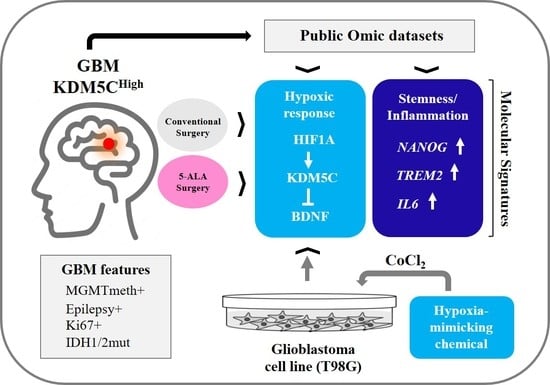
The Chromatin-Oxygen Sensor Gene KDM5C Associates with Novel Hypoxia-Related Signatures in Glioblastoma Multiforme
Abstract
1. Introduction
2. Results
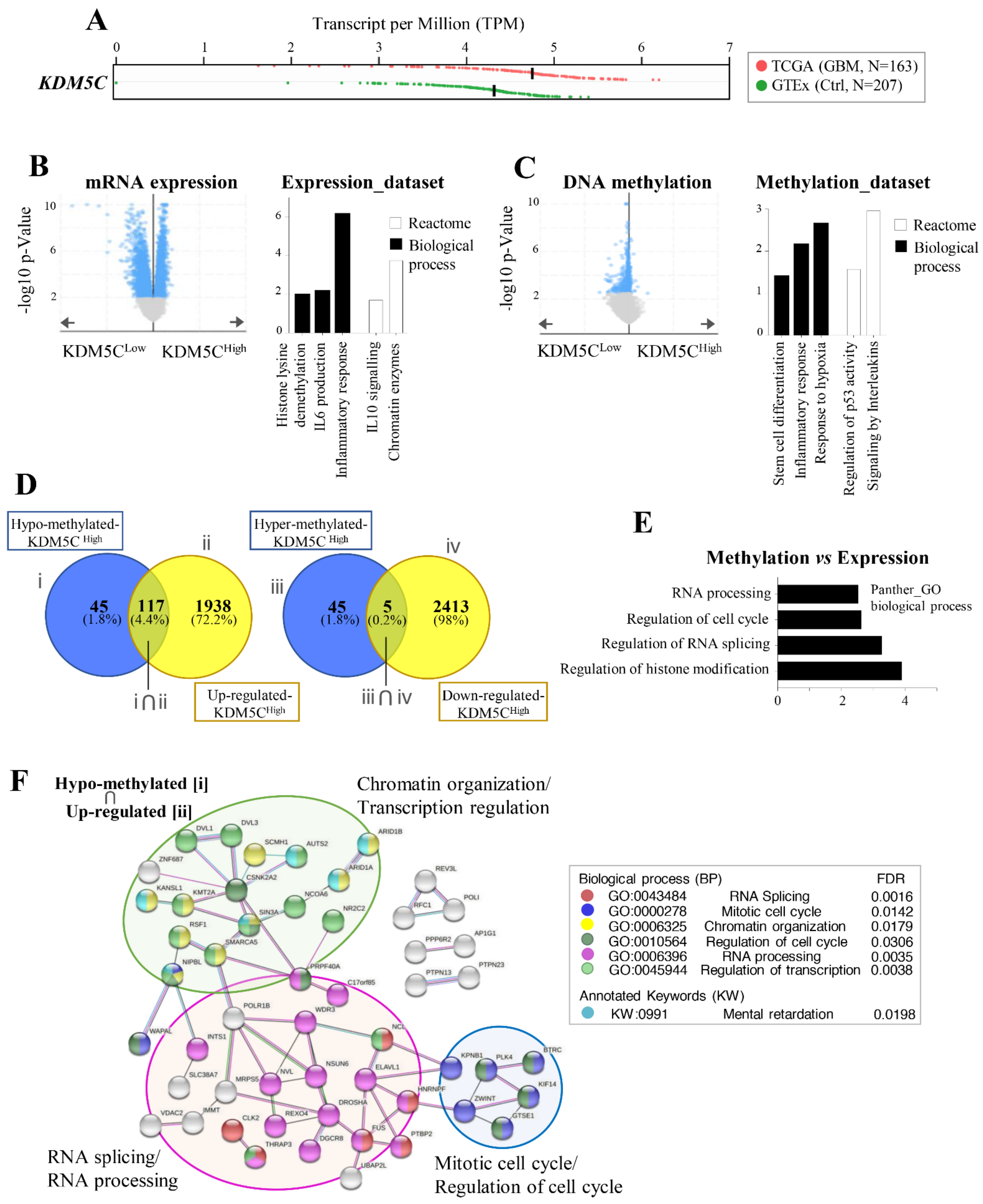
2.1. Stratification of GBM Patients Based on KDM5C Gene Expression Levels
2.2. Hypoxia-Related Signatures Correlate with KDM5C Levels
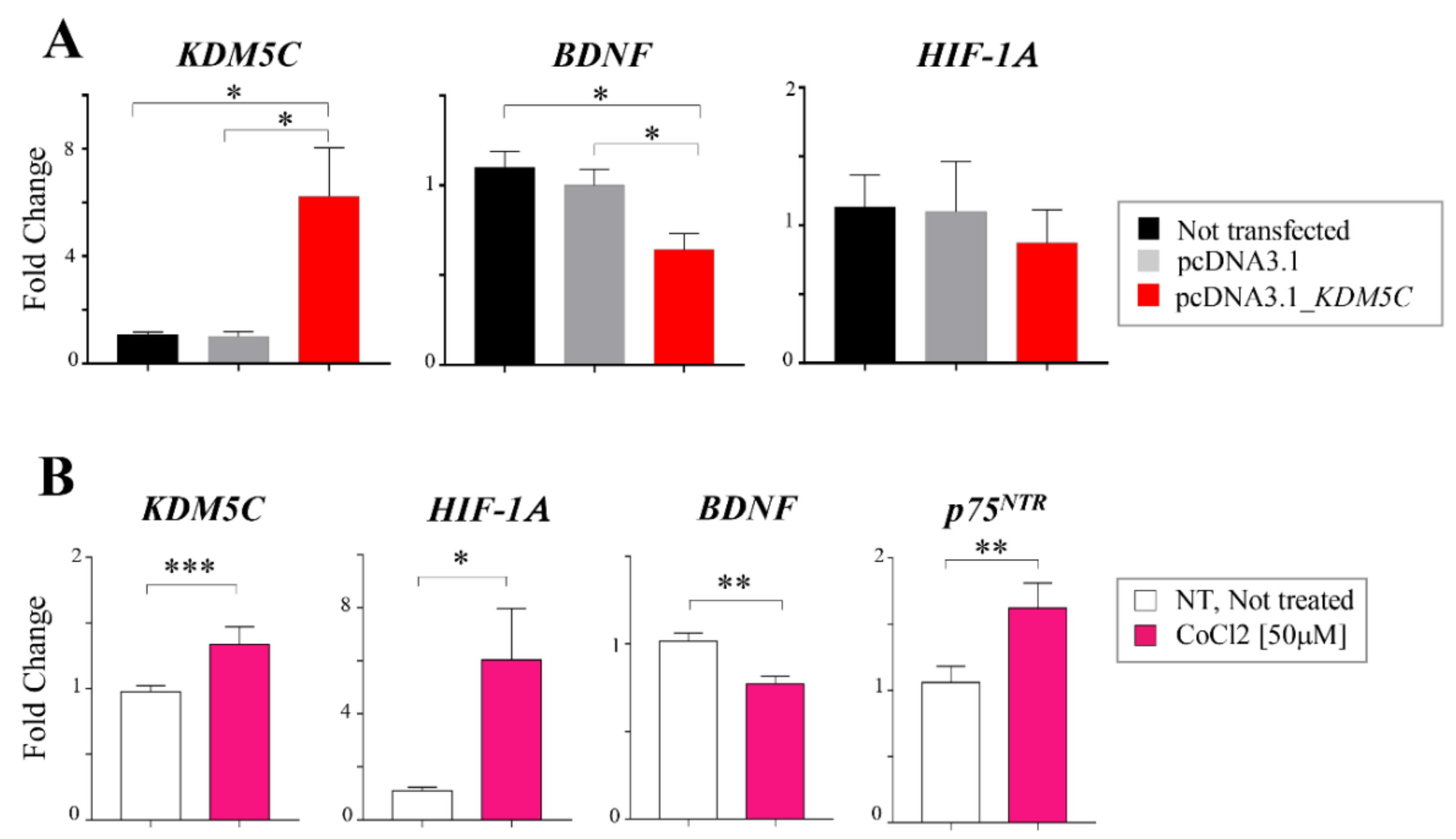
2.3. Hif-1α Stabilization Induces KDM5C Increase and BDNF Repression in T98G Glioblastoma Cell Line
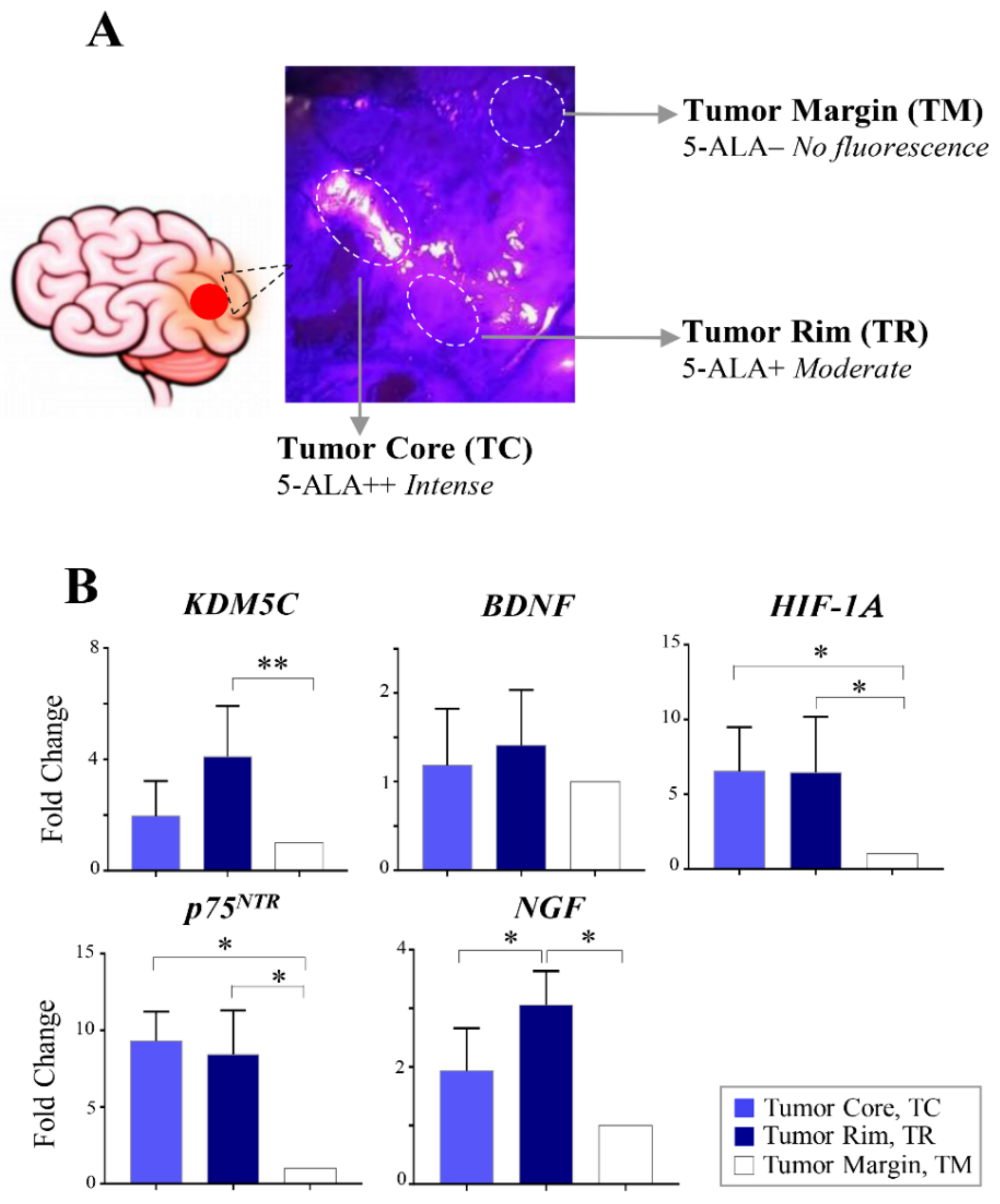
2.4. Regional Heterogeneity of KDM5C and HIF1A Expression Profiles in Distinct GBM Areas Isolated by 5-Aminolevulinic acid Fluorescence-Guided Surgery
2.5. GBMs Expressing High or Low KDM5C Display Markedly Different Expression and Methylation Patterns
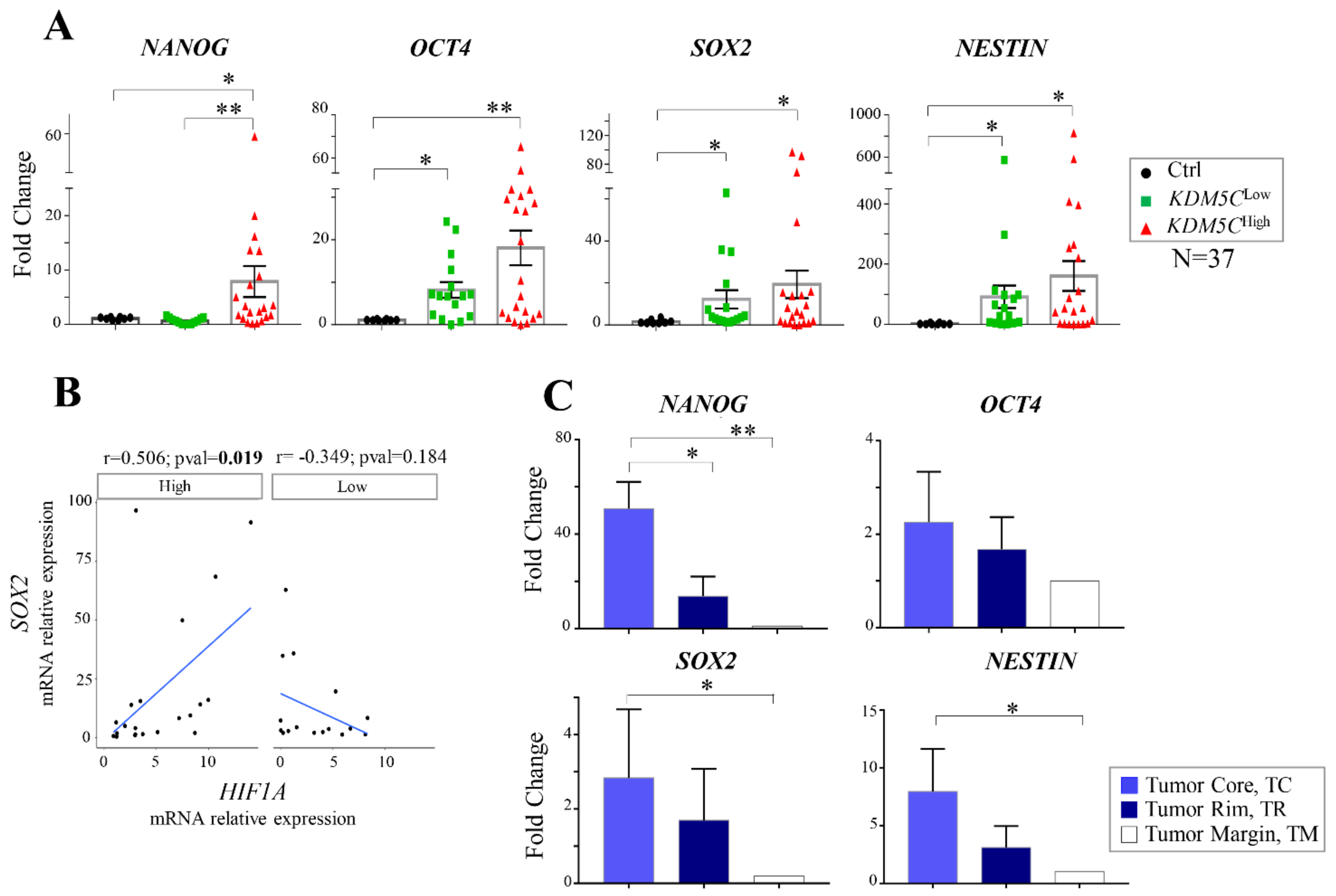
2.6. Analysis of Stem Cell-Associated Genes Expression in the GBM Cohorts
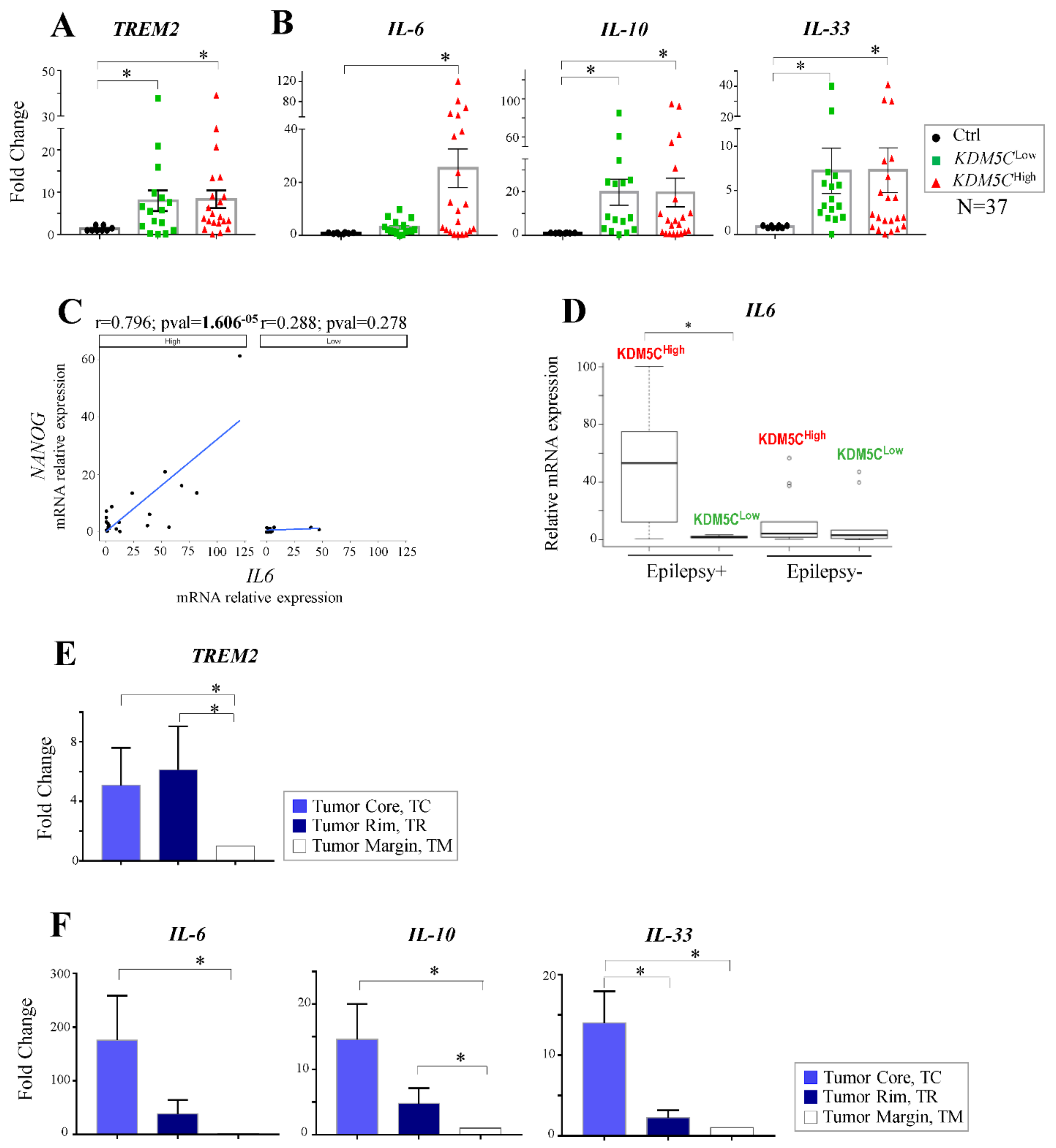
2.7. Expression Profiles of Tumor Progression Markers and Inflammatory Cytokines in GBM Samples
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perry, A.; Wesseling, P. Histologic classification of gliomas. Handb. Clin. Neurol. 2016, 134, 71–95. [Google Scholar] [CrossRef] [PubMed]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Belisario, D.C.; Kopecka, J.; Pasino, M.; Akman, M.; De Smaele, E.; Donadelli, M.; Riganti, C. Hypoxia Dictates Metabolic Rewiring of Tumors: Implications for Chemoresistance. Cells 2020, 9, 2598. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Bastola, S.; Pavlyukov, M.S.; Yamashita, D.; Ghosh, S.; Cho, H.; Kagaya, N.; Zhang, Z.; Minata, M.; Lee, Y.; Sadahiro, H.; et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun. 2020, 11, 4660. [Google Scholar] [CrossRef]
- Ruano, Y.; Mollejo, M.; Ribalta, T.; Fiaño, C.; Camacho, F.I.; Gómez, E.; de Lope, A.R.; Hernández-Moneo, J.L.; Martínez, P.; Meléndez, B. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol. Cancer 2006, 26, 5–39. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Batie, M.; Rocha, S. JmjC histone demethylases act as chromatin oxygen sensors. Mol. Cell. Oncol. 2019, 6, 1608501. [Google Scholar] [CrossRef]
- Xu, L.; Wu, W.; Cheng, G.; Qian, M.; Hu, K.; Yin, G.; Wang, S. Enhancement of Proliferation and Invasion of Gastric Cancer Cell by KDM5C via Decrease in p53 Expression. Technol. Cancer Res. Treat. 2017, 16, 141–149. [Google Scholar] [CrossRef]
- Iwase, S.; Lan, F.; Bayliss, P.; de la Torre-Ubieta, L.; Huarte, M.; Qi, H.H.; Whetstine, J.R.; Bonni, A.; Roberts, T.M.; Shi, Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007, 128, 1077–1088. [Google Scholar] [CrossRef]
- Tahiliani, M.; Mei, P.; Fang, R.; Leonor, T.; Rutenberg, M.; Shimizu, F.; Li, J.; Rao, A.; Shi, Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007, 447, 601–605. [Google Scholar] [CrossRef]
