Exploring the Healthy Eye Microbiota Niche in a Multicenter Study
Abstract
:1. Introduction
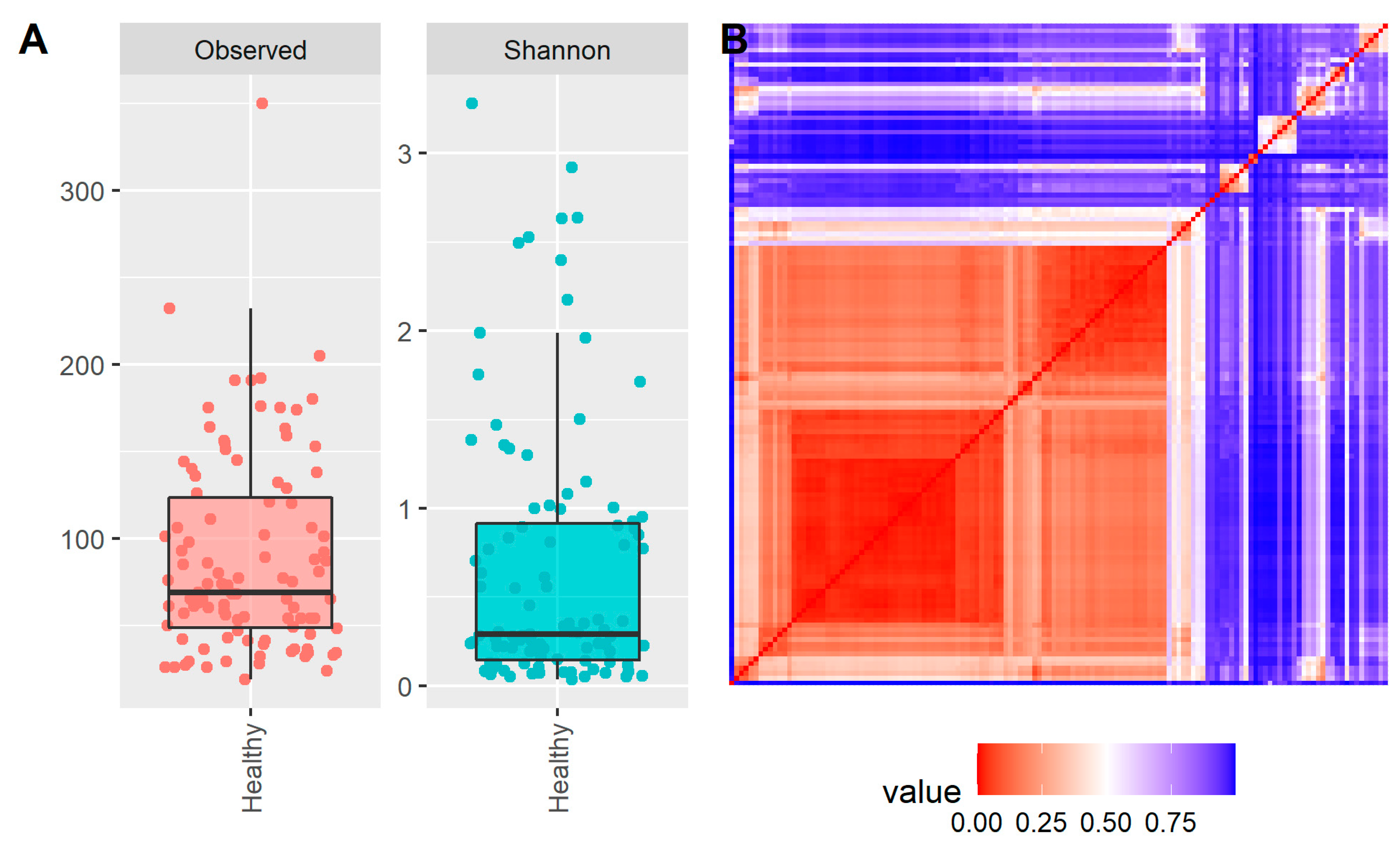
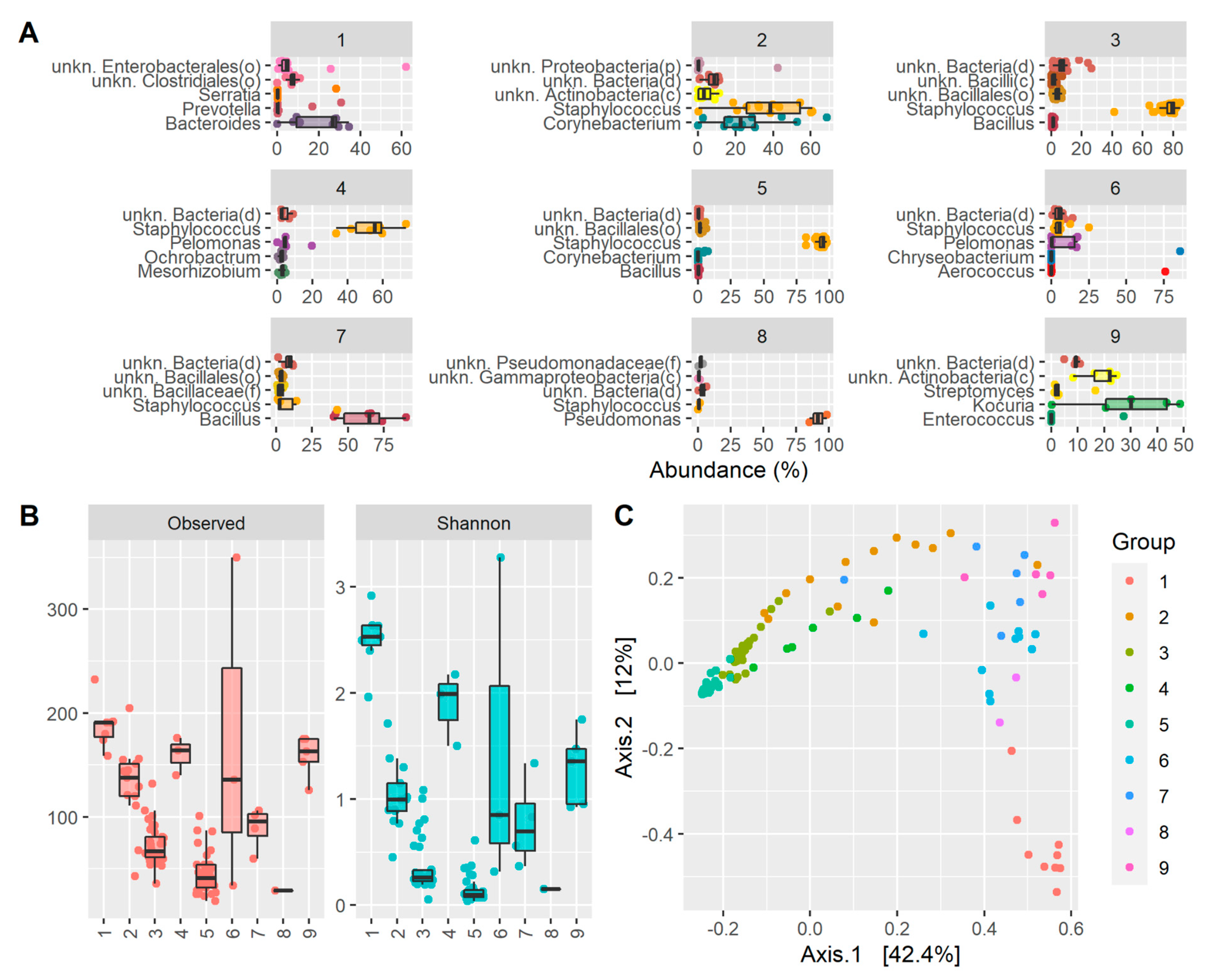
2. Results
3. Discussion
4. Materials and Methods
4.1. Recruitment of Subjects
4.2. Sampling Technique, DNA Extraction, PCR Amplification, Library Preparation, and Amplicon Sequencing
4.3. Data Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shestopalov, V.I.; Antonopoulos, D.A.; Miller, D.; Iovieno, A.; Brulc, J.M.; Bates, B.; Garoutte, A.; Domanus, M.; Ivanov, D.V.; Slepak, V. Metagenomic analysis of bacterial community at the human conjunctiva. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2409. [Google Scholar]
- Aragona, P.; Baudouin, C.; Del Castillo, J.M.B.; Messmer, E.; Barabino, S.; Merayo-Lloves, J.; Brignole-Baudouin, F.; Inferrera, L.; Rolando, M.; Mencucci, R.; et al. The ocular microbiome and microbiota and their effects on ocular surface pathophysiology and disorders. Surv. Ophthalmol. 2021, 66, 907–925. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Yi, S.; Wei, L. Ocular microbiota and intraocular inflammation. Front. Immunol. 2020, 11, 609765. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.Á.P.; Frizon, L.; Demeda, V.F. Ocular surface microbiome in health and disease. Asia Pac. J. Ophthalmol. 2020, 9, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Ung, L.; Bispo, P.J.M.; Shanbhag, S.S.; Gilmore, M.S.; Chodosh, J. The persistent dilemma of microbial keratitis: Global burden, diagnosis, and antimicrobial resistance. Surv. Ophthalmol. 2019, 64, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Taravati, P.; Lam, D.; Van Gelder, R.N. Role of molecular diagnostics in ocular microbiology. Curr. Ophthalmol. Rep. 2013, 1, 181–189. [Google Scholar] [CrossRef]
- Roy, S.; LaFramboise, W.A.; Nikiforov, Y.E.; Nikiforova, M.N.; Routbort, M.J.; Pfeifer, J.; Nagarajan, R.; Carter, A.B.; Pantanowitz, L. Next-Generation sequencing informatics: Challenges and strategies for implementation in a clinical environment. Arch. Pathol. Lab. Med. 2016, 140, 958–975. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Schloss, P.D.; Handelsman, J. Metagenomics: Genomic analysis of microbial communities. Annu. Rev. Genet. 2004, 38, 525–552. [Google Scholar] [CrossRef]
- Tringe, S.G.; Hugenholtz, P. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 2008, 11, 442–446. [Google Scholar] [CrossRef]
