Distinct Responses to Pathogenic and Symbionic Microorganisms: The Role of Plant Immunity
Abstract
:1. Introduction
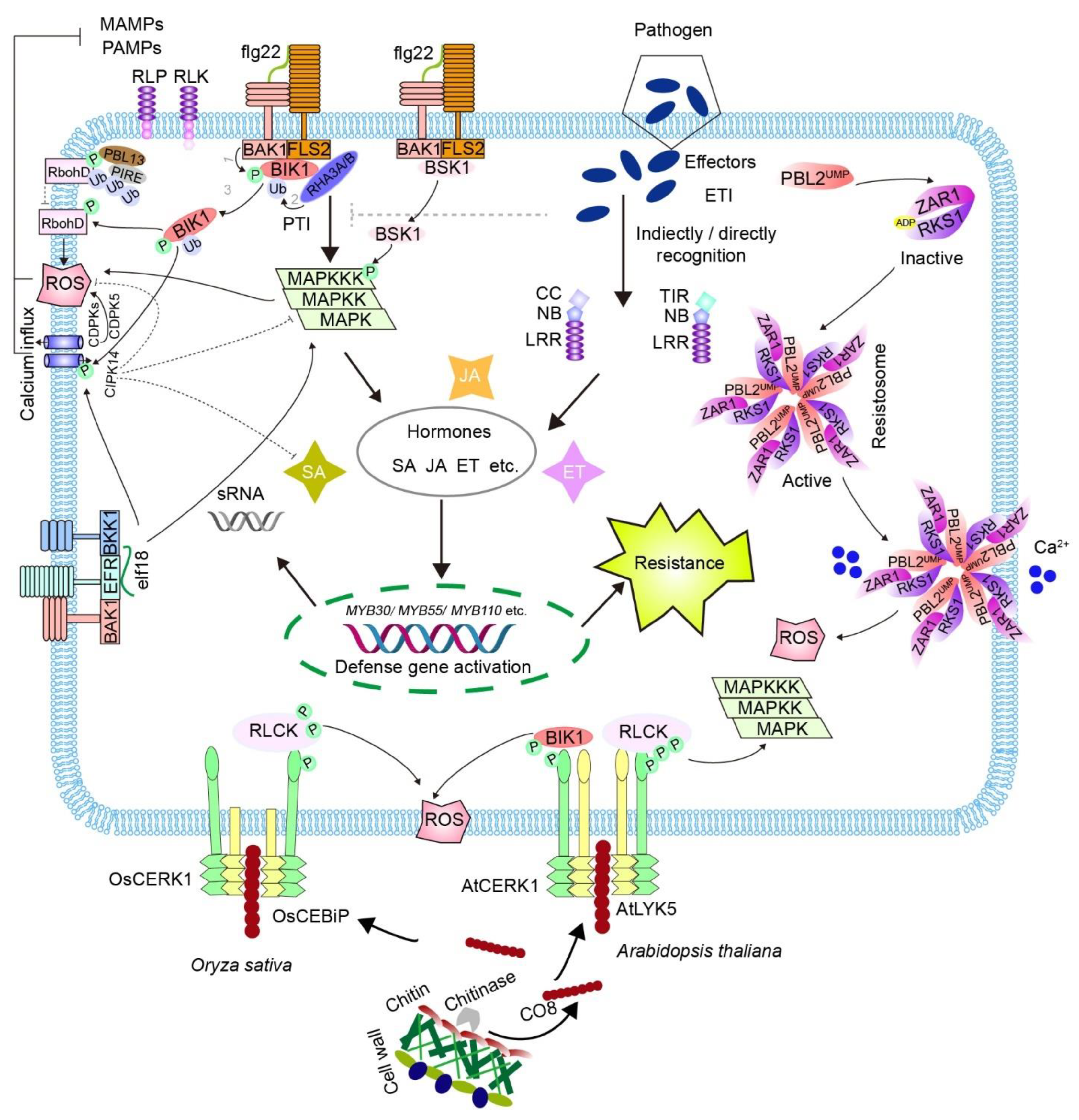
2. The Plant Immune System
3. Interplay between MAMP- and Effector-Triggered Immunity
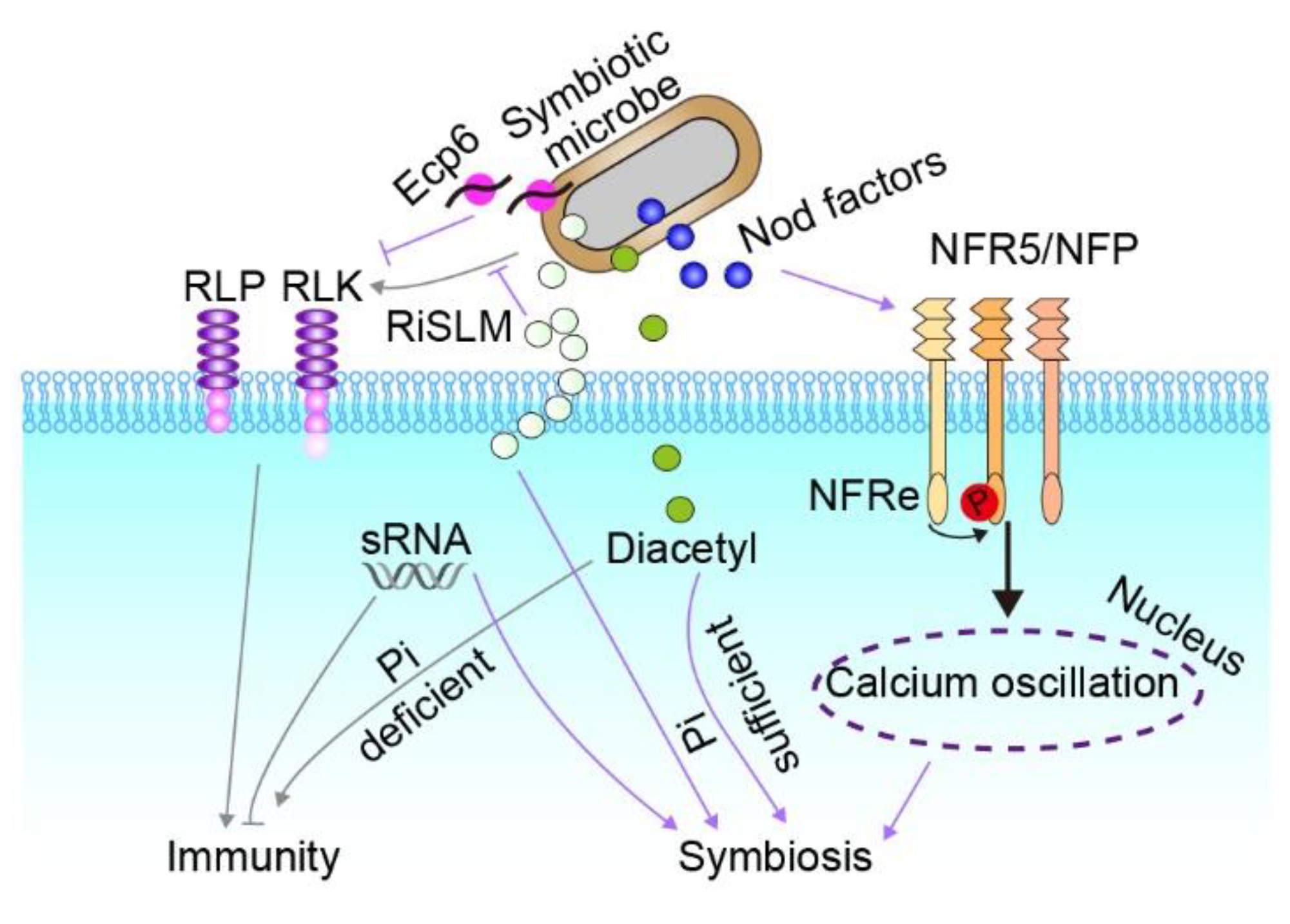
4. Establishment of Symbiosis
5. Tight Regulation of Plant Immunity during Symbiosis
6. Conclusion and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, 2001, re22. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.M.; Woods-Tör, A.; Zipfel, C.; et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Krishnamurthy, N.; Tör, M.; Sjölander, K.V.; Jones, J.D. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005, 138, 611–623. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.W.; et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364, eaav5868. [Google Scholar] [CrossRef]
- Lu, Y.; Tsuda, K. Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol. Plant Microbe Interact. 2021, 34, 3–14. [Google Scholar] [CrossRef]
- Bi, G.; Su, M.; Li, N.; Liang, Y.; Dang, S.; Xu, J.; Hu, M.; Wang, J.; Zou, M.; Deng, Y.; et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 2021, 184, 3528–3541.e3512. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Ma, S.; Lapin, D.; Liu, L.; Sun, Y.; Song, W.; Zhang, X.; Logemann, E.; Yu, D.; Wang, J.; Jirschitzka, J.; et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 2020, 370, eabe3069. [Google Scholar] [CrossRef]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef]
- Zhou, F.; Emonet, A.; Denervaud Tendon, V.; Marhavy, P.; Wu, D.; Lahaye, T.; Geldner, N. Co-incidence of Damage and Microbial Patterns Controls Localized Immune Responses in Roots. Cell 2020, 180, 440–453.e418. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Maier, B.A.; Kiefer, P.; Field, C.M.; Hemmerle, L.; Bortfeld-Miller, M.; Emmenegger, B.; Schäfer, M.; Pfeilmeier, S.; Sunagawa, S.; Vogel, C.M.; et al. A general non-self response as part of plant immunity. Nat. Plants 2021, 7, 696–705. [Google Scholar] [CrossRef]
- Versluys, M.; Tarkowski, L.P.; Van den Ende, W. Fructans as DAMPs or MAMPs: Evolutionary prospects, cross-tolerance, and multistress resistance potential. Front. Plant Sci. 2016, 7, 2061. [Google Scholar] [CrossRef] [Green Version]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor Kinases in Plant-Pathogen Interactions: More Than Pattern Recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Kishi-Kaboshi, M.; Seo, S.; Takahashi, A.; Hirochika, H. The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol. 2018, 59, 903–915. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef]
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef]
- Shi, H.; Shen, Q.; Qi, Y.; Yan, H.; Nie, H.; Chen, Y.; Zhao, T.; Katagiri, F.; Tang, D. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 2013, 25, 1143–1157. [Google Scholar] [CrossRef]