- Højfeldt, J.W.; Agger, K.; Helin, K. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discov. 2013, 12, 917–930. [Google Scholar] [CrossRef]
- Rondinelli, B.; Rosano, D.; Antonini, E.; Frenquelli, M.; Montanini, L.; Huang, D.; Segalla, S.; Yoshihara, K.; Amin, S.B.; Lazarevic, D.; et al. Histone demethylase JARID1C inactivation triggers genomic instability in sporadic renal cancer. J. Clin. Investig. 2016, 126, 4387. [Google Scholar] [CrossRef]
- Varela, I.; Tarpey, P.; Raine, K.; Huang, D.; Ong, C.K.; Stephens, P.; Davies, H.; Jones, D.; Lin, M.L.; Teague, J.; et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011, 469, 539–542. [Google Scholar] [CrossRef]
- Zhan, D.; Zhang, Y.; Xiao, P.; Zheng, X.; Ruan, M.; Zhang, J.; Chen, A.; Zou, Y.; Chen, Y.; Huang, G.; et al. Whole exome sequencing identifies novel mutations of epigenetic regulators in chemorefractory pediatric acute myeloid leukemia. Leuk. Res. 2018, 65, 20–24. [Google Scholar] [CrossRef]
- Lin, H.; Ma, N.; Zhao, L.; Yang, G.; Cao, B. KDM5c Promotes Colon Cancer Cell Proliferation Through the FBXW7-c-Jun Regulatory Axis. Front. Oncol. 2020, 10, 535449. [Google Scholar] [CrossRef]
- DeVaux, R.S.; Herschkowitz, J. Beyond DNA: The role of epigenetics in the premalignant progression of breast cancer. J. Mammary Gland. Biol. Neoplasia 2018, 23, 223–235. [Google Scholar] [CrossRef]
- Chen, X.; Loo, J.X.; Shi, X.; Xiong, W.; Guo, Y.; Ke, H.; Yang, M.; Jiang, Y.; Xia, S.; Zhao, M.; et al. E6 protein expressed by high-risk HPV activates super-enhancers of the EGFR and c-MET oncogenes by destabilizing the histone demethylase KDM5C. Cancer Res. 2018, 78, 1418–1430. [Google Scholar] [CrossRef]
- Stein, J.; Majores, M.; Rohde, M.; Lim, S.; Schneider, S.; Krappe, E.; Ellinger, J.; Dietel, M.; Stephan, C.; Jung, K.; et al. KDM5C is overexpressed in prostate cancer and is a prognostic marker for prostate-specific antigen-relapse following radical prostatectomy. Am. J. Pathol. 2014, 184, 2430–2437. [Google Scholar] [CrossRef]
- Chen, X.J.; Ren, A.Q.; Zheng, L.; Zheng, E.D. Predictive Value of KDM5C Alterations for Immune Checkpoint Inhibitors Treatment Outcomes in Patients with Cancer. Front. Immunol. 2021, 12, 664847. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Zhang, T.; Liao, L.; Zhou, L.; Lindner, D.J.; Zhou, M.; Rini, B.; Yan, Q.; Yang, H. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene 2012, 31, 776–786. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, P.; Zhou, X.; Qiang, Y.; Fan, J.; Lin, Y.; Chen, Y.; Guo, J.; Wang, F.; Xue, H.; et al. Deficiency of the X-inactivation escaping gene KDM5C in clear cell renal cell carcinoma promotes tumorigenicity by reprogramming glycogen metabolism and inhibiting ferroptosis. Theranostics 2021, 11, 8674–8691. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.F.; Zhang, W.J.; Huang, Y.; He, Y.H.; Hu, G.S.; Wang, L.; Peng, B.L.; Yi, J.; Li, T.T.; Rong, R.; et al. The Dual Function of KDM5C in Both Gene Transcriptional Activation and Repression Promotes Breast Cancer Cell Growth and Tumorigenesis. Adv. Sci. 2021, 8, 2004635. [Google Scholar] [CrossRef] [PubMed]
