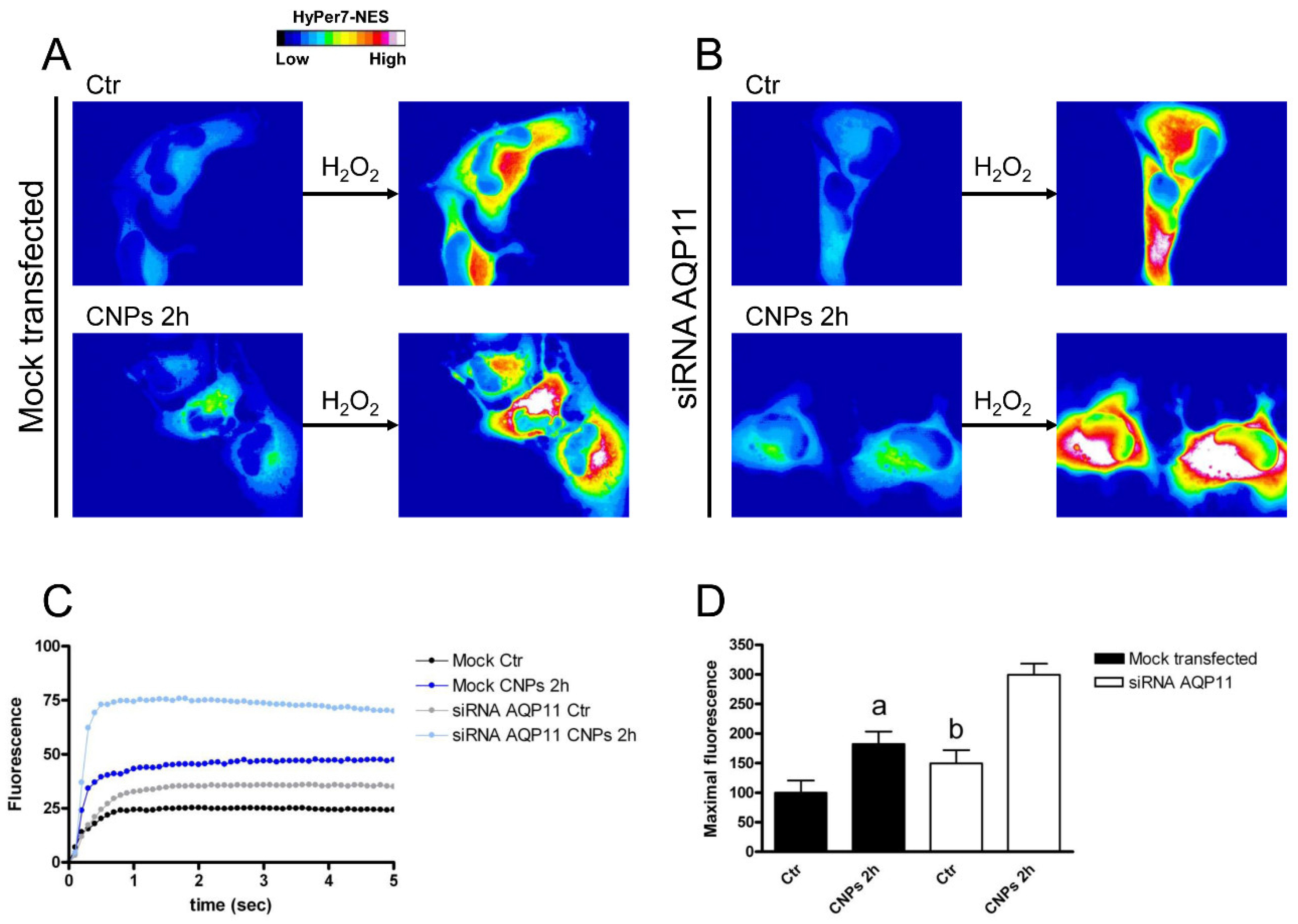
2.4. Effect of CNPs in HeLa Cells with Reduced Expression of Single AQPs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cerium Nanoparticles Preparation and Characterization
4.3. RNA Isolation and RT-qPCR
4.4. SDS-PAGE and Immunoblotting
4.5. Double Immunofluorescence
4.6. Gene Silencing
4.7. Water Permeability Measurements
4.8. Hydrogen Peroxide Indicator Transfection for Optical Imaging
4.9. Intracellular H2O2 Detection by HyPer7-NES Imaging
4.10. Protein Content
4.11. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- King, L.S.; Kozono, D.; Agre, P. From structure to disease: The evolving tale of aquaporin biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Benga, G. The first discovered water channel protein, later called aquaporin 1: Molecular characteristics, functions and medical implications. Mol. Aspects Med. 2012, 33, 518–534. [Google Scholar] [CrossRef] [PubMed]
- Verkman, A.S. More than just water channels: Unexpected cellular roles of aquaporins. J Cell Sci 2005, 118, 3225–3232. [Google Scholar] [CrossRef]
- Laforenza, U.; Bottino, C.; Gastaldi, G. Mammalian aquaglyceroporin function in metabolism. Biochim. Biophys. Acta 2016, 1858, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Saadoun, S.; Papadopoulos, M.C.; Hara-Chikuma, M.; Verkman, A.S. Impairment of angiogenesis and cell migration by targeted aquaporin-1 gene disruption. Nature 2005, 434, 786–792. [Google Scholar] [CrossRef]
- Azad, A.K.; Raihan, T.; Ahmed, J.; Hakim, A.; Emon, T.H.; Chowdhury, P.A. Human Aquaporins: Functional Diversity and Potential Roles in Infectious and Non-infectious Diseases. Front. Genet. 2021, 12, 654865. [Google Scholar] [CrossRef]
- Verkman, A.S.; Anderson, M.O.; Papadopoulos, M.C. Aquaporins: Important but elusive drug targets. Nat. Rev. Drug Discov. 2014, 13, 259–277. [Google Scholar] [CrossRef]
- Medraño-Fernandez, I.; Bestetti, S.; Bertolotti, M.; Bienert, G.P.; Bottino, C.; Laforenza, U.; Rubartelli, A.; Sitia, R. Stress Regulates Aquaporin-8 Permeability to Impact Cell Growth and Survival. Antioxid. Redox Signal. 2016, 24, 1031–1044. [Google Scholar] [CrossRef]
- Bienert, G.P.; Chaumont, F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 2014, 1840, 1596–1604. [Google Scholar] [CrossRef]
- Bestetti, S.; Galli, M.; Sorrentino, I.; Pinton, P.; Rimessi, A.; Sitia, R.; Medraño-Fernandez, I. Human aquaporin-11 guarantees efficient transport of H2O2 across the endoplasmic reticulum membrane. Redox Biol. 2020, 28, 101326. [Google Scholar] [CrossRef]