- Ma, X.; Claus, L.A.N.; Leslie, M.E.; Tao, K.; Wu, Z.; Liu, J.; Yu, X.; Li, B.; Zhou, J.; Savatin, D.V.; et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 2020, 581, 199–203. [Google Scholar] [CrossRef]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, Q.; He, J.; Cao, J.; Liu, Z.; Wang, J.; Yang, Y. The kinase CIPK14 functions as a negative regulator of plant immune responses to Pseudomonas syringae in Arabidopsis. Plant Sci. 2021, 312, 111017. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.M. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Yan, H.; Zhao, Y.; Shi, H.; Li, J.; Wang, Y.; Tang, D. BRASSINOSTEROID-SIGNALING KINASE1 Phosphorylates MAPKKK5 to Regulate Immunity in Arabidopsis. Plant Physiol. 2018, 176, 2991–3002. [Google Scholar] [CrossRef]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tör, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Jeworutzki, E.; Roelfsema, M.R.; Anschutz, U.; Krol, E.; Elzenga, J.T.; Felix, G.; Boller, T.; Hedrich, R.; Becker, D. Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca-associated opening of plasma membrane anion channels. Plant J. 2010, 62, 367–378. [Google Scholar] [CrossRef]
- Bozsoki, Z.; Gysel, K.; Hansen, S.B.; Lironi, D.; Kronauer, C.; Feng, F.; de Jong, N.; Vinther, M.; Kamble, M.; Thygesen, M.B.; et al. Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 2020, 369, 663–670. [Google Scholar] [CrossRef]
- Narusaka, Y.; Shinya, T.; Narusaka, M.; Motoyama, N.; Shimada, H.; Murakami, K.; Shibuya, N. Presence of LYM2 dependent but CERK1 independent disease resistance in Arabidopsis. Plant Signal. Behav. 2013, 8, e25345. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Xue, D.X.; Li, C.L.; Xie, Z.P.; Staehelin, C. LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 5507–5516. [Google Scholar] [CrossRef]
- Wan, J.; Tanaka, K.; Zhang, X.C.; Son, G.H.; Brechenmacher, L.; Nguyen, T.H.; Stacey, G. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef]
- Petutschnig, E.K.; Jones, A.M.; Serazetdinova, L.; Lipka, U.; Lipka, V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef]
- Swiderski, M.R.; Innes, R.W. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001, 26, 101–112. [Google Scholar] [CrossRef]
- Muto, H.; Yabe, N.; Asami, T.; Hasunuma, K.; Yamamoto, K.T. Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol. 2004, 136, 3124–3133. [Google Scholar] [CrossRef]
- Kim, T.W.; Guan, S.; Burlingame, A.L.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Squeglia, F.; Berisio, R.; Shibuya, N.; Kaku, H. Defense against pathogens: Structural insights into the mechanism of chitin induced activation of innate immunity. Curr. Med. Chem. 2017, 24, 3980–3986. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yamada, K.; Ishikawa, K.; Yoshimura, S.; Hayashi, N.; Uchihashi, K.; Ishihama, N.; Kishi-Kaboshi, M.; Takahashi, A.; Tsuge, S.; et al. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 2013, 13, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of chitin-induced MAPK signaling pathways in rice and arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wang, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T.; et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yoshimura, Y.; Nakagawa, S.; Mezaki, H.; Yoshimura, S.; Kawasaki, T. OsDRE2 contributes to chitin-triggered response through its interaction with OsRLCK185. Biosci. Biotechnol. Biochem. 2019, 83, 281–290. [Google Scholar] [CrossRef]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 2018, 23, 498–510.e495. [Google Scholar] [CrossRef]
