Phenanthroindolizidine Alkaloids Isolated from Tylophora ovata as Potent Inhibitors of Inflammation, Spheroid Growth, and Invasion of Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Phenanthroindolizidine Alkaloid (PA) Library
2.2. Phenanthroindolizidine Alkaloids (PAs) Are Potent NFκB-Inhibitors in TNBC
2.3. PAs Reduce Cell Viability in the 3D TNBC Co-Culture Model
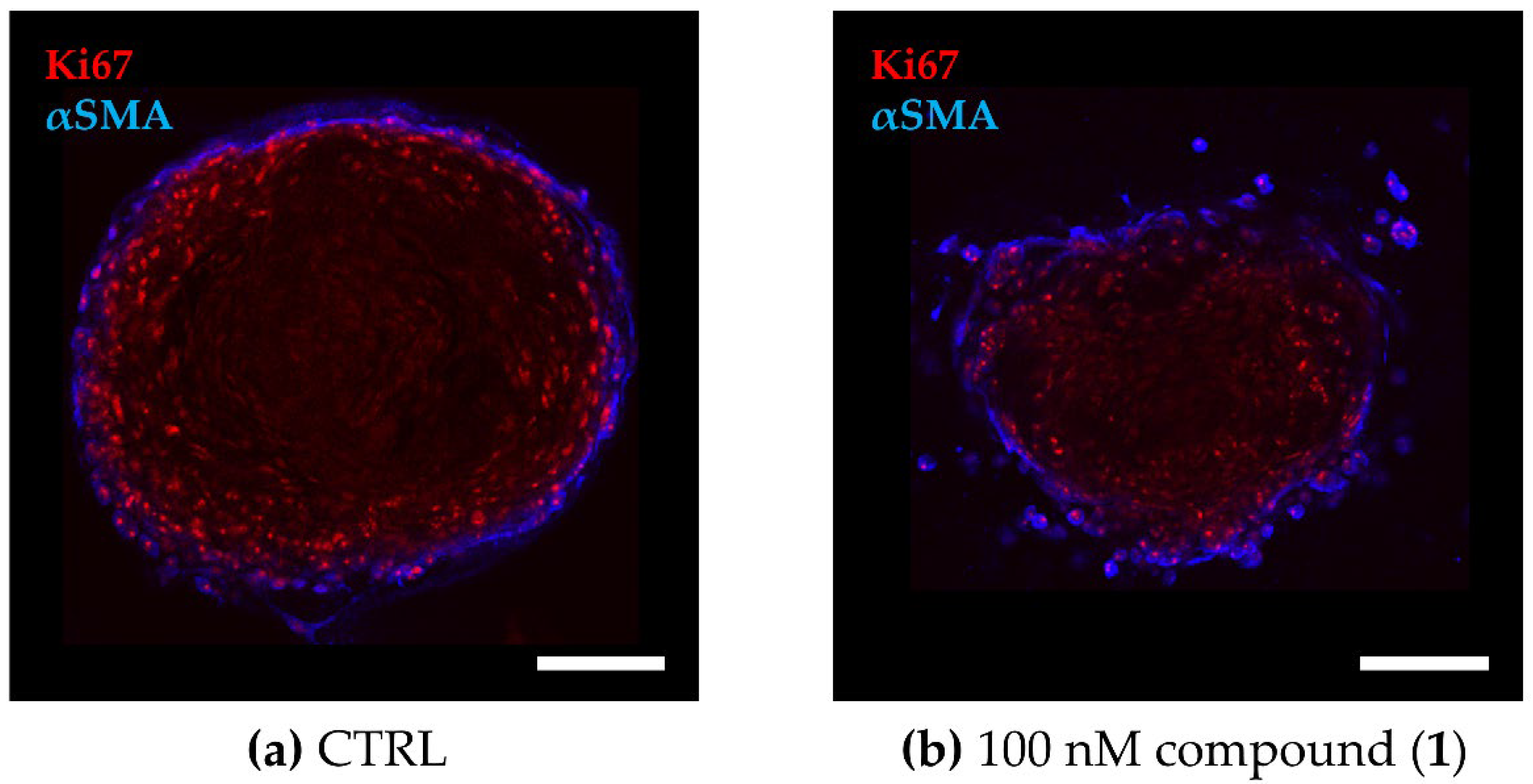
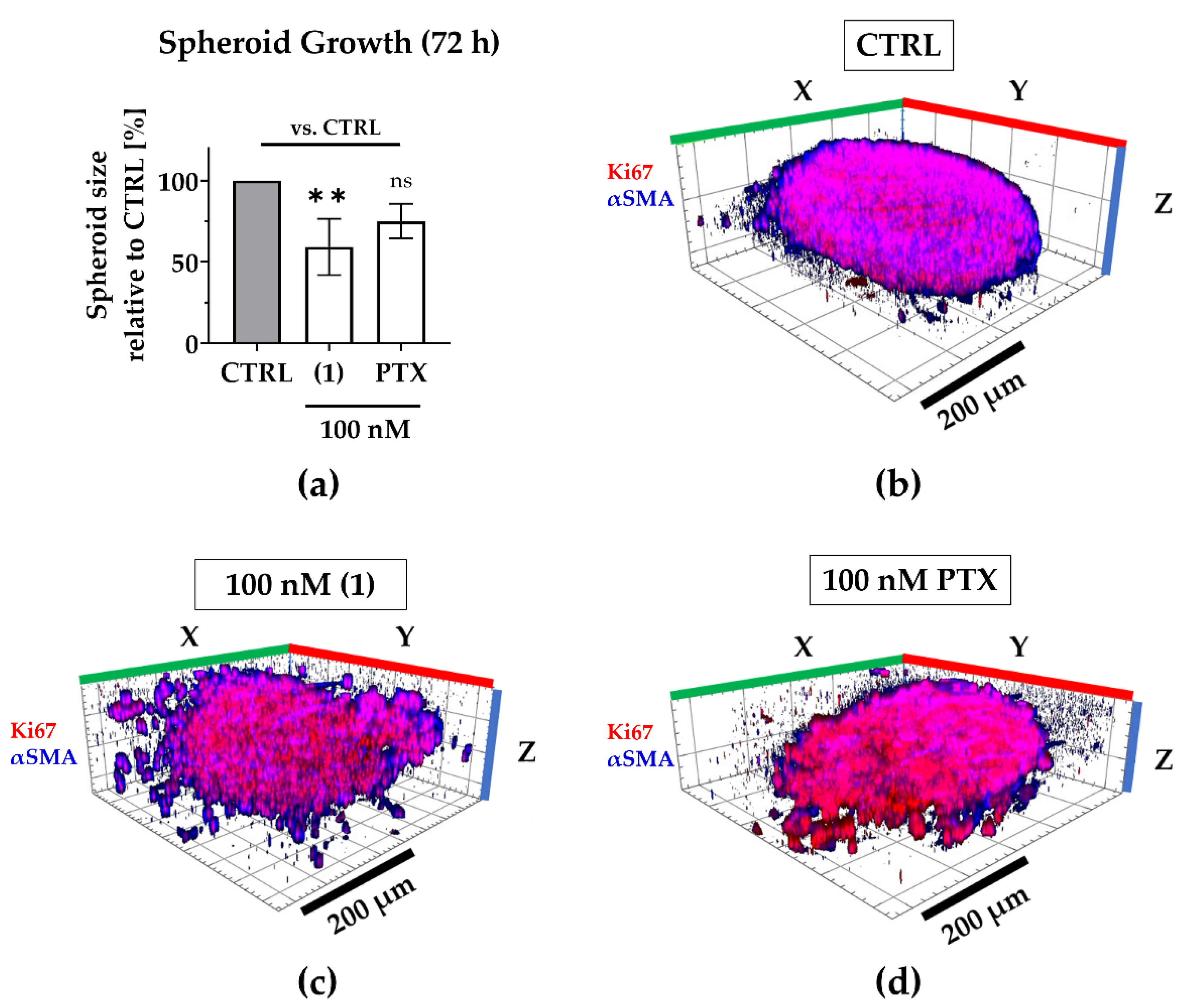
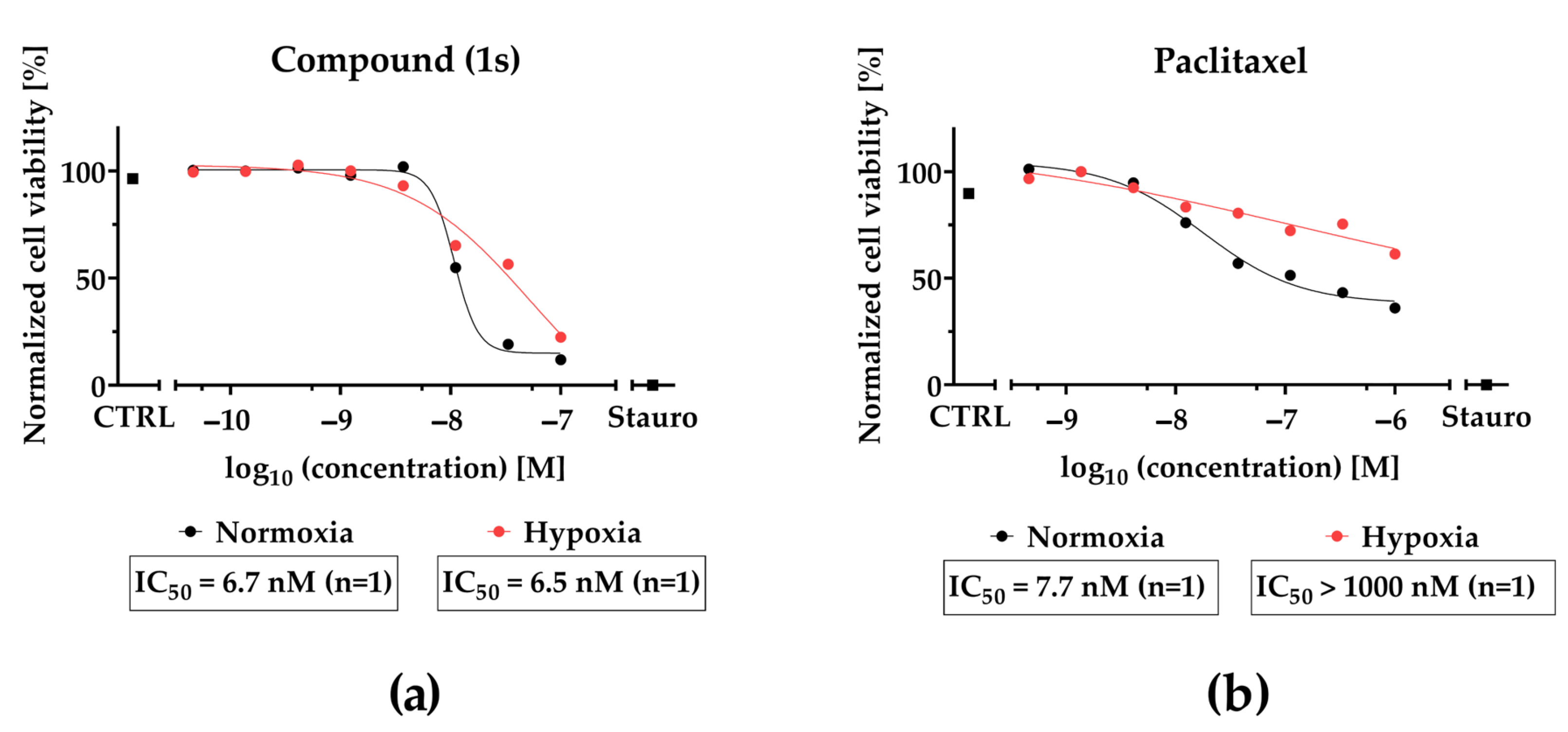
2.4. Compound (1) Exhibits Superior Anti-Tumor Capacity against TNBC Compared to Paclitaxel
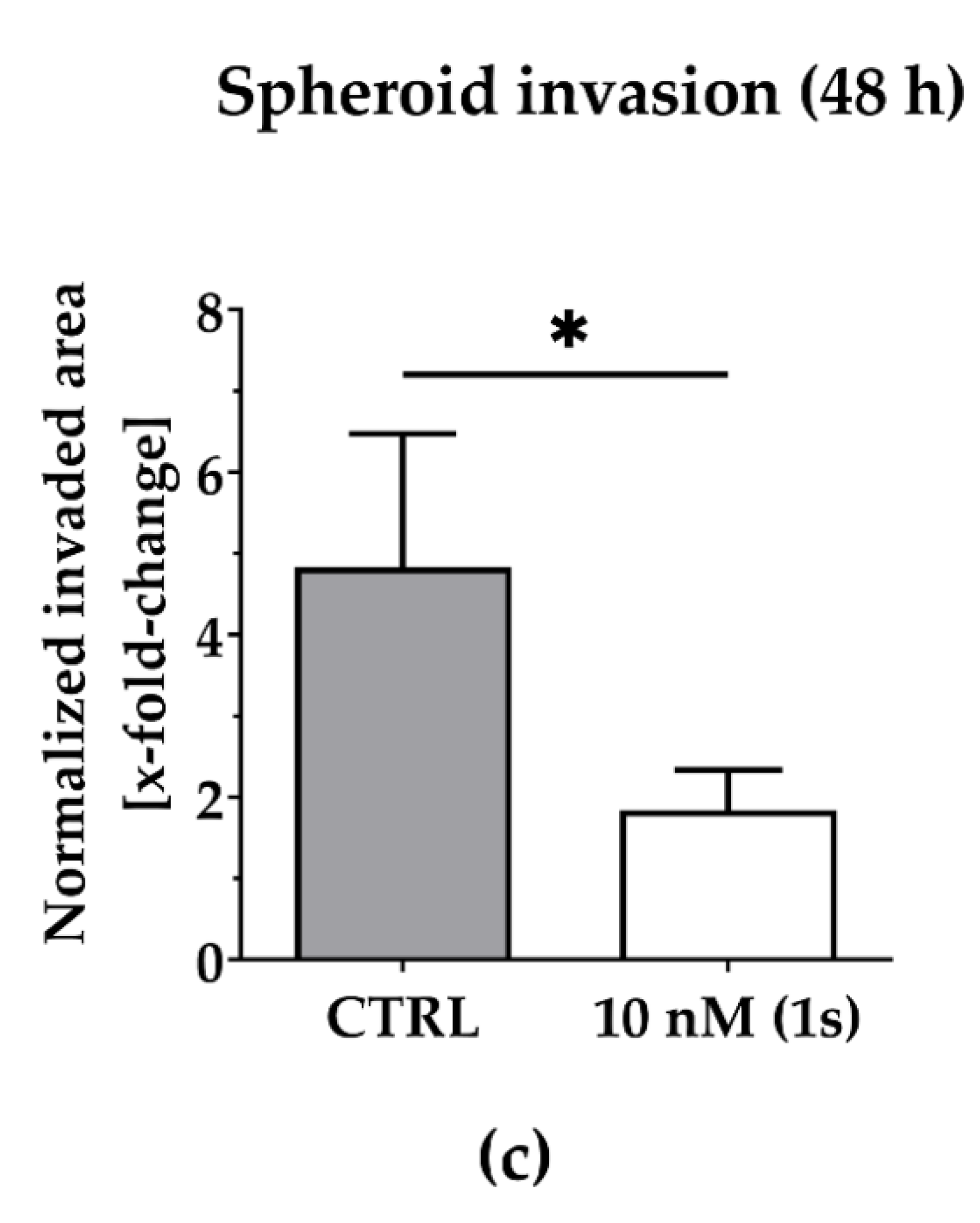
2.5. Compound (1s) Blocks Invasion of TNBC Monoculture 3D Spheroids
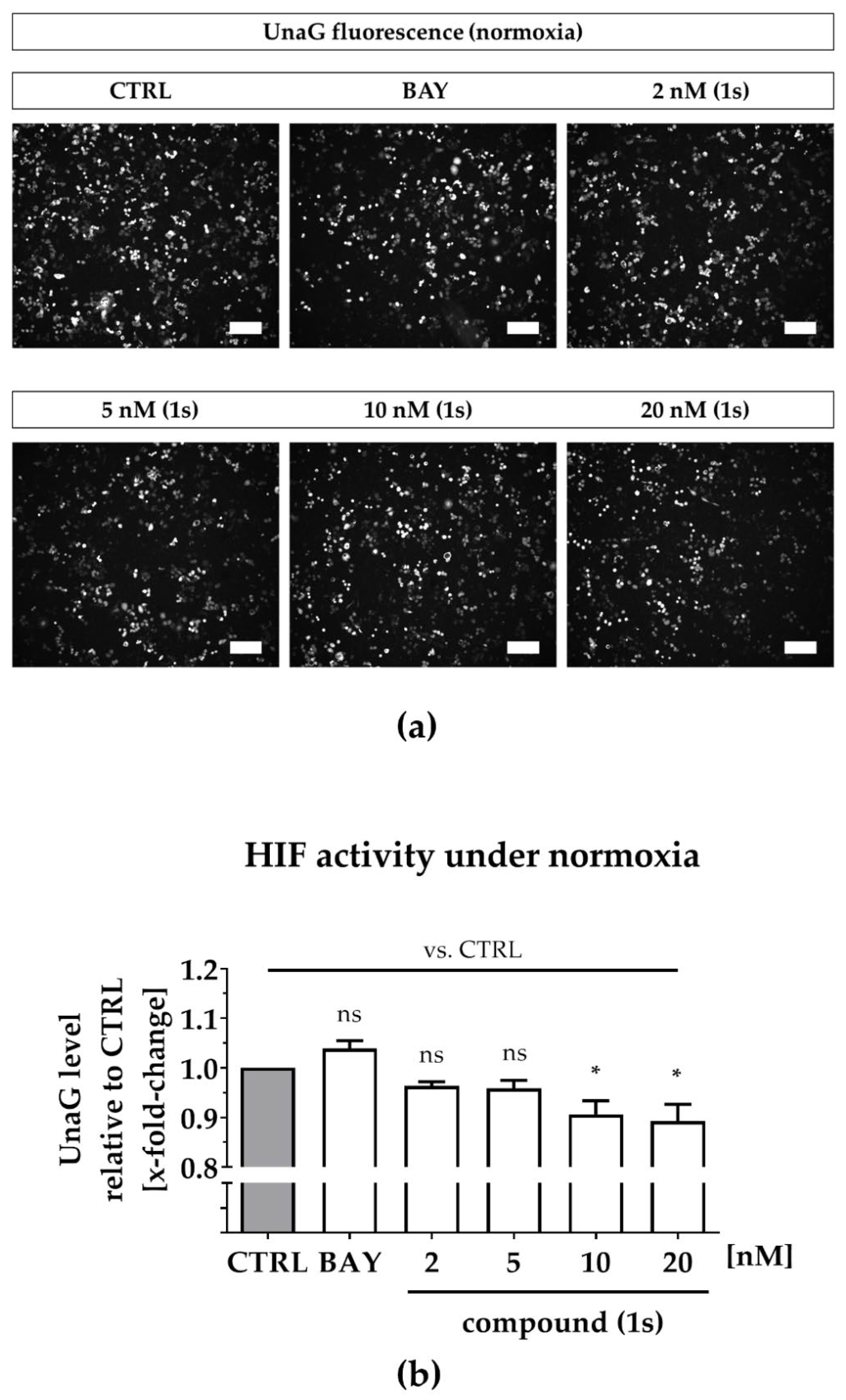
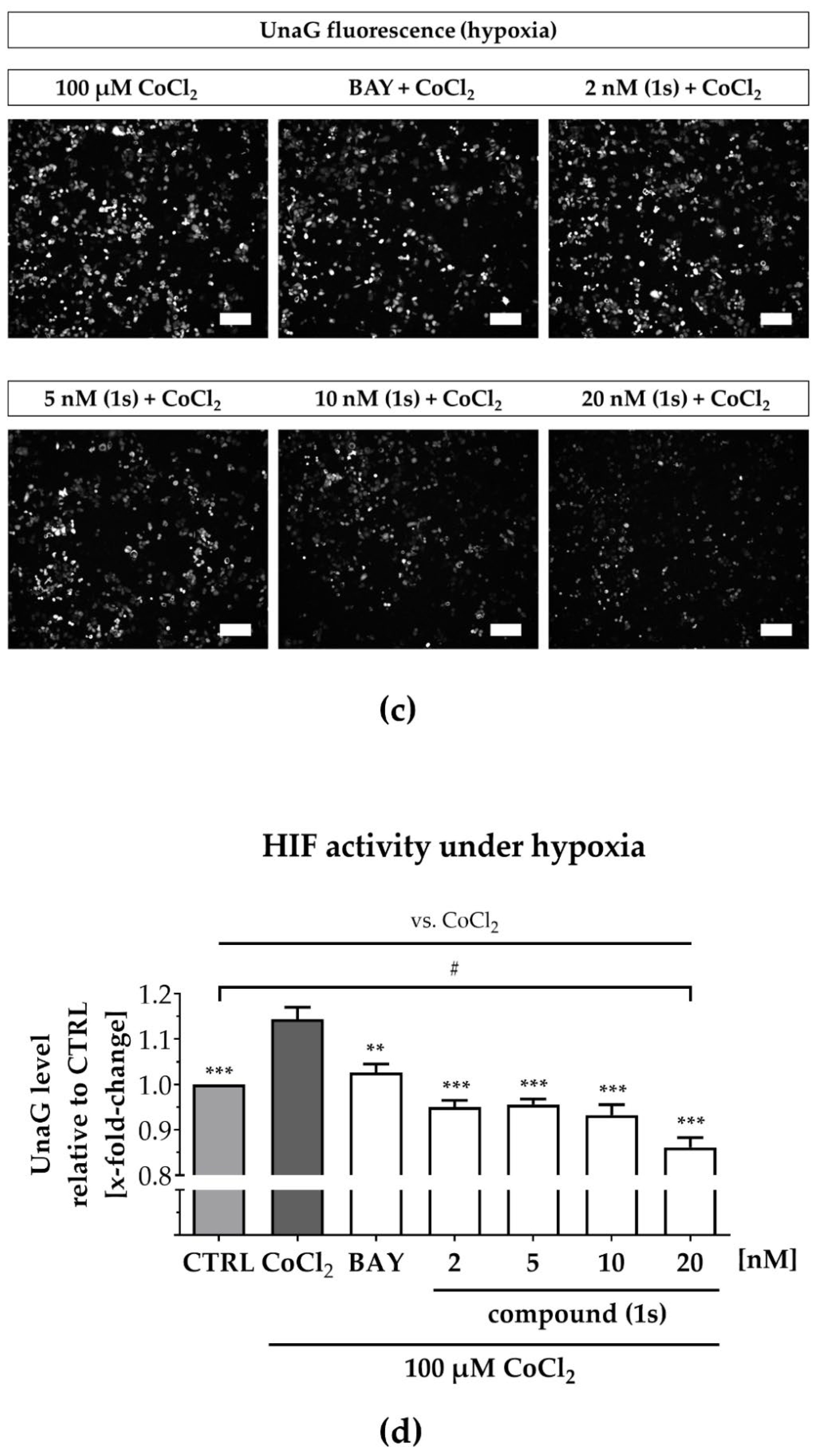
2.6. Compound (1s) Blocks HIF-Regulated Transcription
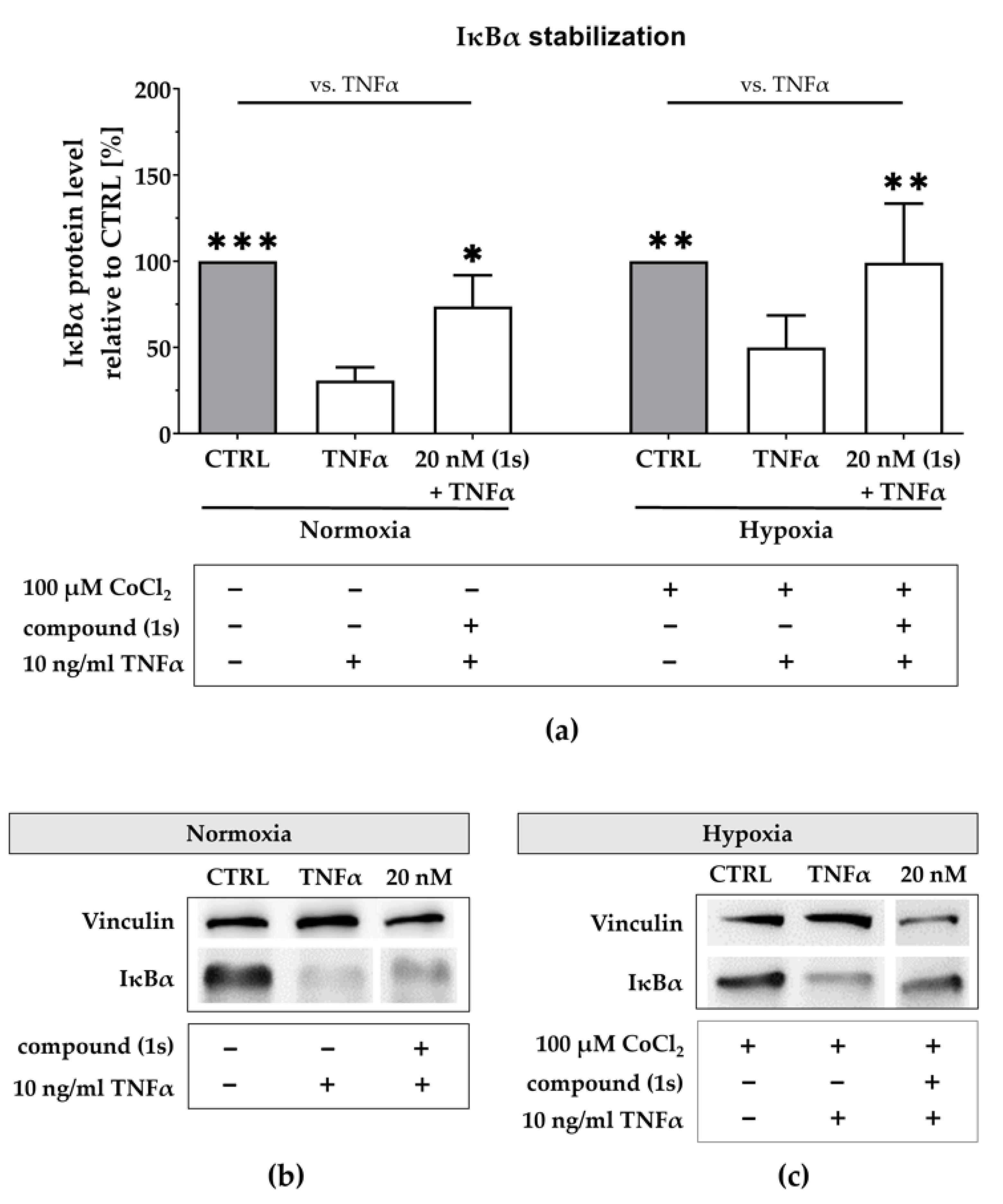
2.7. Compound (1s) Inhibits NFκB Activation Via IκBα Stabilization
2.8. Compound (1s) Blocks Cell Proliferation in the G0/G1 Phase and Delays Cell Cycle Progression
3. Discussion
3.1. PAs Block Inflammation under Hypoxia in TNBC
3.2. PAs Block Proliferation in TNBC through Arrest at the G0/G1-State and Retardation in Cell Cycle Progression
3.3. PAs Maintain Anti-Tumor Potential in a 3D Co-Culture Model Comprising Murine CAFs and Block TNBC Invasion in a 3D Monoculture Spheroid
3.4. Compound (1)/(1s) Exhibits Superior Anti-Tumor Potential Compared to the Chemotherapeutic Agent Paclitaxel
4. Materials and Methods
4.1. Cell Culture and Compounds
4.2. Extraction and Purification of a Phenanthroindolizidines Alkaloids Library
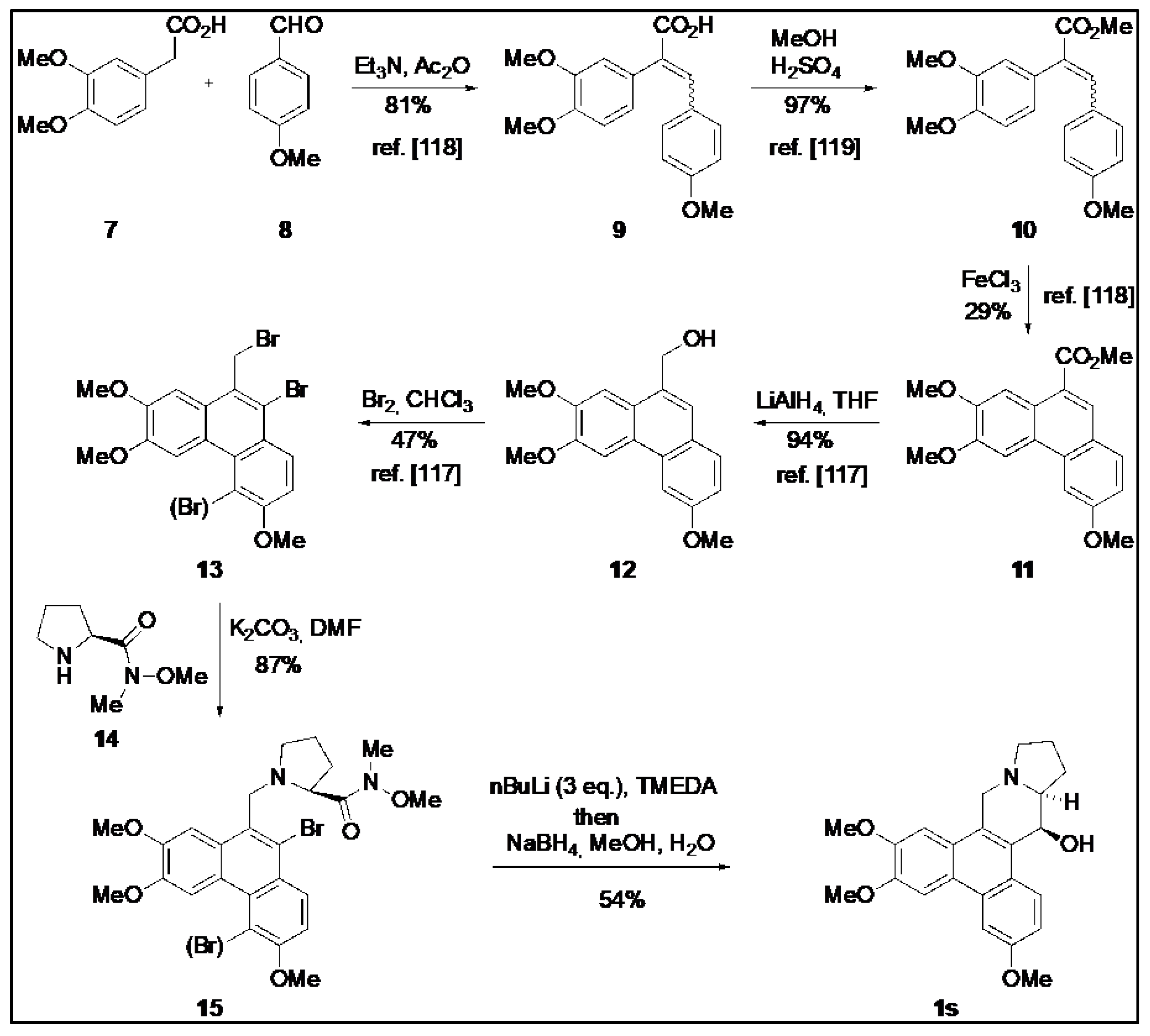
4.3. Chemical Synthesis of O-Methyltylophorinidine (1s)
4.4. Nuclear Factor Kappa B (NFκB) Inhibition Studies
4.4.1. NFκB Inhibition Assay (2 h)
4.4.2. NFκB Inhibition Assay (24 h)
4.5. 2D Cell Viability
4.6. 3D Co-Culture Studies
4.6.1. Spheroid Immunostaining
4.6.2. 3D Cell Viability Studies
4.6.3. 3D Growth Studies
4.7. 3D Invasion Studies
4.8. Hypoxia Studies
4.8.1. HIF Inhibition
4.8.2. Western Blot
4.8.3. Cell Cycle Studies
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guda, C. Integrative exploration of genomic profiles for triple negative breast cancer identifies potential drug targets. Medicine 2016, 95, e4321. [Google Scholar] [CrossRef]
- The American Cancer Society Medical and Editorial Content Team. Triple-Negative Breast Cancer. Available online: https://www.cancer.org/cancer/breast-cancer/about/types-of-breast-cancer/triple-negative.html (accessed on 19 June 2022).
- Hwang, K.-T.; Kim, J.; Jung, J.; Chang, J.H.; Chai, Y.J.; Oh, S.W.; Oh, S.; Kim, Y.A.; Park, S.B.; Hwang, K.R. Impact of Breast Cancer Subtypes on Prognosis of Women with Operable Invasive Breast Cancer: A Population-based Study Using SEER Database. Clin. Cancer Res. 2019, 25, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.K.; Childs, B.H.; Pegram, M. Triple negative breast cancer: Unmet medical needs. Breast Cancer Res. Treat. 2011, 125, 627–636. [Google Scholar] [CrossRef]
- Savard, M.-F.; Khan, O.; Hunt, K.K.; Verma, S. Redrawing the Lines: The Next Generation of Treatment in Metastatic Breast Cancer. Am. Soc. Clin. Oncol. 2019, 39, e8–e21. [Google Scholar] [CrossRef]
- Mani, S.; Hande, A.; Boichuk, S. Triple-Negative Breast Cancer: The Current Aspects of Pathogenesis and Therapies. BioNanoScience 2022, 1–32. [Google Scholar] [CrossRef]
- Lebert, J.M.; Lester, R.; Powell, E.; Seal, M.; McCarthy, J. Advances in the Systemic Treatment of Triple-Negative Breast Cancer. Curr. Oncol. 2018, 25, 142–150. [Google Scholar] [CrossRef]
- Gaptulbarova, K.A.; Tsyganov, M.M.; Pevzner, A.M.; Ibragimova, M.K.; Litviakov, N.V. NF-kB as a potential prognostic marker and a candidate for targeted therapy of cancer. Exp. Oncol. 2020, 42, 263–269. [Google Scholar] [CrossRef]
- Diep, S.; Maddukuri, M.; Yamauchi, S.; Geshow, G.; Delk, N.A. Interleukin-1 and Nuclear Factor Kappa B Signaling Promote Breast Cancer Progression and Treatment Resistance. Cells 2022, 11, 1673. [Google Scholar] [CrossRef]