- Hara-Chikuma, M.; Chikuma, S.; Sugiyama, Y.; Kabashima, K.; Verkman, A.S.; Inoue, S.; Miyachi, Y. Chemokine-dependent T cell migration requires aquaporin-3-mediated hydrogen peroxide uptake. J. Exp. Med. 2012, 209, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.W.; Dickinson, B.C.; Chang, C.J. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 15681–15686. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.; Mósca, A.F.; Martins, A.P.; Nobre, T.; Prista, C.; Antunes, F.; Cipak Gasparovic, A.; Soveral, G. Rat Aquaporin-5 Is pH-Gated Induced by Phosphorylation and Is Implicated in Oxidative Stress. Int. J. Mol. Sci. 2016, 17, 2090. [Google Scholar] [CrossRef] [PubMed]
- Satooka, H.; Hara-Chikuma, M. Aquaporin-3 Controls Breast Cancer Cell Migration by Regulating Hydrogen Peroxide Transport and Its Downstream Cell Signaling. Mol. Cell Biol. 2016, 36, 1206–1218. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Thiagarajah, J.; Zhao, D.; Verkman, A. Impaired enterocyte proliferation in aquaporin-3 deficiency in mouse models of colitis. Gut 2007, 56, 1529–1535. [Google Scholar] [CrossRef]
- Watanabe, S.; Moniaga, C.S.; Nielsen, S.; Hara-Chikuma, M. Aquaporin-9 facilitates membrane transport of hydrogen peroxide in mammalian cells. Biochem. Biophys. Res. Commun. 2016, 471, 191–197. [Google Scholar] [CrossRef]
- Varadaraj, K.; Kumari, S.S. Lens aquaporins function as peroxiporins to facilitate membrane transport of hydrogen peroxide. Biochem. Biophys. Res. Commun. 2020, 524, 1025–1029. [Google Scholar] [CrossRef]
- Laforenza, U.; Pellavio, G.; Marchetti, A.L.; Omes, C.; Todaro, F.; Gastaldi, G. Aquaporin-Mediated Water and Hydrogen Peroxide Transport Is Involved in Normal Human Spermatozoa Functioning. Int. J. Mol. Sci. 2016, 18, 66. [Google Scholar] [CrossRef]
- Pellavio, G.; Rui, M.; Caliogna, L.; Martino, E.; Gastaldi, G.; Collina, S.; Laforenza, U. Regulation of Aquaporin Functional Properties Mediated by the Antioxidant Effects of Natural Compounds. Int. J. Mol. Sci. 2017, 18, 2665. [Google Scholar] [CrossRef]
- Pellavio, G.; Todaro, F.; Alberizzi, P.; Scotti, C.; Gastaldi, G.; Lolicato, M.; Omes, C.; Caliogna, L.; Nappi, R.E.; Laforenza, U. HPV Infection Affects Human Sperm Functionality by Inhibition of Aquaporin-8. Cells 2020, 9, 1241. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, I.; Maggio, A.; Gena, P.; de Bari, O.; Tamma, G.; Portincasa, P.; Calamita, G. Modulation of Aquaporins by dietary patterns and plant bioactive compounds. Curr. Med. Chem. 2019, 26, 3457–3470. [Google Scholar] [CrossRef] [PubMed]
- Abir-Awan, M.; Kitchen, P.; Salman, M.M.; Conner, M.T.; Conner, A.C.; Bill, R.M. Inhibitors of Mammalian Aquaporin Water Channels. Int. J. Mol. Sci. 2019, 20, 1589. [Google Scholar] [CrossRef] [PubMed]
- Tesse, A.; Grossini, E.; Tamma, G.; Brenner, C.; Portincasa, P.; Marinelli, R.A.; Calamita, G. Aquaporins as Targets of Dietary Bioactive Phytocompounds. Front. Mol. Biosci. 2018, 5, 30. [Google Scholar] [CrossRef]