- Ade, J.; DeYoung, B.J.; Golstein, C.; Innes, R.W. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 2007, 104, 2531–2536. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J.; Staskawicz, B.J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 2003, 112, 369–377. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Mucyn, T.S.; Clemente, A.; Andriotis, V.M.; Balmuth, A.L.; Oldroyd, G.E.; Staskawicz, B.J.; Rathjen, J.P. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 2006, 18, 2792–2806. [Google Scholar] [CrossRef] [PubMed]
- Laflamme, B.; Dillon, M.M.; Martel, A.; Almeida, R.N.D.; Desveaux, D.; Guttman, D.S. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 2020, 367, 763–768. [Google Scholar] [CrossRef]
- Wang, J.; Chern, M.; Chen, X. Structural dynamics of a plant NLR resistosome: Transition from autoinhibition to activation. Sci. China Life Sci. 2020, 63, 617–619. [Google Scholar] [CrossRef]
- Lee, D.; Lal, N.K.; Lin, Z.D.; Ma, S.; Liu, J.; Castro, B.; Toruno, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1838. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Lang, J.; Genot, B.; Bigeard, J.; Colcombet, J. MAP kinases 3 and 6 control salicylic acid signaling by upregulating NLR receptors during pattern- and effector-triggered immunity. J. Exp. Bot. 2022, 73, 2190–2205. [Google Scholar] [CrossRef]
- Tian, H.; Wu, Z.; Chen, S.; Ao, K.; Huang, W.; Yaghmaiean, H.; Sun, T.; Xu, F.; Zhang, Y.; Wang, S.; et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature 2021, 598, 500–503. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Locci, F.; Wanke, F.; Zhang, L.; Saile, S.C.; Joe, A.; Karelina, D.; Hua, C.; Frohlich, K.; Wan, W.L.; et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 2021, 598, 495–499. [Google Scholar] [CrossRef]
- Zhai, K.; Liang, D.; Li, H.; Jiao, F.; Yan, B.; Liu, J.; Lei, Z.; Huang, L.; Gong, X.; Wang, X.; et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 2022, 601, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; Nomura, K.; Aung, K.; Velásquez, A.C.; Yao, J.; Boutrot, F.; Chang, J.H.; Zipfel, C.; He, S.Y. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 2016, 539, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Spaink, H.P. The molecular basis of infection and nodulation by rhizobia: The ins and outs of sympathogenesis. Annu. Rev. Phytopathol. 1995, 33, 345–368. [Google Scholar] [CrossRef]
- Peters, N.K.; Frost, J.W.; Long, S.R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 1986, 233, 977–980. [Google Scholar] [CrossRef]
- Dénarié, J.; Cullimore, J. Lipo-oligosaccharide nodulation factors: A minireview new class of signaling molecules mediating recognition and morphogenesis. Cell 1993, 74, 951–954. [Google Scholar] [CrossRef]