- Wang, X.; Belguise, K.; Kersual, N.; Kirsch, K.H.; Mineva, N.D.; Galtier, F.; Chalbos, D.; Sonenshein, G.E. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat. Cell Biol. 2007, 9, 470–478. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Pielka, E.; Lipinski, A.; Jelen, M.; Kielan, W.; Agrawal, S. Clinical validation of nuclear factor kappa B expression in invasive breast cancer. Tumor Biol. 2018, 40, 1010428317750929. [Google Scholar] [CrossRef]
- Arora, R.; Yates, C.; Gary, B.D.; McClellan, S.; Tan, M.; Xi, Y.; Reed, E.; Piazza, G.A.; Owen, L.B.; Dean-Colomb, W. Panepoxydone Targets NF-kB and FOXM1 to Inhibit Proliferation, Induce Apoptosis and Reverse Epithelial to Mesenchymal Transition in Breast Cancer. PLoS ONE 2014, 9, e98370. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Lyu, Y.L.; Cai, L. NF-κB Affects Proliferation and Invasiveness of Breast Cancer Cells by Regulating CD44 Expression. PLoS ONE 2014, 9, e106966. [Google Scholar] [CrossRef] [PubMed]
- Prajoko, Y.W.; Aryandono, T. Expression of Nuclear Factor Kappa B (NF-κB) as a Predictor of Poor Pathologic Response to Chemotherapy in Patients with Locally Advanced Breast Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 595–598. [Google Scholar] [CrossRef]
- Ossovskaya, V.; Wang, Y.; Budoff, A.; Xu, Q.; Lituev, A.; Potapova, O.; VanSant, G.; Monforte, J.; Daraselia, N. Exploring Molecular Pathways of Triple-Negative Breast Cancer. Genes Cancer 2011, 2, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Cogswell, P.C.; Guttridge, D.C.; Funkhouser, W.K.; Baldwin, A.S., Jr. Selective activation of NF-κB subunits in human breast cancer: Potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 2000, 19, 1123–1131. [Google Scholar] [CrossRef]
- Xia, L.; Tan, S.; Zhou, Y.; Lin, J.; Wang, H.; Oyang, L.; Tian, Y.; Liu, L.; Su, M.; Wang, H.; et al. Role of the NFκB-signaling pathway in cancer. OncoTargets Ther. 2018, 11, 2063–2073. [Google Scholar] [CrossRef] [PubMed]
- Esparza-López, J.; Longoria, O.; De La Cruz-Escobar, E.N.; Garibay-Díaz, J.C.; León-Rodríguez, E.; Ibarra-Sánchez, M.D.J. Paclitaxel resistance is mediated by NF-κB on mesenchymal primary breast cancer cells. Oncol. Lett. 2022, 23, 50. [Google Scholar] [CrossRef]
- Hartman, Z.C.; Poage, G.M.; Den Hollander, P.; Tsimelzon, A.; Hill, J.; Panupinthu, N.; Zhang, Y.; Mazumdar, A.; Hilsenbeck, S.G.; Mills, G.B.; et al. Growth of Triple-Negative Breast Cancer Cells Relies upon Coordinate Autocrine Expression of the Proinflammatory Cytokines IL-6 and IL-8. Cancer Res. 2013, 73, 3470–3480. [Google Scholar] [CrossRef]
- Jayatilaka, H.; Tyle, P.; Chen, J.J.; Kwak, M.; Ju, J.; Kim, H.J.; Lee, J.S.H.; Wu, P.-H.; Gilkes, D.M.; Fan, R.; et al. Synergistic IL-6 and IL-8 paracrine signalling pathway infers a strategy to inhibit tumour cell migration. Nat. Commun. 2017, 8, 15584. [Google Scholar] [CrossRef] [PubMed]
- Pires, B.R.B.; Mencalha, A.L.; Ferreira, G.M.; De Souza, W.F.; Morgado-Díaz, J.A.; Maia, A.M.; Corrêa, S.; Abdelhay, E.S.F.W. NF-kappaB Is Involved in the Regulation of EMT Genes in Breast Cancer Cells. PLoS ONE 2017, 12, e0169622. [Google Scholar] [CrossRef]
- Sperlich, J.; Teusch, N. Pseudopterosin Inhibits Proliferation and 3D Invasion in Triple-Negative Breast Cancer by Agonizing Glucocorticoid Receptor Alpha. Molecules 2018, 23, 1992. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chu, Y.; Sun, S.; Li, G.; Xu, S.; Zhang, X.; Jiang, Y.; Gao, S.; Wang, Q.; Zhang, J.; et al. Hypoxia-Mediated Complement 1q Binding Protein Regulates Metastasis and Chemoresistance in Triple-Negative Breast Cancer and Modulates the PKC-NF-κB-VCAM-1 Signaling Pathway. Front. Cell Dev. Biol. 2021, 9, 607142. [Google Scholar] [CrossRef]
- Vaupel, P. Hypoxia and Aggressive Tumor Phenotype: Implications for Therapy and Prognosis. Oncologist 2008, 13, 21–26. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Gkegka, A.G.; Pouliliou, S.; Biziota, E.; Kakolyris, S.; Koukourakis, M. Hypoxia and anaerobic metabolism relate with immunologically cold breast cancer and poor prognosis. Breast Cancer Res. Treat. 2022, 194, 13–23. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Wang, M.; Schmid, T.; Xin, Z.; Kozhuharova, L.; Yu, W.-K.; Huang, Y.; Cai, F.; Biskup, E. Hypoxia in Breast Cancer—Scientific Translation to Therapeutic and Diagnostic Clinical Applications. Front. Oncol. 2021, 11, 652266. [Google Scholar] [CrossRef]