- Portincasa, P.; Calamita, G. Phytocompounds modulating Aquaporins: Clinical benefits are anticipated. Food Chem. 2019, 274, 642–650. [Google Scholar] [CrossRef]
- Pellavio, G.; Rossino, G.; Gastaldi, G.; Rossi, D.; Linciano, P.; Collina, S.; Laforenza, U. Sigma-1 Receptor Agonists Acting on Aquaporin-Mediated H2O2 Permeability: New Tools for Counteracting Oxidative Stress. Int. J. Mol. Sci. 2021, 22, 9790. [Google Scholar] [CrossRef]
- Tarnuzzer, R.W.; Colon, J.; Patil, S.; Seal, S. Vacancy engineered ceria nanostructures for protection from radiation-induced cellular damage. Nano Lett. 2005, 5, 2573–2577. [Google Scholar] [CrossRef]
- Schubert, D.; Dargusch, R.; Raitano, J.; Chan, S.W. Cerium and yttrium oxide nanoparticles are neuroprotective. Biochem. Biophys. Res. Commun. 2006, 342, 86–91. [Google Scholar] [CrossRef]
- Estevez, A.Y.; Pritchard, S.; Harper, K.; Aston, J.W.; Lynch, A.; Lucky, J.J.; Ludington, J.S.; Chatani, P.; Mosenthal, W.P.; Leiter, J.C.; et al. Neuroprotective mechanisms of cerium oxide nanoparticles in a mouse hippocampal brain slice model of ischemia. Free Radic. Biol. Med. 2011, 51, 1155–1163. [Google Scholar] [CrossRef]
- Dowding, J.M.; Song, W.; Bossy, K.; Karakoti, A.; Kumar, A.; Kim, A.; Bossy, B.; Seal, S.; Ellisman, M.H.; Perkins, G.; et al. Cerium oxide nanoparticles protect against Aβ-induced mitochondrial fragmentation and neuronal cell death. Cell Death Differ. 2014, 21, 1622–1632. [Google Scholar] [CrossRef]
- Wason, M.S.; Zhao, J. Cerium oxide nanoparticles: Potential applications for cancer and other diseases. Am. J. Transl. Res. 2013, 5, 126–131. [Google Scholar] [PubMed]
- Fiorani, L.; Passacantando, M.; Santucci, S.; Di Marco, S.; Bisti, S.; Maccarone, R. Cerium Oxide Nanoparticles Reduce Microglial Activation and Neurodegenerative Events in Light Damaged Retina. PLoS ONE 2015, 10, e0140387. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.J.; Cha, M.Y.; Kim, D.; Kim, D.K.; Soh, M.; Shin, K.; Hyeon, T.; Mook-Jung, I. Mitochondria-Targeting Ceria Nanoparticles as Antioxidants for Alzheimer’s Disease. A.C.S. Nano. 2016, 10, 2860–2870. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, D.; Tredici, I.G.; Ghigna, P.; Castillo-Michel, H.; Falqui, A.; Di Benedetto, C.; Alberti, G.; Ricci, V.; Anselmi-Tamburini, U.; Sommi, P. Dependence of the Ce(iii)/Ce(iv) ratio on intracellular localization in ceria nanoparticles internalized by human cells. Nanoscale 2017, 9, 1527–1538. [Google Scholar] [CrossRef]
- Sommi, P.; Vitali, A.; Coniglio, S.; Callegari, D.; Barbieri, S.; Casu, A.; Falqui, A.; Vigano’, L.; Vigani, B.; Ferrari, F.; et al. Microvilli Adhesion: An Alternative Route for Nanoparticle Cell Internalization. A.C.S. Nano. 2021, 15, 15803–15814. [Google Scholar] [CrossRef]