- Outchkourov, N.S.; Muiño, J.M.; Kaufmann, K.; van Ijcken, W.F.; Groot Koerkamp, M.J.; van Leenen, D.; de Graaf, P.; Holstege, F.C.; Grosveld, F.G.; Timmers, H.T. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep. 2013, 3, 1071–1079. [Google Scholar] [CrossRef]
- Vallianatos, C.N.; Farrehi, C.M.; Friez, M.J.; Burmeister, M.; Keegan, C.E.; Iwase, S. Altered gene-regulatory function of KDM5C by a novel mutation associated with autism and intellectual disability. Front. Mol. Neurosci. 2018, 11, 104. [Google Scholar] [CrossRef]
- Xu, J.; Deng, X.; Disteche, C.M. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS ONE 2008, 3, e2553. [Google Scholar] [CrossRef]
- Iwase, S.; Brookes, E.; Agarwal, S.; Badeaux, A.I.; Ito, H.; Vallianatos, C.N.; Tomassy, G.S.; Kasza, T.; Lin, G.; Thompson, A.; et al. A Mouse Model of X-linked Intellectual Disability Associated with Impaired Removal of Histone Methylation. Cell Rep. 2016, 14, 1000–1009. [Google Scholar] [CrossRef]
- Poeta, L.; Fusco, F.; Drongitis, D.; Shoubridge, C.; Manganelli, G.; Filosa, S.; Paciolla, M.; Courtney, M.; Collombat, P.; Lioi, M.B.; et al. A Regulatory Path Associated with X-Linked Intellectual Disability and Epilepsy Links KDM5C to the Polyalanine Expansions in ARX. Am. J. Hum. Genet. 2013, 92, 114–125. [Google Scholar] [CrossRef]
- Poeta, L.; Padula, A.; Attianese, B.; Valentino, M.; Verrillo, L.; Filosa, S.; Shoubridge, C.; Barra, A.; Schwartz, C.E.; Christensen, J.; et al. Histone demethylase KDM5C is a SAHA-sensitive central hub at the crossroads of transcriptional axes involved in multiple neurodevelopmental disorders. Hum. Mol. Genet. 2019, 28, 4089–4102. [Google Scholar] [CrossRef]
- Poeta, L.; Padula, A.; Lioi, M.B.; van Bokhoven, H.; Miano, M.G. Analysis of a Set of KDM5C Regulatory Genes Mutated in Neurodevelopmental Disorders Identifies Temporal Coexpression Brain Signatures. Genes 2021, 12, 1088. [Google Scholar] [CrossRef]
- Hu, X.; Miao, W.; Zou, Y.; Zhang, W.; Zhang, Y.; Liu, H. Expression of p53, epidermal growth factor receptor, Ki-67 and O6-methylguanine-DNA methyltransferase in human gliomas. Oncol. Lett. 2013, 6, 130–134. [Google Scholar] [CrossRef][Green Version]
- Colucci-D’Amato, L.; Speranza, L.; Volpicelli, F. Neurotrophic Factor BDNF, Physiological Functions and Therapeutic Potential in Depression, Neurodegeneration and Brain Cancer. Int. J. Mol. Sci. 2020, 21, 7777. [Google Scholar] [CrossRef]