- D’Ignazio, L.; Batie, M.; Rocha, S. Hypoxia and Inflammation in Cancer, Focus on HIF and NF-κB. Biomedicines 2017, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, X.; Fan, D. Ginsenoside CK Inhibits Hypoxia-Induced Epithelial–Mesenchymal Transformation through the HIF-1α/NF-κB Feedback Pathway in Hepatocellular Carcinoma. Foods 2021, 10, 1195. [Google Scholar] [CrossRef]
- Zhang, H.; Steed, A.; Co, M.; Chen, X. Cancer stem cells, epithelial-mesenchymal transition, ATP and their roles in drug resistance in cancer. Cancer Drug Resist 2021, 4, 684–709. [Google Scholar] [CrossRef]
- Fultang, N.; Chakraborty, M.; Peethambaran, B. Regulation of cancer stem cells in triple negative breast cancer. Cancer Drug Resist 2021, 4, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Kuo, Y.C.; Ho, Y.S.; Huang, Y.H. Triple-negative breast cancer: Current understanding and future therapeutic breakthrough targeting cancer stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef]
- You, L.; Wu, W.; Wang, X.; Fang, L.; Adam, V.; Nepovimova, E.; Wu, Q.; Kuca, K. The role of hypoxia-inducible factor 1 in tumor immune evasion. Med. Res. Rev. 2021, 41, 1622–1643. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Zhang, X.H.-F. One microenvironment does not fit all: Heterogeneity beyond cancer cells. Cancer Metastasis Rev. 2016, 35, 601–629. [Google Scholar] [CrossRef]
- Gupta, S.; Roy, A.; Dwarakanath, B.S. Metabolic Cooperation and Competition in the Tumor Microenvironment: Implications for Therapy. Front. Oncol. 2017, 7, 68. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- De Andrés, J.L.; Griñán-Lisón, C.; Jiménez, G.; Marchal, J.A. Cancer stem cell secretome in the tumor microenvironment: A key point for an effective personalized cancer treatment. J. Hematol. Oncol. 2020, 13, 136. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.M.; Vink, R.; Heijmans, H.J.; van der Palen, J.; Kouwenhoven, E.A. The prognostic value of tumour-stroma ratio in triple-negative breast cancer. Eur. J. Surg. Oncol. 2012, 38, 307–313. [Google Scholar] [CrossRef]
- Yavuz, B.G.; Gunaydin, G.; Gedik, M.E.; Kosemehmetoglu, K.; Karakoc, D.; Ozgür, F.F.; Guc, D. Cancer associated fibroblasts sculpt tumour microenvironment by recruiting monocytes and inducing immunosuppressive PD-1+ TAMs. Sci. Rep. 2019, 9, 3172. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018, 33, 463–479.e10. [Google Scholar] [CrossRef]
- Mehraj, U.; Dar, A.H.; Wani, N.A.; Mir, M.A. Tumor microenvironment promotes breast cancer chemoresistance. Cancer Chemother. Pharmacol. 2021, 87, 147–158. [Google Scholar] [CrossRef]
- Hendrayani, S.-F.; Al-Harbi, B.; Al-Ansari, M.M.; Silva, G.; Aboussekhra, A. The inflammatory/cancer-related IL-6/STAT3/NF-κB positive feedback loop includes AUF1 and maintains the active state of breast myofibroblasts. Oncotarget 2016, 7, 41974–41985. [Google Scholar] [CrossRef]
- Deepak, K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; Nagini, S.; Rao, D.N.; Malla, R.R. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020, 153, 104683. [Google Scholar] [CrossRef] [PubMed]
- Mollah, F.; Varamini, P. Overcoming Therapy Resistance and Relapse in TNBC: Emerging Technologies to Target Breast Cancer-Associated Fibroblasts. Biomedicines 2021, 9, 1921. [Google Scholar] [CrossRef]
- Mehraj, U.; Ganai, R.A.; Macha, M.A.; Hamid, A.; Zargar, M.A.; Bhat, A.A.; Nasser, M.W.; Haris, M.; Batra, S.K.; Alshehri, B.; et al. The tumor microenvironment as driver of stemness and therapeutic resistance in breast cancer: New challenges and therapeutic opportunities. Cell. Oncol. 2021, 44, 1209–1229. [Google Scholar] [CrossRef]
- Liubomirski, Y.; Lerrer, S.; Meshel, T.; Rubinstein-Achiasaf, L.; Morein, D.; Wiemann, S.; Körner, C.; Ben-Baruch, A. Tumor-Stroma-Inflammation Networks Promote Pro-metastatic Chemokines and Aggressiveness Characteristics in Triple-Negative Breast Cancer. Front. Immunol. 2019, 10, 757. [Google Scholar] [CrossRef]