- Parekh, M.; Borroni, D.; Romano, V.; Kaye, S.B.; Camposampiero, D.; Ponzin, D.; Ferrari, S. Next-generation sequencing for the detection of microorganisms present in human donor corneal preservation medium. BMJ Open Ophthalmol. 2019, 4, e000246. [Google Scholar] [CrossRef]
- Cole, J.R.; Chai, B.; Marsh, T.L.; Farris, R.J.; Wang, Q.; Kulam, S.A.; Chandra, S.; McGarrell, D.M.; Schmidt, T.M.; Garrity, G.M.; et al. The ribosomal database project (RDP-II): Previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003, 31, 442–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar, A.; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Li, J.J.; Zou, Y.; Zou, B.; Wei, L. Microbiota and ocular diseases. Front. Cell. Infect. Microbiol. 2021, 11, 759333. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, J.; Nielsen, S.; Diez-Vives, C.; Coroneo, M.; Thomas, T.; Willcox, M. Temporal stability and composition of the ocular surface microbiome. Sci. Rep. 2017, 7, 9880. [Google Scholar] [CrossRef]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of bacteria at healthy human conjunctiva. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5408–5413. [Google Scholar] [CrossRef]
- Miller, D.; Iovieno, A. The role of microbial flora on the ocular surface. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 466–470. [Google Scholar] [CrossRef]
- Borroni, D.; Romano, V.; Kaye, S.B.; Somerville, T.; Napoli, L.; Fasolo, A.; Gallon, P.; Ponzin, D.; Esposito, A.; Ferrari, S. Metagenomics in ophthalmology: Current findings and future prospectives. BMJ Open Ophthalmol. 2019, 4, e000248. [Google Scholar] [CrossRef]
- Ozkan, J.; Willcox, M.D. The ocular microbiome: Molecular characterisation of a unique and low microbial environment. Curr. Eye Res. 2019, 44, 685–694. [Google Scholar] [CrossRef]
- Okonkwo, A.; Rimmer, V.; Walkden, A.; Brahma, A.; Carley, F.; McBain, A.J.; Radhakrishnan, H. Next-Generation sequencing of the ocular surface microbiome: In health, contact lens wear, diabetes, trachoma, and dry eye. Eye Contact Lens 2020, 46, 254–261. [Google Scholar] [CrossRef]