- Chédeville, A.L.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Radiotherapy Resistance. Cancers 2021, 13, 542. [Google Scholar] [CrossRef]
- Liu, O.H.; Kiema, M.; Beter, M.; Ylä-Herttuala, S.; Laakkonen, J.P.; Kaikkonen, M.U. Hypoxia-Mediated Regulation of Histone Demethylases Affects Angiogenesis-Associated Functions in Endothelial Cells. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2665–2677. [Google Scholar] [CrossRef]
- Peng, X.; Gao, H.; Xu, R.; Wang, H.; Mei, J.; Liu, C. The interplay between HIF-1α and noncoding RNAs in cancer. J. Exp. Clin. Cancer Res. 2020, 39, 27. [Google Scholar] [CrossRef]
- Lo Dico, A.; Martelli, C.; Diceglie, C.; Lucignani, G.; Ottobrini, L. Hypoxia-Inducible Factor-1α Activity as a Switch for Glioblastoma Responsiveness to Temozolomide. Front. Oncol. 2018, 8, 249. [Google Scholar] [CrossRef]
- Kanwar, J.R.; Kamalapuram, S.K.; Kanwar, R.K. Targeting survivin in cancer: The cell-signalling perspective. Drug Discov. Today 2011, 16, 485–494. [Google Scholar] [CrossRef]
- Tong, B.; Pantazopoulou, V.; Johansson, E.; Pietras, A. The p75 neurotrophin receptor enhances HIF-dependent signaling in glioma. Exp. Cell Res. 2018, 371, 122–129. [Google Scholar] [CrossRef]
- Johnston, A.L.; Lun, X.; Rahn, J.J.; Liacini, A.; Wang, L.; Hamilton, M.G.; Parney, I.F.; Hempstead, B.L.; Robbins, S.M.; Forsyth, P.A.; et al. The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biol. 2007, 5, e212. [Google Scholar] [CrossRef]
- Rana, N.K.; Singh, P.; Koch, B. CoCl2 simulated hypoxia induce cell proliferation and alter the expression pattern of hypoxia associated genes involved in angiogenesis and apoptosis. Biol. Res. 2019, 52, 12. [Google Scholar] [CrossRef]
- Kitamuro, T.; Takahashi, K.; Ogawa, K.; Udono-Fujimori, R.; Takeda, K.; Furuyama, K.; Nakayama, M.; Sun, J.; Fujita, H.; Hida, W.; et al. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J. Biol. Chem. 2003, 278, 9125–9133. [Google Scholar] [CrossRef]
- Manini, I.; Caponnetto, F.; Dalla, E.; Ius, T.; Della Pepa, G.M.; Pegolo, E.; Bartolini, A.; La Rocca, G.; Menna, G.; Di Loreto, C.; et al. Heterogeneity Matters: Different Regions of Glioblastoma Are Characterized by Distinctive Tumor-Supporting Pathways. Cancers 2020, 12, 2960. [Google Scholar] [CrossRef]
- Yagi, R.; Kawabata, S.; Ikeda, N.; Nonoguchi, N.; Furuse, M.; Katayama, Y.; Kajimoto, Y.; Kuroiwa, T. Intraoperative 5-aminolevulinic acid-induced photodynamic diagnosis of metastatic brain tumors with histopathological analysis. World J. Surg. Oncol. 2017, 15, 179. [Google Scholar] [CrossRef]
- Kim, J.H.; Jeong, K.; Li, J.; Murphy, J.M.; Vukadin, L.; Stone, J.K.; Richard, A.; Tran, J.; Gillespie, G.Y.; Flemington, E.K.; et al. SON drives oncogenic RNA splicing in glioblastoma by regulating PTBP1/PTBP2 switching and RBFOX2 activity. Nat. Commun. 2021, 12, 5551. [Google Scholar] [CrossRef]