- Sperlich, J.; Kerr, R.; Teusch, N. The Marine Natural Product Pseudopterosin Blocks Cytokine Release of Triple-Negative Breast Cancer and Monocytic Leukemia Cells by Inhibiting NF-κB Signaling. Mar. Drugs 2017, 15, 262. [Google Scholar] [CrossRef]
- Erkisa, M.; Sariman, M.; Geyik, O.G.; Geyik, C.; Stanojkovic, T.; Ulukaya, E. Natural Products as a Promising Therapeutic Strategy to Target Cancer Stem Cells. Curr. Med. Chem. 2022, 29, 741–783. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Hemsley, H.; Happ, H.; Léveillé, H.H.; Hemsley, N. 39. Tylophora. Flora China 1995, 1995, 253–262. [Google Scholar]
- Endress, M.E.; Bruyns, P.V. A revised classification of the Apocynaceae s.l. Bot. Rev. 2000, 66, 1–56. [Google Scholar] [CrossRef]
- Raina, V.; Raina, S. The responsiveness of leukocyte adenyl cyclase to tylophorine in asthmatic subjects. Biochem. Biophys. Res. Commun. 1980, 94, 1074–1077. [Google Scholar] [CrossRef]
- Bashir, A.; Ali, N.; Bashir, S.; Choudhary, M. Biological activities of aerial parts of Tylophora hirsuta Wall. Afr. J. Biotechnol. 2009, 8, 4627–4631. [Google Scholar]
- Balasubramanian, B.; Dhanabal, M.; Perumal, A.; George, S.D. Studies on the antibac-terial activity and phytochemical screening of Tylophora indica linnon opportunistic bacterial pathogens coinfected with HIV. Drug Invent. Today 2010, 2, 402–404. [Google Scholar]
- Dhiman, M.; Parab, R.R.; Manju, S.L.; Desai, D.C.; Mahajan, G.B. Antifungal Activity of Hydrochloride Salts of Tylophorinidine and Tylophorinine. Nat. Prod. Commun. 2012, 7, 1934578X1200700. [Google Scholar] [CrossRef]
- Yang, C.-W.; Lee, Y.-Z.; Hsu, H.-Y.; Shih, C.; Chao, Y.-S.; Chang, H.-Y.; Lee, S.-J. Targeting Coronaviral Replication and Cellular JAK2 Mediated Dominant NF-κB Activation for Comprehensive and Ultimate Inhibition of Coronaviral Activity. Sci. Rep. 2017, 7, 4105. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-W.; Chen, W.-L.; Wu, P.-L.; Tseng, H.-Y.; Lee, S.-J. Anti-Inflammatory Mechanisms of Phenanthroindolizidine Alkaloids. Mol. Pharmacol. 2006, 69, 749–758. [Google Scholar] [CrossRef]
- Joa, H.; Blažević, T.; Grojer, C.; Zeller, I.; Heiss, E.H.; Atanasov, A.G.; Feldler, I.; Gruzdaitis, P.; Czaloun, C.; Proksch, P.; et al. Tylophorine reduces protein biosynthesis and rapidly decreases cyclin D1, inhibiting vascular smooth muscle cell proliferation in vitro and in organ culture. Phytomedicine 2019, 60, 152938. [Google Scholar] [CrossRef]
- Saraswati, S.; Kanaujia, P.K.; Kumar, S.; Kumar, R.; Alhaider, A.A. Tylophorine, a phenan-thraindolizidine alkaloid isolated from Tylophora indica exerts antiangiogenic and antitumor activity by targeting vascular endothelial growth factor receptor 2mediated angiogenesis. Mol. Cancer 2013, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Lam, W.; Zhong, S.; Kaczmarek, C.; Baker, D.C.; Cheng, Y.-C. Novel Mode of Action of Tylophorine Analogs as Antitumor Compounds. Cancer Res. 2004, 64, 678–688. [Google Scholar] [CrossRef]
- Gao, W.; Bussom, S.; Grill, S.P.; Gullen, E.A.; Hu, Y.-C.; Huang, X.; Zhong, S.; Kaczmarek, C.; Gutierrez, J.; Francis, S.; et al. Structure–activity studies of phenanthroindolizidine alkaloids as potential antitumor agents. Bioorg. Med. Chem. Lett. 2007, 17, 4338–4342. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Zhu, G.-Y.; Wang, J.-R.; Jiang, Z.-H. Phenanthroindolizidine alkaloids from Tylophora atrofolliculata with hypoxia-inducible factor-1 (HIF-1) inhibitory activity. RSC Adv. 2016, 6, 79958–79967. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, H.; Li, H.; Zhang, Y.; Zhang, H.; Su, F.; Xu, S.; Li, Y.; Si, Y.; Yu, S.; et al. Interaction Studies of an Anticancer Alkaloid, (+)-(13aS)-Deoxytylophorinine, with Calf Thymus DNA and Four Repeated Double-Helical DNAs. Chemotherapy 2011, 57, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-J.; Lv, H.-N.; Li, H.-Y.; Zhang, Y.; Zhang, H.-J.; Su, F.-Q.; Si, Y.-K.; Yu, S.-S.; Chen, X.-G. Anticancer effect and neurotoxicity of S-(+)-deoxytylophorinidine, a new phenanthroindolizidine alkaloid that interacts with nucleic acids. J. Asian Nat. Prod. Res. 2011, 13, 400–408. [Google Scholar] [CrossRef] [PubMed]