- Long, S.R. Rhizobium symbiosis: Nod factors in perspective. Plant Cell 1996, 8, 1885–1898. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Igolkina, A.A.; Bazykin, G.A.; Chizhevskaya, E.P.; Provorov, N.A.; Andronov, E.E. Matching population diversity of rhizobial nodA and legume NFR5 genes in plant-microbe symbiosis. Ecol. Evol. 2019, 9, 10377–10386. [Google Scholar] [CrossRef] [PubMed]
- Broghammer, A.; Krusell, L.; Blaise, M.; Sauer, J.; Sullivan, J.T.; Maolanon, N.; Vinther, M.; Lorentzen, A.; Madsen, E.B.; Jensen, K.J.; et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 2012, 109, 13859–13864. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Oldroyd, G.E. Nuclear calcium signaling in plants. Plant Physiol 2013, 163, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Murakami, E.; Cheng, J.; Gysel, K.; Bozsoki, Z.; Kawaharada, Y.; Hjuler, C.T.; Sorensen, K.K.; Tao, K.; Kelly, S.; Venice, F.; et al. Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. eLife 2018, 7, e33506. [Google Scholar] [CrossRef]
- Tampakaki, A.P. Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front. Plant Sci. 2014, 5, 114. [Google Scholar] [CrossRef]
- Portaliou, A.G.; Tsolis, K.C.; Loos, M.S.; Zorzini, V.; Economou, A. Type III Secretion: Building and Operating a Remarkable Nanomachine. Trends Biochem. Sci. 2016, 41, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Lara-Tejero, M.; Kato, J.; Wagner, S.; Liu, X.; Galán, J.E. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 2011, 331, 1188–1191. [Google Scholar] [CrossRef] [PubMed]
- Bartsev, A.V.; Deakin, W.J.; Boukli, N.M.; McAlvin, C.B.; Stacey, G.; Malnoë, P.; Broughton, W.J.; Staehelin, C. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 2004, 134, 871–879. [Google Scholar] [CrossRef]
- Xin, D.W.; Liao, S.; Xie, Z.P.; Hann, D.R.; Steinle, L.; Boller, T.; Staehelin, C. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 2012, 8, e1002707. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Okazaki, S. How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 2017, 38, 148–154. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pages, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Becard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 190–202. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; Andre, O.; Puech-Pages, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Miyata, K.; Hayafune, M.; Kobae, Y.; Kaku, H.; Nishizawa, Y.; Masuda, Y.; Shibuya, N.; Nakagawa, T. Evaluation of the role of the LysM receptor-like kinase, OsNFR5/OsRLK2 for AM symbiosis in rice. Plant Cell Physiol. 2016, 57, 2283–2290. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.E.; Wang, E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef]