- Awah, C.U.; Winter, J.; Mazdoom, C.M.; Ogunwobi, O.O. NSUN6, an RNA methyltransferase of 5-mC controls glioblastoma response to temozolomide (TMZ) via NELFB and RPS6KB2 interaction. Cancer Biol. Ther. 2021, 22, 587–597. [Google Scholar] [CrossRef]
- Kalscheuer, V.M.; FitzPatrick, D.; Tommerup, N.; Bugge, M.; Niebuhr, E.; Neumann, L.M.; Tzschach, A.; Shoichet, S.A.; Menzel, C.; Erdogan, F.; et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum. Genet. 2007, 121, 501–509. [Google Scholar] [CrossRef]
- Li, D.; Wang, Q.; Gong, N.N.; Kurolap, A.; Feldman, H.B.; Boy, N.; Brugger, M.; Grand, K.; McWalter, K.; Guillen Sacoto, M.J.; et al. Pathogenic variants in SMARCA5, a chromatin remodeler, cause a range of syndromic neurodevelopmental features. Sci. Adv. 2021, 7, eabf2066. [Google Scholar] [CrossRef] [PubMed]
- Zollino, M.; Marangi, G.; Ponzi, E.; Orteschi, D.; Ricciardi, S.; Lattante, S.; Murdolo, M.; Battaglia, D.; Contaldo, I.; Mercuri, E.; et al. Intragenic KANSL1 mutations and chromosome 17q21.31 deletions: Broadening the clinical spectrum and genotype-phenotype correlations in a large cohort of patients. J. Med. Genet. 2015, 52, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.D.; Dafou, D.; McEntagart, M.; Woollard, W.J.; Elmslie, F.V.; Holder-Espinasse, M.; Irving, M.; Saggar, A.K.; Smithson, S.; Trembath, R.C.; et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 2012, 91, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.L.V.; Gomes, I.N.F.; Carloni, A.C.; Rosa, M.N.; da Silva, L.S.; Evangelista, A.F.; Reis, R.M.; Silva, V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 2021, 12, 206. [Google Scholar] [CrossRef] [PubMed]
- Prager, B.C.; Bhargava, S.; Mahadev, V.; Hubert, C.G.; Rich, J.N. Glioblastoma Stem Cells: Driving Resilience through Chaos. Trends Cancer 2020, 6, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Bae, J.M.; Wen, X.; Jung, S.; Kim, Y.; Kim, K.J.; Cho, N.Y.; Kim, J.H.; Han, S.W.; Kim, T.Y.; et al. p53 expression status is associated with cancer-specific survival in stage III and high-risk stage II colorectal cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Br. J. Cancer 2019, 120, 797–805. [Google Scholar] [CrossRef]
- Molgora, M.; Esaulova, E.; Vermi, W.; Hou, J.; Chen, Y.; Luo, J.; Brioschi, S.; Bugatti, M.; Omodei, A.S.; Ricci, B.; et al. TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 2020, 182, 886–900. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Shao, Z.; Wen, X.; Jiang, J.; Ma, Q.; Wang, Y.; Huang, L.; Ding, X.; Zhang, L. TREM2: Keeping Pace With Immune Checkpoint Inhibitors in Cancer Immunotherapy. Front. Immunol. 2021, 12, 716710. [Google Scholar] [CrossRef]