- MDA-MB-231/ATCC Breast Cell Line. AID 95—NCI Human Tumor Cell Line Growth Inhibition Assay. Data for the MDA-MB-231/ATCC Breast Cell Line—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/95 (accessed on 22 July 2019).
- PubChem. AID—83 NCI Human Tumor Cell Line Growth Inhibition Assay. Data for the MCF7 Breast Cell Line. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/83 (accessed on 22 July 2019).
- PubChem. AID 91—NCI Human Tumor Cell Line Growth Inhibition Assay. Data for the T-47D Breast Cell Line—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/91 (accessed on 22 July 2019).
- Pratama, N.P.; Wulandari, S.; Nugroho, A.E.; Fakhrudin, N.; Astuti, P. Sudarsono Tylophorine Abrogates G2/M Arrest Induced by Doxorubicine and Promotes Increased Apoptosis in T47D Breast Cancer Cells. Asian Pac. J. Cancer Prev. 2018, 19, 3065–3069. [Google Scholar] [CrossRef]
- Fatherree, J.P.; Guarin, J.R.; McGinn, R.A.; Naber, S.P.; Oudin, M.J. Chemotherapy-Induced Collagen IV Drives Cancer Cell Motility through Activation of Src and Focal Adhesion Kinase. Cancer Res. 2022, 82, 2031–2044. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Erapaneedi, R.; Belousov, V.V.; Schäfers, M.; Kiefer, F. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J. 2016, 35, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Rana, N.K.; Singh, P.; Koch, B. CoCl2 simulated hypoxia induce cell proliferation and alter the expression pattern of hypoxia associated genes involved in angiogenesis and apoptosis. Biol. Res. 2019, 52, 12. [Google Scholar] [CrossRef] [PubMed]
- Befani, C.; Mylonis, I.; Gkotinakou, I.-M.; Georgoulias, P.; Hu, C.-J.; Simos, G.; Liakos, P. Cobalt stimulates HIF-1-dependent but inhibits HIF-2-dependent gene expression in liver cancer cells. Int. J. Biochem. Cell Biol. 2013, 45, 2359–2368. [Google Scholar] [CrossRef]
- Yuan, Y.; Hilliard, G.; Ferguson, T.; Millhorn, D.E. Cobalt Inhibits the Interaction between Hypoxia-inducible Factor-α and von Hippel-Lindau Protein by Direct Binding to Hypoxia-inducible Factor-α. J. Biol. Chem. 2003, 278, 15911–15916. [Google Scholar] [CrossRef]
- Xie, J.; Xiao, Y.; Zhu, X.; Ning, Z.; Xu, H.; Wu, H. Hypoxia regulates stemness of breast cancer MDA-MB-231 cells. Med. Oncol. 2016, 33, 42. [Google Scholar] [CrossRef] [PubMed]
- Sunters, A.; de Mattos, S.F.; Stahl, M.; Brosens, J.J.; Zoumpoulidou, G.; Saunders, C.A.; Coffer, P.J.; Medema, R.H.; Coombes, R.C.; Lam, E.W.-F. FoxO3a Transcriptional Regulation of Bim Controls Apoptosis in Paclitaxel-treated Breast Cancer Cell Lines. J. Biol. Chem. 2003, 278, 49795–49805. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yu, S.-S.; Du, D.; Wang, Y.-D. Bioactive constituents from toxic seed plants in China. RSC Adv. 2013, 3, 10078–10102. [Google Scholar] [CrossRef]
- Yang, C.-W.; Lee, Y.-Z.; Hsu, H.-Y.; Wu, C.-M.; Chang, H.-Y.; Chao, Y.-S.; Lee, S.-J. c-Jun-mediated anticancer mechanisms of tylophorine. Carcinogenesis 2013, 34, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Lu, L.; Liao, Y.; Wang, X.; Zhang, Y.; Liu, Y.; Huang, S.; Sun, H.; Li, Z.; Zhao, L. Influence of c-Src on hypoxic resistance to paclitaxel in human ovarian cancer cells and reversal of FV-429. Cell Death Dis. 2018, 8, e3178. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Yaegashi, T.; Matsuzaki, T.; Yamazaki, R.; Hashimoto, S.; Sawada, S. Synthesis of phenanthroindolizidine alkaloids and evaluation of their antitumor activities and toxicities. Bioorg. Med. Chem. Lett. 2011, 21, 5978–5981. [Google Scholar] [CrossRef]
- Niphakis, M.J.; Gay, B.C.; Hong, K.H.; Bleeker, N.P.; Georg, G.I. Synthesis and evaluation of the anti-proliferative and NF-κB activities of a library of simplified tylophorine analogs. Bioorg. Med. Chem. 2012, 20, 5893–5900. [Google Scholar] [CrossRef]
- Huang, X.; Gao, S.; Fan, L.; Yu, S.; Liang, X. Cytotoxic Alkaloids from the Roots of Tylophora atrofolliculata. Planta Med. 2004, 70, 441–445. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Bai, L.-P.; Ke, Z.-F.; Liu, Y.; Wang, J.-R.; Jiang, Z.-H. G-Quadruplex DNA-binding quaternary alkaloids from Tylophora atrofolliculata. RSC Adv. 2016, 6, 114135–114142. [Google Scholar] [CrossRef]