- Pellavio, G.; Martinotti, S.; Patrone, M.; Ranzato, E.; Laforenza, U. Aquaporin-6 May Increase the Resistance to Oxidative Stress of Malignant Pleural Mesothelioma Cells. Cells 2022, 11, 1892. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- van Steensel, B.; van Binnendijk, E.P.; Hornsby, C.D.; van der Voort, H.T.; Krozowski, Z.S.; de Kloet, E.R.; van Driel, R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 1996, 109, 787–792. [Google Scholar] [CrossRef]
- Manders, E.M.M.; Verbeek, F.J.; Aten, J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993, 169, 375–382. [Google Scholar] [CrossRef]
- Pellavio, G.; Laforenza, U. Human sperm functioning is related to the aquaporin-mediated water and hydrogen peroxide transport regulation. Biochimie 2021, 188, 45–51. [Google Scholar] [CrossRef]
- Veal, E.A.; Day, A.M.; Morgan, B.A. Hydrogen peroxide sensing and signaling. Mol. Cell 2007, 26, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Zhou, T.; Pannell, B.K.; Ziegler, A.C.; Best, T.M. Biological and physiological role of reactive oxygen species--the good, the bad and the ugly. Acta Physiol. (Oxf) 2015, 214, 329–348. [Google Scholar] [CrossRef] [PubMed]
- Tamma, G.; Valenti, G.; Grossini, E.; Donnini, S.; Marino, A.; Marinelli, R.A.; Calamita, G. Aquaporin Membrane Channels in Oxidative Stress, Cell Signaling, and Aging: Recent Advances and Research Trends. Oxid. Med. Cell. Longev. 2018, 2018, 1501847. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Schjoerring, J.K.; Jahn, T.P. Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 2006, 1758, 994–1003. [Google Scholar] [CrossRef]
- Thanan, R.; Oikawa, S.; Hiraku, Y.; Ohnishi, S.; Ma, N.; Pinlaor, S.; Yongvanit, P.; Kawanishi, S.; Murata, M. Oxidative stress and its significant roles in neurodegenerative diseases and cancer. Int. J. Mol. Sci. 2014, 16, 193–217. [Google Scholar] [CrossRef]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef]
- Pimpão, C.; Wragg, D.; da Silva, I.V.; Casini, A.; Soveral, G. Aquaglyceroporin Modulators as Emergent Pharmacological Molecules for Human Diseases. Front. Mol. Biosci. 2022, 9, 845237. [Google Scholar] [CrossRef]
- Rzigalinski, B.A.; Carfagna, C.S.; Ehrich, M. Cerium oxide nanoparticles in neuroprotection and considerations for efficacy and safety. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, 10. [Google Scholar] [CrossRef]
- Hu, X.N.; Liu, J.B.; Hou, S.; Wen, T.; Liu, W.Q.; Zhang, K.; He, W.W.; Ji, Y.L.; Ren, H.X.; Wang, Q.; et al. Research progress of nanoparticles as enzyme mimetics. Sci. China Phys. Mech. Astron. 2011, 54, 1749–1756. [Google Scholar] [CrossRef]
- Mandoli, C.; Pagliari, F.; Pagliari, S.; Forte, G.; Di Nardo, P.; Licoccia, S.; Traversa, E. Stem Cells Aligned Growth Induced by CeO2 Nanoparticles in PLGA Scaffolds with Improved Bioactivity for Regenerative Medicine. Adv. Funct. Mater. 2010, 20, 1617–1624. [Google Scholar] [CrossRef]