- West, A.J.; Tsui, V.; Stylli, S.S.; Nguyen, H.; Morokoff, A.P.; Kaye, A.H.; Luwor, R.B. The role of interleukin-6-STAT3 signalling in glioblastoma. Oncol. Lett. 2018, 16, 4095–4104. [Google Scholar] [CrossRef] [PubMed]
- Widodo, S.S.; Dinevska, M.; Furst, L.M.; Stylli, S.S.; Mantamadiotis, T. IL-10 in glioma. Br. J. Cancer 2021, 125, 1466–1476. [Google Scholar] [CrossRef]
- De Boeck, A.; Ahn, B.Y.; D’Mello, C.; Lun, X.; Menon, S.V.; Alshehri, M.M.; Szulzewsky, F.; Shen, Y.; Khan, L.; Dang, N.H.; et al. Glioma-derived IL-33 orchestrates an inflammatory brain tumor microenvironment that accelerates glioma progression. Nat. Commun. 2020, 11, 4997. [Google Scholar] [CrossRef]
- Tong, X.; Yang, P.; Wang, K.; Liu, Y.; Liu, X.; Shan, X.; Huang, R.; Zhang, K.; Wang, J. Survivin is a prognostic indicator in glioblastoma and may be a target of microRNA-218. Oncol. Lett. 2019, 18, 359–367. [Google Scholar] [CrossRef]
- Kumar, R.; Jain, V.; Kushwah, N.; Dheer, A.; Mishra, K.P.; Prasad, D.; Singh, S.B. Role of DNA Methylation in Hypobaric Hypoxia-Induced Neurodegeneration and Spatial Memory Impairment. Ann. Neurosci. 2018, 4, 191–200. [Google Scholar] [CrossRef]
- Muthukrishnan, S.D.; Alvarado, A.G.; Kornblum, H.I. Building Bonds: Cancer Stem Cells Depend on Their Progeny to Drive Tumor Progression. Cell Stem Cell 2018, 4, 473–474. [Google Scholar] [CrossRef]
- Tafani, M.; Di Vito, M.; Frati, A.; Pellegrini, L.; De Santis, E.; Sette, G.; Eramo, A.; Sale, P.; Mari, E.; Santoro, A.; et al. Pro-inflammatory gene expression in solid glioblastoma microenvironment and in hypoxic stem cells from human glioblastoma. J. Neuroinflamm. 2011, 8, 32. [Google Scholar] [CrossRef]
- Lu, H.; Lyu, Y.; Tran, L.; Lan, J.; Xie, Y.; Yang, Y.; Murugan, N.L.; Wang, Y.J.; Semenza, G.L. HIF-1 recruits NANOG as a coactivator for TERT gene transcription in hypoxic breast cancer stem cells. Cell Rep. 2021, 36, 109757. [Google Scholar] [CrossRef]
- Castillo-Rodríguez, R.A.; Trejo-Solís, C.; Cabrera-Cano, A.; Gómez-Manzo, S.; Dávila-Borja, V.M. Hypoxia as a Modulator of Inflammation and Immune Response in Cancer. Cancers 2022, 14, 2291. [Google Scholar] [CrossRef]
- Yan, S.F.; Tritto, I.; Pinsky, D.; Liao, H.; Huang, J.; Fuller, G.; Brett, J.; May, L.; Stern, D. Induction of interleukin 6 (IL-6) by hypoxia in vascular cells. Central role of the binding site for nuclear factor-IL-6. J. Biol. Chem. 1995, 270, 11463–11471. [Google Scholar] [CrossRef]
- Radharani, N.N.V.; Yadav, A.S.; Nimma, R.; Kumar, T.; Bulbule, A.; Chanukuppa, V.; Kumar, D.; Patnaik, S.; Rapole, S.; Kundu, G.C. Tumor-associated macrophage derived IL-6 enriches cancer stem cell population and promotes breast tumor progression via Stat-3 pathway. Cancer Cell Int. 2022, 22, 122. [Google Scholar] [CrossRef]