- Shiah, H.-S.; Gao, W.; Baker, D.C.; Cheng, Y.-C. Inhibition of cell growth and nuclear factor-κB activity in pancreatic cancer cell lines by a tylophorine analogue, DCB-3503. Mol. Cancer Ther. 2006, 5, 2484–2493. [Google Scholar] [CrossRef]
- Yoshida, T.; Hashimura, M.; Mastumoto, T.; Tazo, Y.; Inoue, H.; Kuwata, T.; Saegusa, M. Transcriptional upregulation of HIF-1α by NF-κB/p65 and its associations with β-catenin/p300 complexes in endometrial carcinoma cells. Lab. Investig. 2013, 93, 1184–1193. [Google Scholar] [CrossRef]
- Ferrari, P.; Scatena, C.; Ghilli, M.; Bargagna, I.; Lorenzini, G.; Nicolini, A. Molecular Mechanisms, Biomarkers and Emerging Therapies for Chemotherapy Resistant TNBC. Int. J. Mol. Sci. 2022, 23, 1665. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Q.; Lu, D.; Pang, X.; Yin, S.; Wang, K.; Wang, R.; Yang, S.; Zhang, Y.; Qiu, Y.; et al. HTBPI, an active phenanthroindolizidine alkaloid, inhibits liver tumorigenesis by targeting Akt. FASEB J. 2020, 34, 12255–12268. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lam, W.; Chen, S.-R.; Guan, F.-L.; Dutchman, G.E.; Francis, S.; Baker, D.C.; Cheng, Y.-C. Tylophorine Analog DCB-3503 Inhibited Cyclin D1 Translation through Allosteric Regulation of Heat Shock Cognate Protein 70. Sci. Rep. 2016, 6, 32832. [Google Scholar] [CrossRef]
- Mostafa, E.M.; Mohammed, H.A.; Musa, A.; Abdelgawad, M.A.; Al-Sanea, M.M.; Almahmoud, S.A.; Ghoneim, M.M.; Gomaa, H.A.M.; Rahman, F.E.-Z.S.A.; Shalaby, K.; et al. In Vitro Anti-Proliferative, and Kinase Inhibitory Activity of Phenanthroindolizidine Alkaloids Isolated from Tylophora indica. Plants 2022, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef]
- Li, Z.; Tang, X.; Luo, Y.; Chen, B.; Zhou, C.; Wu, X.; Tang, Z.; Qi, X.; Cao, G.; Hao, J.; et al. NK007 helps in mitigating paclitaxel resistance through p38MAPK activation and HK2 degradation in ovarian cancer. J. Cell. Physiol. 2019, 234, 16178–16190. [Google Scholar] [CrossRef]
- Wu, C.-M.; Yang, C.-W.; Lee, Y.-Z.; Chuang, T.-H.; Wu, P.-L.; Chao, Y.-S.; Lee, S.-J. Tylophorine arrests carcinoma cells at G1 phase by downregulating cyclin A2 expression. Biochem. Biophys. Res. Commun. 2009, 386, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef]
- Kampan, N.C.; Madondo, M.T.; McNally, O.M.; Quinn, M.; Plebanski, M. Paclitaxel and Its Evolving Role in the Management of Ovarian Cancer. BioMed Res. Int. 2015, 2015, 413076. [Google Scholar] [CrossRef]
- Gully, C.P.; Velazquez-Torres, G.; Shin, J.-H.; Fuentes-Mattei, E.; Wang, E.; Carlock, C.; Chen, J.; Rothenberg, D.; Adams, H.P.; Choi, H.H.; et al. Aurora B kinase phosphorylates and instigates degradation of p53. Proc. Natl. Acad. Sci. USA 2012, 109, E1513–E1522. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.; Venkatachalam, S. Inhibition of dihydrofolate reductase and cell growth activity by the phenanthroindolizidine alkaloids pergularinine and tylophorinidine: The in vitro cytotoxicity of these plant alkaloids and their potential as antimicrobial and anticancer agents. Toxicol. Vitr. 2000, 14, 53–59. [Google Scholar] [CrossRef]
- Rao, K.N.; Bhattacharya, R.; Venkatachalam, S. Thymidylate synthase activity in leukocytes from patients with chronic myelocytic leukemia and acute lymphocytic leukemia and its inhibition by phenanthroindolizidine alkaloids pergularinine and tylophorinidine. Cancer Lett. 1998, 128, 183–188. [Google Scholar] [CrossRef]
- Ueno, S.; Yamazaki, R.; Ikeda, T.; Yaegashi, T.; Matsuzaki, T. Antitumor effect of a novel phenanthroindolizidine alkaloid derivative through inhibition of protein synthesis. Anticancer Res. 2014, 34, 3391–3398. [Google Scholar] [PubMed]
- Wang, Y.; Gao, W.; Svitkin, Y.V.; Chen, A.P.-C.; Cheng, Y.-C. DCB-3503, a Tylophorine Analog, Inhibits Protein Synthesis through a Novel Mechanism. PLoS ONE 2010, 5, e11607. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.S.; Barros, A.S.; Costa, E.C.; Moreira, A.F.; Correia, I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019, 116, 206–226. [Google Scholar] [CrossRef] [PubMed]
- Abu-Jamous, B.; Buffa, F.M.; Harris, A.L.; Nandi, A.K. In vitro downregulated hypoxia transcriptome is associated with poor prognosis in breast cancer. Mol. Cancer 2017, 16, 105. [Google Scholar] [CrossRef]