- Yasui, M.; Hazama, A.; Kwon, T.H.; Nielsen, S.; Guggino, W.B.; Agre, P. Rapid gating and anion permeability of an intracellular aquaporin. Nature 1999, 402, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Celardo, I.; Pedersen, J.Z.; Traversa, E.; Ghibelli, L. Pharmacological potential of cerium oxide nanoparticles. Nanoscale 2011, 3, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Korsvik, C.; Patil, S.; Seal, S.; Self, W.T. Superoxide dismutase mimetic properties exhibited by vacancy engineered ceria nanoparticles. Chem. Commun. 2007, 10, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Dong, H.; Lyu, G.M.; Zhang, H.Y.; Ke, J.; Kang, L.Q.; Teng, J.L.; Sun, L.D.; Si, R.; Zhang, J.; et al. Engineering the defect state and reducibility of ceria based nanoparticles for improved anti-oxidation performance. Nanoscale 2015, 7, 13981–13990. [Google Scholar] [CrossRef] [PubMed]
- Pimpão, C.; da Silva, I.V.; Mósca, A.F.; Pinho, J.O.; Gaspar, M.M.; Gumerova, N.I.; Rompel, A.; Aureliano, M.; Soveral, G. The Aquaporin-3-Inhibiting Potential of Polyoxotungstates. Int. J. Mol. Sci. 2020, 21, 2467. [Google Scholar] [CrossRef]
- Tiwari, P.M.; Eroglu, E.; Bawage, S.S.; Vig, K.; Miller, M.E.; Pillai, S.; Dennis, V.A.; Singh, S.R. Enhanced intracellular translocation and biodistribution of gold nanoparticles functionalized with a cell-penetrating peptide (VG-21) from vesicular stomatitis virus. Biomaterials 2014, 35, 9484–9494. [Google Scholar] [CrossRef]
- Chen, C.Y.; Liao, P.L.; Tsai, C.H.; Chan, Y.J.; Cheng, Y.W.; Hwang, L.L.; Lin, K.H.; Yen, T.L.; Li, C.H. Inhaled gold nanoparticles cause cerebral edema and upregulate endothelial aquaporin 1 expression, involving caveolin 1 dependent repression of extracellular regulated protein kinase activity. Part. Fibre Toxicol. 2019, 16, 37. [Google Scholar] [CrossRef]
- Laforenza, U.; Miceli, E.; Gastaldi, G.; Scaffino, M.F.; Ventura, U.; Fontana, J.M.; Orsenigo, M.N.; Corazza, G.R. Solute transporters and aquaporins are impaired in celiac disease. Biol. Cell. 2010, 102, 457–467. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Yeung, Y.G.; Stanley, E.R. A solution for stripping antibodies from polyvinylidene fluoride immunoblots for multiple reprobing. Anal. Biochem. 2009, 389, 89–91. [Google Scholar] [CrossRef]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Martínez Gache, S.A.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653.e646. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]







| Gene | Primer Sequences | Size (bp) | Accession Number | |
|---|---|---|---|---|
| AQP1 a | Forward | 5’-ACACCTCCTGGCTATTGACTACAC-3’ | 134 | NM_198098; variants 1, 5 |