- Wang, T.; Song, P.; Zhong, T.; Wang, X.; Xiang, X.; Liu, Q.; Chen, H.; Xia, T.; Liu, H.; Niu, Y.; et al. The inflammatory cytokine IL-6 induces FRA1 deacetylation promoting colorectal cancer stem-like properties. Oncogene 2019, 38, 4932–4947. [Google Scholar] [CrossRef]
- Deng, L.; Zhang, X.; Xiang, X.; Xiong, R.; Xiao, D.; Chen, Z.; Liu, K.; Feng, G. NANOG Promotes Cell Proliferation, Invasion, and Stemness via IL-6/STAT3 Signaling in Esophageal Squamous Carcinoma. Technol. Cancer Res. Treat. 2021, 20, 1–9. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, X.; Nie, K.; Cheng, L.; Zhang, Z.; Hu, Y.; Peng, W. Systematic Pan-Cancer Analysis Identifies TREM2 as an Immunological and Prognostic Biomarker. Front. Immunol. 2021, 12, 646523. [Google Scholar] [CrossRef]
- Jin, Y.; Xiao, W.; Song, T.; Feng, G.; Dai, Z. Expression and Prognostic Significance of p53 in Glioma Patients: A Meta-analysis. Neurochem. Res. 2016, 41, 1723–1731. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, Y.S.; Kang, K.N.; Kim, K.H.; Park, Y.J.; Kim, C.W. Multiple microRNAs as biomarkers for early breast cancer diagnosis. Mol. Clin. Oncol. 2021, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xiang, W.; Yanhui, L.; Ruofei, L.; Shuang, L.; Yingjun, F.; Qiao, Z.; Yanwu, Y.; Qing, M. Ki-67 overexpression in WHO grade II gliomas is associated with poor postoperative seizure control. Seizure 2013, 22, 877–881. [Google Scholar] [CrossRef] [PubMed]
- You, G.; Sha, Z.; Jiang, T. The pathogenesis of tumor-related epilepsy and its implications for clinical treatment. Seizure 2012, 21, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Kleihues, P.; Cavenee, W.K. Pathology and Genetics of Tumours of the Nervous System; International Agency for Research on Cancer (IARC) Press: Lyon, France, 2000. [Google Scholar]
- Hadjipanayis, C.G.; Stummer, W. 5-ALA and FDA approval for glioma surgery. J. Neurooncol. 2019, 141, 479–486. [Google Scholar] [CrossRef]
- Li, N.; Carrel, L. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc. Natl. Acad. Sci. USA 2008, 105, 17055–17060. [Google Scholar] [CrossRef]
- Kruidenier, L.; Chung, C.W.; Cheng, Z.; Liddle, J.; Che, K.; Joberty, G.; Bantscheff, M.; Bountra, C.; Bridges, A.; Diallo, H.; et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012, 488, 404–408. [Google Scholar] [CrossRef]
- Hashizume, R.; Andor, N.; Ihara, Y.; Lerner, R.; Gan, H.; Chen, X.; Fang, D.; Huang, X.; Tom, M.W.; Ngo, V.; et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat. Med. 2014, 12, 1394–1396. [Google Scholar] [CrossRef]
- Sui, A.; Xu, Y.; Li, Y.; Hu, Q.; Wang, Z.; Zhang, H.; Yang, J.; Guo, X.; Zhao, W. The pharmacological role of histone demethylase JMJD3 inhibitor GSK-J4 on glioma cells. Oncotarget 2017, 40, 68591–68598. [Google Scholar] [CrossRef]