- Kopp, S.; Sahana, J.; Islam, T.; Petersen, A.G.; Bauer, J.; Corydon, T.J.; Schulz, H.; Saar, K.; Huebner, N.; Slumstrup, L.; et al. The role of NFκB in spheroid formation of human breast cancer cells cultured on the Random Positioning Machine. Sci. Rep. 2018, 8, 921. [Google Scholar] [CrossRef]
- Wang, W.; Mani, A.M.; Wu, Z.-H. DNA damage-induced nuclear factor-kappa B activation and its roles in cancer progression. J. Cancer Metastasis Treat. 2017, 3, 45–59. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Xu, Z.; Qian, P.; Sun, W.; Wang, X.; Jian, Z.; Xia, T.; Xu, Y.; Tang, J. RelB sustains endocrine resistant malignancy: An insight of noncanonical NF-κB pathway into breast Cancer progression. Cell Commun. Signal. 2020, 18, 128. [Google Scholar] [CrossRef]
- Haryanti, S.; Murwanti, R.; Putri, H.; Lmawati, G.P.N.; Pramono, S.; Meiyanto, E. Different 4T1 Cells Mi-gration under Caesalpinia sappan L. and Ficus septica Burm.f Ethanolic Extracts. Indones. J. Cancer Chemopre-Vention 2017, 8, 21–26. [Google Scholar] [CrossRef][Green Version]
- Kim, C.-H.; Kim, D.-E.; Kim, D.-H.; Min, G.-H.; Park, J.-W.; Kim, Y.-B.; Sung, C.K.; Yim, H. Mitotic protein kinase-driven crosstalk of machineries for mitosis and metastasis. Exp. Mol. Med. 2022, 54, 414–425. [Google Scholar] [CrossRef]
- Wang, M.; Feng, R.; Chen, Z.; Shi, W.; Li, C.; Liu, H.; Wu, K.; Li, D.; Li, X. Identification of Cancer-Associated Fibroblast Subtype of Triple-Negative Breast Cancer. J. Oncol. 2022, 2022, 6452636. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Kauppila, S.; Stenbäck, F.; Risteli, J.; Jukkola, A.; Risteli, L. Aberrant type I and type III collagen gene expression in human breast cancerin vivo. J. Pathol. 1998, 186, 262–268. [Google Scholar] [CrossRef]
- Li, Q.; Li, M.; Zheng, K.; Tang, S.; Ma, S. Expression pattern analysis and drug differential sensitivity of cancer-associated fibroblasts in triple-negative breast cancer. Transl. Oncol. 2021, 14, 100891. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, M.; Fushida, S.; Tsukada, T.; Kinoshita, J.; Oyama, K.; Miyashita, T.; Ninomiya, I.; Harada, S.; Ohta, T. The effect of HIF-1α and PKM1 expression on acquisition of chemoresistance. Cancer Manag. Res. 2018, 10, 1865–1874. [Google Scholar] [CrossRef]
- Notte, A.; Ninane, N.; Arnould, T.; Michiels, C. Hypoxia counteracts taxol-induced apoptosis in MDA-MB-231 breast cancer cells: Role of autophagy and JNK activation. Cell Death Dis. 2013, 4, e638. [Google Scholar] [CrossRef]
- Wen, J.; Yeo, S.; Wang, C.; Chen, S.; Sun, S.; Haas, M.A.; Tu, W.; Jin, F.; Guan, J.-L. Autophagy inhibition re-sensitizes pulse stimulation-selected paclitaxel-resistant triple negative breast cancer cells to chemotherapy-induced apoptosis. Breast Cancer Res. Treat. 2015, 149, 619–629. [Google Scholar] [CrossRef]
- Lin, J.-C.; Yang, S.-C.; Hong, T.-M.; Yu, S.-L.; Shi, Q.; Wei, L.; Chen, H.-Y.; Yang, P.-C.; Lee, K.-H. Phenanthrene-Based Tylophorine-1 (PBT-1) Inhibits Lung Cancer Cell Growth through the Akt and NF-κB Pathways. J. Med. Chem. 2009, 52, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, M.; Manju, S.L.; Khanna, A. A new phenanthroindolizidine alkaloid from Tylophora indica. Chem. Pap. 2013, 67, 245–248. [Google Scholar] [CrossRef]
- Jungwirth, U.; van Weverwijk, A.; Jenkins, L.; Alexander, J.; Vicente, D.; Gao, Q.; Haider, S.; Iravani, M.; Isacke, C.M. Im-pairment of a distinct cancer-associated fibroblast population limits tumour growth and metastasis. Nat. Commun. 2021, 12, 3516. [Google Scholar] [CrossRef] [PubMed]
- Govindachari, T.; Viswanathan, N.; Radhakrishnan, J.; Pai, B.; Natarajan, S.; Subramaniam, P. Minor alkaloids of Tylophora asthmatica: Revised structure of tylophorinidine. Tetrahedron 1973, 29, 891–897. [Google Scholar] [CrossRef]
- Govindachari, T.R.; Viswanathan, N.; Radhakrishnan, J.; Charubala, R.; Rao, N.N.; Pai, B.R. Quaternary alkaloids from Ty-lophora asthmatica. Indian J. Chem. 1973, 1215–1216. [Google Scholar]