- Kugadas, A.; Wright, Q.; Geddes-McAlister, J.; Gadjeva, M. Role of microbiota in strengthening ocular mucosal barrier function through secretory IgA. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4593–4600. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sheppard, J.D. Gut microbiome and its influence on ocular surface and ocular surface diseases. Eye Contact Lens 2022, 48, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Zilliox, M.J.; Gange, W.S.; Kuffel, G.; Mores, C.R.; Joyce, C.; de Bustros, P.; Bouchard, C.S. Assessing the ocular surface microbiome in severe ocular surface diseases. Ocul. Surf. 2020, 18, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, D.; Ge, C.; Zhang, L.; Reinach, P.S.; Tian, X.; Tao, C.; Zhao, Z.; Zhao, C.; Fu, W.; et al. Metagenomic profiling of ocular surface microbiome changes in meibomian gland dysfunction. Investig. Ophthalmol. Vis. Sci. 2020, 61, 22. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J.; Vogt, J.K.; Dalgaard, M.D.; Pedersen, O.; Holmgaard, K.; Heegaard, S. Ocular surface microbiota in patients with aqueous tear-deficient dry eye. Ocul. Surf. 2021, 19, 210–217. [Google Scholar] [CrossRef]
- Kang, Y.; Tian, L.; Gu, X.; Chen, Y.; Ma, X.; Lin, S.; Li, Z.; Lou, Y.; Zheng, M. Characterization of the Ocular surface microbiome in keratitis patients after repeated ophthalmic antibiotic exposure. Microbiol. Spectr. 2022, 10, e0216221. [Google Scholar] [CrossRef]
- Shin, H.; Price, K.; Albert, L.; Dodick, J.; Park, L.; Dominguez-Bello, M.G. Changes in the eye microbiota associated with contact lens wearing. mBio 2016, 7, e00198. [Google Scholar] [CrossRef]
- Boost, M.; Cho, P.; Wang, Z. Disturbing the balance: Effect of contact lens use on the ocular proteome and microbiome. Clin. Exp. Optom. 2017, 100, 459–472. [Google Scholar] [CrossRef]
- Udomwech, L.; Karnjana, K.; Jewboonchu, J.; Rattanathamma, P.; Narkkul, U.; Juhong, J.; Mordmuang, A. Bacterial microbiota of the contact lens surface and associated care behaviours. Heliyon 2022, 8, e09038. [Google Scholar] [CrossRef]
- Andersson, J.; Vogt, J.K.; Dalgaard, M.D.; Pedersen, O.; Holmgaard, K.; Heegaard, S. Ocular surface microbiota in contact lens users and contact-lens-associated bacterial keratitis. Vision 2021, 5, 27. [Google Scholar] [CrossRef]
- Stapleton, F.; Shrestha, G.S.; Vijay, A.K.; Carnt, N. Epidemiology, microbiology, and genetics of contact lens-related and non-contact lens-related infectious keratitis. Eye Contact Lens 2022, 48, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar Moeller, C.T.; Branco, B.C.; Yu, M.C.Z.; Farah, M.E.; Santos, M.A.; Höfling-Lima, A.L. Evaluation of normal ocular bacterial flora with two different culture media. Can. J. Ophthalmol. 2005, 40, 448–453. [Google Scholar] [CrossRef]