- Carotenuto, G.; Chabaud, M.; Miyata, K.; Capozzi, M.; Takeda, N.; Kaku, H.; Shibuya, N.; Nakagawa, T.; Barker, D.G.; Genre, A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017, 214, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Leppyanen, I.V.; Shakhnazarova, V.Y.; Shtark, O.Y.; Vishnevskaya, N.A.; Tikhonovich, I.A.; Dolgikh, E.A. Receptor-like kinase LYK9 in pisum sativum L. is the CERK1-like receptor that controls both plant immunity and AM symbiosis development. Int. J. Mol. Sci. 2017, 19, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibelin-Viala, C.; Amblard, E.; Puech-Pages, V.; Bonhomme, M.; Garcia, M.; Bascaules-Bedin, A.; Fliegmann, J.; Wen, J.; Mysore, K.S.; le Signor, C.; et al. The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 2019, 223, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Kozaki, T.; Kouzai, Y.; Ozawa, K.; Ishii, K.; Asamizu, E.; Okabe, Y.; Umehara, Y.; Miyamoto, A.; Kobae, Y.; et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014, 55, 1864–1872. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, C.; Dai, H.; Liu, H.; Zhang, X.; Yang, J.; Chen, X.; Zhu, Y.; Wang, D.; Qi, X.; et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 2019, 12, 1561–1576. [Google Scholar] [CrossRef]
- Zhang, C.; He, J.; Dai, H.; Wang, G.; Zhang, X.; Wang, C.; Shi, J.; Chen, X.; Wang, D.; Wang, E. Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2023738118. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Choi, J.; Summers, W.; Paszkowski, U. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annu. Rev. Phytopathol. 2018, 56, 135–160. [Google Scholar] [CrossRef]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA 2017, 114, e8118–e8127. [Google Scholar] [CrossRef]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; van den Berg, W.; Gasciolli, V.; Cottaz, S.; Fort, S.; Thomma, B.; Bono, J.J.; et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2020, 225, 448–460. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef]
- Arthikala, M.K.; Sánchez-López, R.; Nava, N.; Santana, O.; Cárdenas, L.; Quinto, C. RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 2014, 202, 886–900. [Google Scholar] [CrossRef]
- Campos-Soriano, L.; García-Martínez, J.; San Segundo, B. The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol. Plant Pathol. 2012, 13, 579–592. [Google Scholar] [CrossRef]
- Jones, K.M.; Sharopova, N.; Lohar, D.P.; Zhang, J.Q.; VandenBosch, K.A.; Walker, G.C. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc. Natl. Acad. Sci. USA 2008, 105, 704–709. [Google Scholar] [CrossRef]
- Aslam, S.N.; Newman, M.A.; Erbs, G.; Morrissey, K.L.; Chinchilla, D.; Boller, T.; Jensen, T.T.; De Castro, C.; Ierano, T.; Molinaro, A.; et al. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr. Biol. 2008, 18, 1078–1083. [Google Scholar] [CrossRef]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.; Qiu, J.; Stacey, G. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 2013, 341, 1384–1387. [Google Scholar] [CrossRef]
- Morcillo, R.J.; Singh, S.K.; He, D.; An, G.; Vilchez, J.I.; Tang, K.; Yuan, F.; Sun, Y.; Shao, C.; Zhang, S.; et al. Rhizobacterium-derived diacetyl modulates plant immunity in a phosphate-dependent manner. EMBO J. 2020, 39, e102602. [Google Scholar] [CrossRef]
- Block, A.; Alfano, J.R. Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr. Opin. Microbiol. 2011, 14, 39–46. [Google Scholar] [CrossRef]