| Clinicopathological Parameters | No. of Patients (Frequency) | |
|---|---|---|
| Gender | Male | 20 (54%) |
| Female | 17 (46%) | |
| Epilepsy | Yes | 13 (35%) |
| No | 24 (65%) | |
| Location of lesion | Parietal | 4 (10.8 %) |
| Frontal | 7 (18.9%) | |
| Temporal | 5 (13.6%) | |
| Rolandic | 3 (8.1%) | |
| Corpus Callosum | 2 (5.4%) | |
| Frontotemporal | 4 (10.8%) | |
| Temporo-Parietal | 3 (8.1%) | |
| Frontoparietal | 3 (8.1%) | |
| Parieto-Occipital | 2 (5.4%) | |
| Others | 4 (10.8%) | |
| Lobe localization | Right hemisphere | 17 (45.9%) |
| Left hemisphere | 17 (45.9%) | |
| Others | 3 (8.2%) | |
| Lesion number | Single | 33 (89%) |
| Multifocal | 4 (11%) | |
| Surgical approach | Total resection with standard method | 25 (68%) |
| Total resection with 5-ALA | 9 (24%) | |
| Biopsy | 3 (8%) | |
| Radiotherapy | Yes | 28 (75.6%) |
| No | 9 (24.4%) | |
| Chemotherapy | Yes | 34 (92%) |
| No | 3 (8%) | |
| MGMT methylation | Yes | 33 (89.2%) |
| No | 4 (10.8%) | |
| IDH1/2 mutation | WT | 14 (37.8%) |
| MUT | 23 (62.2%) | |
| p53 expression | Yes | 32 (86.5%) |
| No | 5 (13.5%) | |
| Ki67 expression | Yes | 37 (100%) |
| Survival | -- | 17 (46%) |
| Mortality rate | -- | 20 (54%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drongitis, D.; Verrillo, L.; De Marinis, P.; Orabona, P.; Caiola, A.; Turitto, G.; Alfieri, A.; Bruscella, S.; Gentile, M.; Moriello, V.; et al. The Chromatin-Oxygen Sensor Gene KDM5C Associates with Novel Hypoxia-Related Signatures in Glioblastoma Multiforme. Int. J. Mol. Sci. 2022, 23, 10250. https://doi.org/10.3390/ijms231810250
Drongitis D, Verrillo L, De Marinis P, Orabona P, Caiola A, Turitto G, Alfieri A, Bruscella S, Gentile M, Moriello V, et al. The Chromatin-Oxygen Sensor Gene KDM5C Associates with Novel Hypoxia-Related Signatures in Glioblastoma Multiforme. International Journal of Molecular Sciences. 2022; 23(18):10250. https://doi.org/10.3390/ijms231810250
Chicago/Turabian StyleDrongitis, Denise, Lucia Verrillo, Pasqualino De Marinis, Pasquale Orabona, Agnese Caiola, Giacinto Turitto, Alessandra Alfieri, Sara Bruscella, Marisa Gentile, Vania Moriello, and et al. 2022. "The Chromatin-Oxygen Sensor Gene KDM5C Associates with Novel Hypoxia-Related Signatures in Glioblastoma Multiforme" International Journal of Molecular Sciences 23, no. 18: 10250. https://doi.org/10.3390/ijms231810250
APA StyleDrongitis, D., Verrillo, L., De Marinis, P., Orabona, P., Caiola, A., Turitto, G., Alfieri, A., Bruscella, S., Gentile, M., Moriello, V., Sannino, E., Di Muccio, I., Costa, V., Miano, M. G., & de Bellis, A. (2022). The Chromatin-Oxygen Sensor Gene KDM5C Associates with Novel Hypoxia-Related Signatures in Glioblastoma Multiforme. International Journal of Molecular Sciences, 23(18), 10250. https://doi.org/10.3390/ijms231810250