- Wang, Z.; Li, Z.; Wang, K.; Wang, Q. Efficient and Chirally Specific Synthesis of Phenanthro-Indolizidine Alkaloids by Parham-Type Cycloacylation. Eur. J. Org. Chem. 2010, 2010, 292–299. [Google Scholar] [CrossRef]
- Wang, K.-L.; Lü, M.-Y.; Wang, Q.-M.; Huang, R.-Q. Iron(III) chloride-based mild synthesis of phenanthrene and its application to total synthesis of phenanthroindolizidine alkaloids. Tetrahedron 2008, 64, 7504–7510. [Google Scholar] [CrossRef]
- Su, C.-R.; Damu, A.G.; Chiang, P.-C.; Bastow, K.F.; Morris-Natschke, S.L.; Lee, K.-H.; Wu, T.-S. Total synthesis of phenanthroindolizidine alkaloids (±)-antofine, (±)-deoxypergularinine, and their dehydro congeners and evaluation of their cytotoxic activity. Bioorg. Med. Chem. 2008, 16, 6233–6241. [Google Scholar] [CrossRef] [PubMed]
- Subik, K.; Lee, J.-F.; Baxter, L.; Strzepek, T.; Costello, D.; Crowley, P.; Xing, L.; Hung, M.-C.; Bonfiglio, T.; Hicks, D.G.; et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer 2010, 4, 35–41. [Google Scholar] [CrossRef]
- Hawsawi, N.M.; Ghebeh, H.; Hendrayani, S.-F.; Tulbah, A.; Al-Eid, M.; Al-Tweigeri, T.; Ajarim, D.; Alaiya, A.; Dermime, S.; Aboussekhra, A. Breast Carcinoma–Associated Fibroblasts and Their Counterparts Display Neoplastic-Specific Changes. Cancer Res. 2008, 68, 2717–2725. [Google Scholar] [CrossRef]
- Xie, B.; Hänsel, J.; Mundorf, V.; Betz, J.; Reimche, I.; Erkan, M.; Büdeyri, I.; Gesell, A.; Kerr, R.G.; Ariantari, N.P.; et al. Pseudopterosin and O-Methyltylophorinidine Suppress Cell Growth in a 3D Spheroid Co-Culture Model of Pancreatic Ductal Adenocarcinoma. Bioengineering 2020, 7, 57. [Google Scholar] [CrossRef]

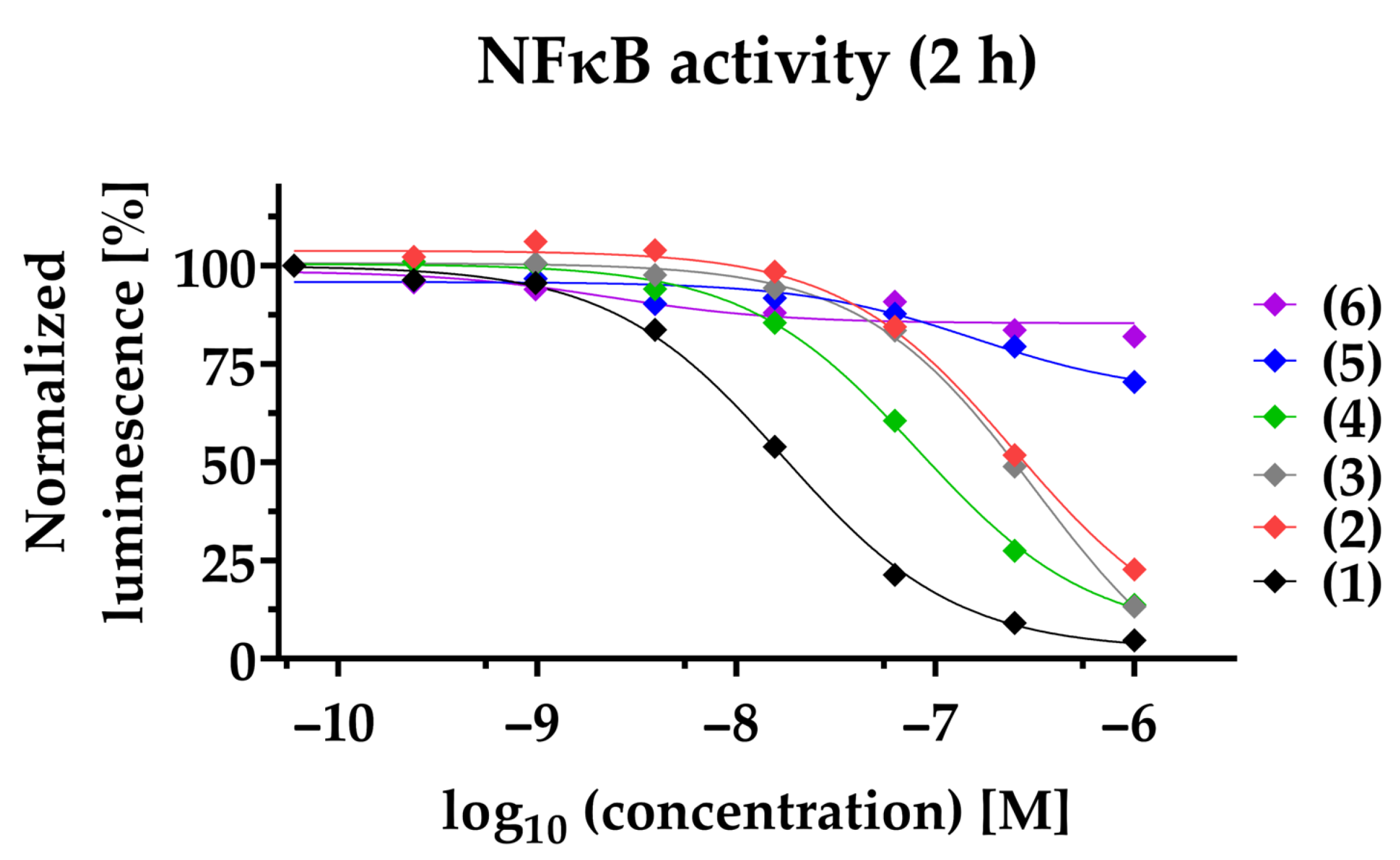




| NFκB Inhibition | Reduction of Cell Viability | ||
|---|---|---|---|
| Compound | 2 h 1 IC50 [nM] 4 | 24 h 2 IC50 [nM] 4 | 24 h 3 IC50 [nM] 4,5,6 |
| (1) | 17.1 ± 2.0 | 3.7 ± 1 | 13.6 ± 0.4 5 |
| (2) | 211.8 ± 69.9 | 38.2 ± 14.2 | 117.9 ± 35.0 4 |
| (3) | 284.9 ± 60.4 | 114.5 ± 17.9 | 55.3 ± 13.2 4 |
| (4) | 83.0 ± 14.7 | 28.3 ± 5.6 | 127 ± 21.4 4 |
| (5) | >1000 | >1000 | >1000 6 |
| (6) | >1000 | >1000 | >1000 6 |
| Paclitaxel | >1000 | >1000 | 37.4 ± 8.6 4 |
| Compound | Reduction of Cell Viability 72 h 1 IC50 [nM] 2 |
|---|---|
| (1) | 21.7 ± 2.5 |
| (2) | 441.5 ± 70.8 |
| (3) | 476.7 ± 160.4 |
| (4) | 517.7 ± 76.8 |
| (5) | >1000 |
| (6) | >1000 |
| Paclitaxel | 43 ± 14.3 |
| Compound | Dilution Factor | Concentration Range |
|---|---|---|
| (1), (2), (3), (4), (5), (6) | 4 | 1000 nM to 0.06 nM |
| (1s) | 3 | 100 nM to 0.05 nM |
| Paclitaxel | 3 | 1000 nM to 0.5 nM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reimche, I.; Yu, H.; Ariantari, N.P.; Liu, Z.; Merkens, K.; Rotfuß, S.; Peter, K.; Jungwirth, U.; Bauer, N.; Kiefer, F.; et al. Phenanthroindolizidine Alkaloids Isolated from Tylophora ovata as Potent Inhibitors of Inflammation, Spheroid Growth, and Invasion of Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2022, 23, 10319. https://doi.org/10.3390/ijms231810319
Reimche I, Yu H, Ariantari NP, Liu Z, Merkens K, Rotfuß S, Peter K, Jungwirth U, Bauer N, Kiefer F, et al. Phenanthroindolizidine Alkaloids Isolated from Tylophora ovata as Potent Inhibitors of Inflammation, Spheroid Growth, and Invasion of Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2022; 23(18):10319. https://doi.org/10.3390/ijms231810319
Chicago/Turabian StyleReimche, Irene, Haiqian Yu, Ni Putu Ariantari, Zhen Liu, Kay Merkens, Stella Rotfuß, Karin Peter, Ute Jungwirth, Nadine Bauer, Friedemann Kiefer, and et al. 2022. "Phenanthroindolizidine Alkaloids Isolated from Tylophora ovata as Potent Inhibitors of Inflammation, Spheroid Growth, and Invasion of Triple-Negative Breast Cancer" International Journal of Molecular Sciences 23, no. 18: 10319. https://doi.org/10.3390/ijms231810319
APA StyleReimche, I., Yu, H., Ariantari, N. P., Liu, Z., Merkens, K., Rotfuß, S., Peter, K., Jungwirth, U., Bauer, N., Kiefer, F., Neudörfl, J.-M., Schmalz, H.-G., Proksch, P., & Teusch, N. (2022). Phenanthroindolizidine Alkaloids Isolated from Tylophora ovata as Potent Inhibitors of Inflammation, Spheroid Growth, and Invasion of Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 23(18), 10319. https://doi.org/10.3390/ijms231810319