| Reverse | 5’-CCGATGAATGGCCCCACCCAGAA-3’ | |||
| AQP2 a | Forward | 5’-CACCTCCTTGGGATCCATTACACC-3’ | 95 | NM_000486 |
| Reverse | 5’-ACCCAGTGGTCATCAAATTTGCC-3’ | |||
| AQP3 b | Forward | 5’-CCTGGTGATGTTTGGCTGTGGCTC-3’ | 147 | NM_004925; variants 1, 2 |
| Reverse | 5’-TTCAGGTGGGCCCCAGAGACC-3’ | |||
| AQP4 | Forward | 5’-GGAGTCACCATGGTTCATGGAA-3’ | 123 | NM_001650; variants 1–3 |
| Reverse | 5’-AGTGACATCAGTCCGTTTGGAA-3’ | |||
| AQP5 | Forward | 5’-GGTGGTGGAGCTGATTCTGA-3’ | 142 | NM_001651 |
| Reverse | 5’-GAAGTAGATTCCGACAAGGTGG-3’ | |||
| AQP6 | Forward | 5’-CACCTCATTGGGATCCACTTC-3’ | 103 | NM_ 001652; variants 1, 2 |
| Reverse | 5’-CCCAGAAGACCCAGTGGACT-3’ | |||
| AQP7 | Forward | 5’-GGACAGCTGATGGTGACCGG-3’ | 104 | NM_001170; variants 1–4 |
| Reverse | 5’-AGCCACGCCTCATTCAGGAA-3’ | |||
| AQP8 | Forward | 5’-TGGAGAGATAGCCATGTGTGAG-3’ | 106 | NM_001169 |
| Reverse | 5’-TGGCTGCACAAACCGTTCGT-3’ | |||
| AQP9 | Forward | 5’-CCCAGCTGTGTCTTTAGCAA-3’ | 133 | NM_020980; variants 1–3 |
| Reverse | 5’-AAGTCCATCATAGTAAATGCCAAA-3’ | |||
| AQP10 | Forward | 5’-CCTATGTTCTCTACCATGATGCCC-3’ | 137 | NM_080429 |
| Reverse | 5’-CTGATCCAGGAAGCCATTGTTC-3’ | |||
| AQP11 | Forward | 5’-TTTCTCTTCCACAGCGCTCT-3’ | 115 | NM_173039; variant 1 |
| Reverse | 5’-CTCCTGTTAGACTTCCTCCTGC-3’ | |||
| B2M | Hs_B2M_1_SG QuantiTect Primer Assay QT00088935, Qiagen | 98 | NM_004048 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellavio, G.; Sommi, P.; Anselmi-Tamburini, U.; DeMichelis, M.P.; Coniglio, S.; Laforenza, U. Cerium Oxide Nanoparticles Regulate Oxidative Stress in HeLa Cells by Increasing the Aquaporin-Mediated Hydrogen Peroxide Permeability. Int. J. Mol. Sci. 2022, 23, 10837. https://doi.org/10.3390/ijms231810837
Pellavio G, Sommi P, Anselmi-Tamburini U, DeMichelis MP, Coniglio S, Laforenza U. Cerium Oxide Nanoparticles Regulate Oxidative Stress in HeLa Cells by Increasing the Aquaporin-Mediated Hydrogen Peroxide Permeability. International Journal of Molecular Sciences. 2022; 23(18):10837. https://doi.org/10.3390/ijms231810837
Chicago/Turabian StylePellavio, Giorgia, Patrizia Sommi, Umberto Anselmi-Tamburini, Maria Paola DeMichelis, Stefania Coniglio, and Umberto Laforenza. 2022. "Cerium Oxide Nanoparticles Regulate Oxidative Stress in HeLa Cells by Increasing the Aquaporin-Mediated Hydrogen Peroxide Permeability" International Journal of Molecular Sciences 23, no. 18: 10837. https://doi.org/10.3390/ijms231810837
APA StylePellavio, G., Sommi, P., Anselmi-Tamburini, U., DeMichelis, M. P., Coniglio, S., & Laforenza, U. (2022). Cerium Oxide Nanoparticles Regulate Oxidative Stress in HeLa Cells by Increasing the Aquaporin-Mediated Hydrogen Peroxide Permeability. International Journal of Molecular Sciences, 23(18), 10837. https://doi.org/10.3390/ijms231810837