- Kittipibul, T.; Puangsricharern, V.; Chatsuwan, T. Comparison of the ocular microbiome between chronic Stevens-Johnson syndrome patients and healthy subjects. Sci. Rep. 2020, 10, 4353. [Google Scholar] [CrossRef] [PubMed]
- Delbeke, H.; Younas, S.; Casteels, I.; Joossens, M. Current knowledge on the human eye microbiome: A systematic review of available amplicon and metagenomic sequencing data. Acta Ophthalmol. 2021, 99, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.E.; Moore, J.E.; Jiru, X.; Moore, J.E.; Goodall, E.A.; Dooley, J.S.G.; Hayes, V.E.A.; Dartt, D.A.; Downes, C.S.; Moore, T.C.B. Ocular pathogen or commensal: A PCR-based study of surface bacterial flora in normal and dry eyes. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5616–5623. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.; Akileswaran, L.; Andersen, D.; Johnson, B.; Ko, N.; Shrestha, A.; Shestopalov, V.; Lee, C.S.; Lee, A.Y.; Van Gelder, R.N. Paucibacterial microbiome and resident DNA virome of the healthy conjunctiva. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5116–5126. [Google Scholar] [CrossRef]
- Zhou, Y.; Holland, M.J.; Makalo, P.; Joof, H.; Mabey, D.C.; Bailey, R.L.; Burton, M.J.; Weinstock, G.M.; Burr, S.E. The conjunctival microbiome in health and trachomatous disease: A case control study. Genome Med. 2014, 6, 99. [Google Scholar] [CrossRef]
- Mitreva, M. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Li, K.; Bihan, M.; Yooseph, S.; Methé, B.A. Analyses of the microbial diversity across the human microbiome. PLoS ONE 2012, 7, e32118. [Google Scholar] [CrossRef]
- Cavuoto, K.M.; Galor, A.; Banerjee, S. Anatomic characterization of the ocular surface microbiome in children. Microorganisms 2019, 7, 259. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, D.H.; Jung, J.Y.; Kim, J.C.; Jeon, C.O. Comparative ocular microbial communities in humans with and without blepharitis. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5585–5593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ham, B.; Hwang, H.B.; Jung, S.H.; Chang, S.; Kang, K.D.; Kwon, M.J. Distribution and diversity of ocular microbial communities in diabetic patients compared with healthy subjects. Curr. Eye Res. 2018, 43, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Shalabi, N.M.; Tuzhikov, A.I.; Galor, A.; Dong, Q.; Panchin, A.U.; Thanathanee, O.; Van Gelder, R.; O’Brien, T.P.; Shestopalov, V. Experimental approaches in the analysis of microbial community in ocular samples. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1976. [Google Scholar]