- Tyler, B.M. Entering and breaking: Virulence effector proteins of oomycete plant pathogens. Cell Microbiol. 2009, 11, 13–20. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef]
- Bolton, M.D.; van Esse, H.P.; Vossen, J.H.; de Jonge, R.; Stergiopoulos, I.; Stulemeijer, I.J.; van den Berg, G.C.; Borras-Hidalgo, O.; Dekker, H.L.; de Koster, C.G.; et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 2008, 69, 119–136. [Google Scholar] [CrossRef]
- de Jonge, R.; Thomma, B.P. Fungal LysM effectors: Extinguishers of host immunity? Trends Microbiol. 2009, 17, 151–157. [Google Scholar] [CrossRef]
- de Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; van der Krol, S.; Shibuya, N.; Joosten, M.H.; Thomma, B.P. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef]
- Feng, Y.; Wu, P.; Liu, C.; Peng, L.; Wang, T.; Wang, C.; Tan, Q.; Li, B.; Ou, Y.; Zhu, H.; et al. Suppression of LjBAK1-mediated immunity by SymRK promotes rhizobial infection in Lotus japonicus. Mol. Plant 2021, 14, 1935–1950. [Google Scholar] [CrossRef] [PubMed]
- Benezech, C.; Berrabah, F.; Jardinaud, M.F.; Le Scornet, A.; Milhes, M.; Jiang, G.; George, J.; Ratet, P.; Vailleau, F.; Gourion, B. Medicago-sinorhizobium-ralstonia co-infection reveals legume nodules as pathogen confined infection sites developing weak defenses. Curr. Biol. 2020, 30, 351–358.e354. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Reynoso, M.A.; Blanco, F.A.; Bailey-Serres, J.; Crespi, M.; Zanetti, M.E. Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J. 2013, 73, 289–301. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Lauressergues, D.; Andre, O.; Gutjahr, C.; Guillotin, B.; Becard, G.; Combier, J.P. Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 2017, 21, 106–112. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, S.; Liu, F.; Wang, W.; Wang, X.; Han, G.; Cheng, B. Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. Int. J. Mol. Sci. 2018, 19, 3201. [Google Scholar] [CrossRef]
- Ren, B.; Wang, X.; Duan, J.; Ma, J. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef]
- Paiva, S.L.; Crews, C.M. Targeted protein degradation: Elements of PROTAC design. Curr. Opin. Chem. Biol. 2019, 50, 111–119. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, L.; Yang, X.; Qi, F. Distinct Responses to Pathogenic and Symbionic Microorganisms: The Role of Plant Immunity. Int. J. Mol. Sci. 2022, 23, 10427. https://doi.org/10.3390/ijms231810427
Ji L, Yang X, Qi F. Distinct Responses to Pathogenic and Symbionic Microorganisms: The Role of Plant Immunity. International Journal of Molecular Sciences. 2022; 23(18):10427. https://doi.org/10.3390/ijms231810427
Chicago/Turabian StyleJi, Li, Xiangrui Yang, and Feifei Qi. 2022. "Distinct Responses to Pathogenic and Symbionic Microorganisms: The Role of Plant Immunity" International Journal of Molecular Sciences 23, no. 18: 10427. https://doi.org/10.3390/ijms231810427
APA StyleJi, L., Yang, X., & Qi, F. (2022). Distinct Responses to Pathogenic and Symbionic Microorganisms: The Role of Plant Immunity. International Journal of Molecular Sciences, 23(18), 10427. https://doi.org/10.3390/ijms231810427