- Cavuoto, K.M.; Mendez, R.; Miller, D.; Galor, A.; Banerjee, S. Effect of clinical parameters on the ocular surface microbiome in children and adults. Clin. Ophthalmol. 2018, 12, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Cavuoto, K.M.; Banerjee, S.; Miller, D.; Galor, A. Composition and comparison of the ocular surface microbiome in infants and older children. Transl. Vis. Sci. Technol. 2018, 7, 16. [Google Scholar] [CrossRef]
- Zhong, Y.; Fang, X.; Wang, X.; Lin, Y.-A.; Wu, H.; Li, C. Effects of sodium hyaluronate eye drops with or without preservatives on ocular surface bacterial microbiota. Front. Med. 2022, 9, 793565. [Google Scholar] [CrossRef]
- Paytuví, A.; Battista, E.; Scippacercola, F.; Aiese Cigliano, R.; Sanseverino, W. GAIA: An integrated metagenomics suite. bioRxiv 2019. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borroni, D.; Paytuví-Gallart, A.; Sanseverino, W.; Gómez-Huertas, C.; Bonci, P.; Romano, V.; Giannaccare, G.; Rechichi, M.; Meduri, A.; Oliverio, G.W.; et al. Exploring the Healthy Eye Microbiota Niche in a Multicenter Study. Int. J. Mol. Sci. 2022, 23, 10229. https://doi.org/10.3390/ijms231810229
Borroni D, Paytuví-Gallart A, Sanseverino W, Gómez-Huertas C, Bonci P, Romano V, Giannaccare G, Rechichi M, Meduri A, Oliverio GW, et al. Exploring the Healthy Eye Microbiota Niche in a Multicenter Study. International Journal of Molecular Sciences. 2022; 23(18):10229. https://doi.org/10.3390/ijms231810229
Chicago/Turabian StyleBorroni, Davide, Andreu Paytuví-Gallart, Walter Sanseverino, Carmen Gómez-Huertas, Paola Bonci, Vito Romano, Giuseppe Giannaccare, Miguel Rechichi, Alessandro Meduri, Giovanni William Oliverio, and et al. 2022. "Exploring the Healthy Eye Microbiota Niche in a Multicenter Study" International Journal of Molecular Sciences 23, no. 18: 10229. https://doi.org/10.3390/ijms231810229
APA StyleBorroni, D., Paytuví-Gallart, A., Sanseverino, W., Gómez-Huertas, C., Bonci, P., Romano, V., Giannaccare, G., Rechichi, M., Meduri, A., Oliverio, G. W., Rocha-de-Lossada, C., & on behalf of LUCY Consortium. (2022). Exploring the Healthy Eye Microbiota Niche in a Multicenter Study. International Journal of Molecular Sciences, 23(18), 10229. https://doi.org/10.3390/ijms231810229









