Bisdemethoxycurcumin (BDC)-Loaded H-Ferritin-Nanocages Mediate the Regulation of Inflammation in Alzheimer’s Disease Patients
Abstract
:1. Introduction
2. Results
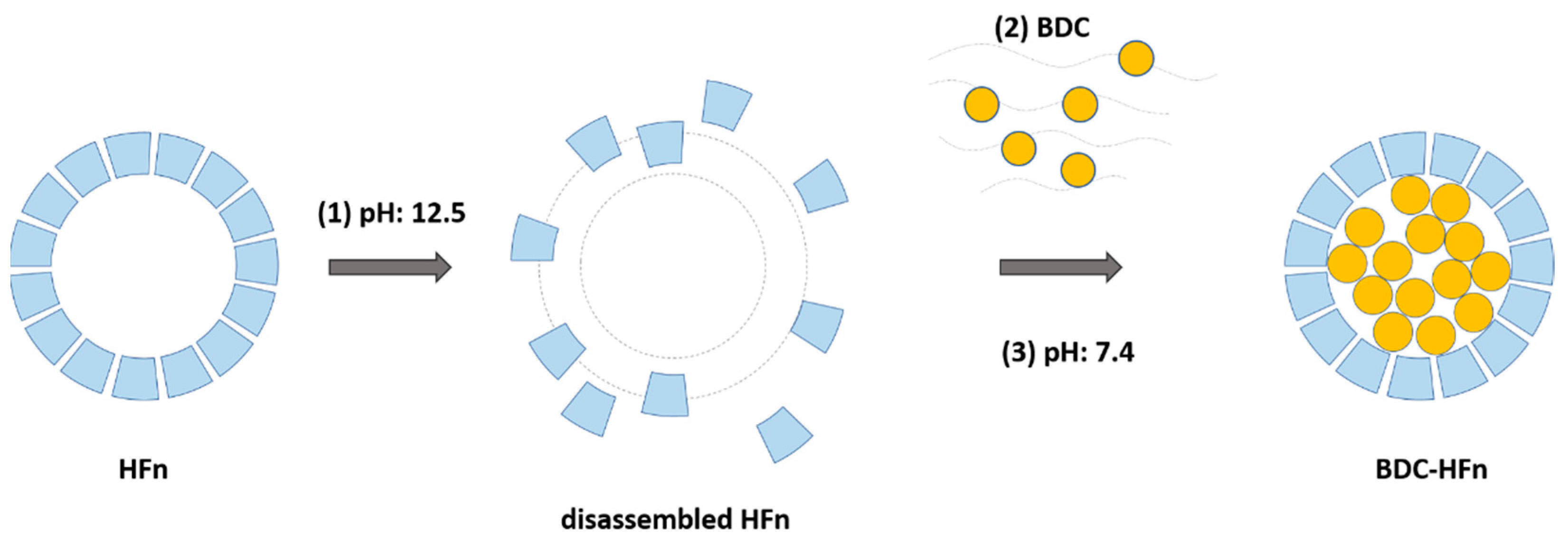
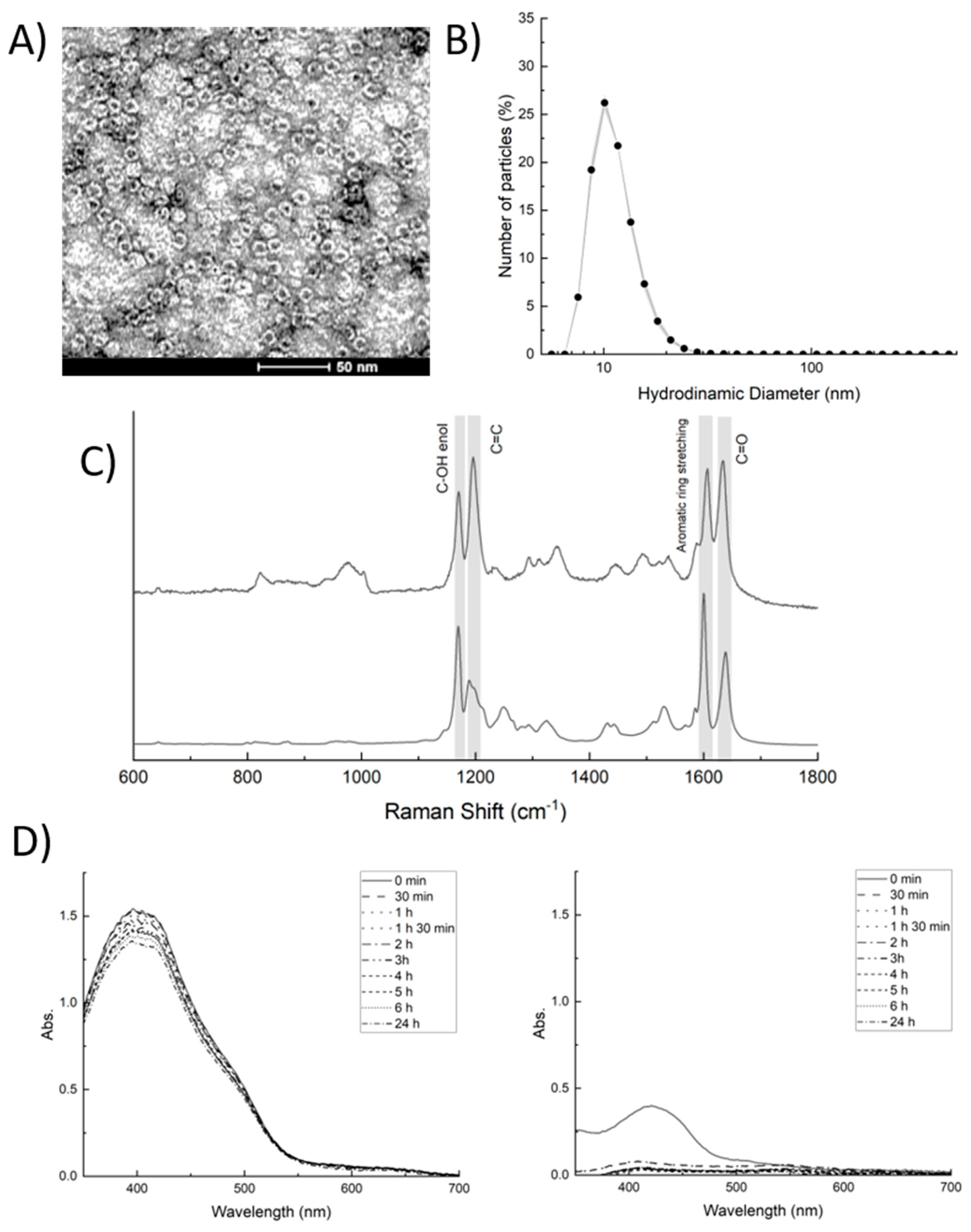
2.1. Hfn-BDC Nanoformulation
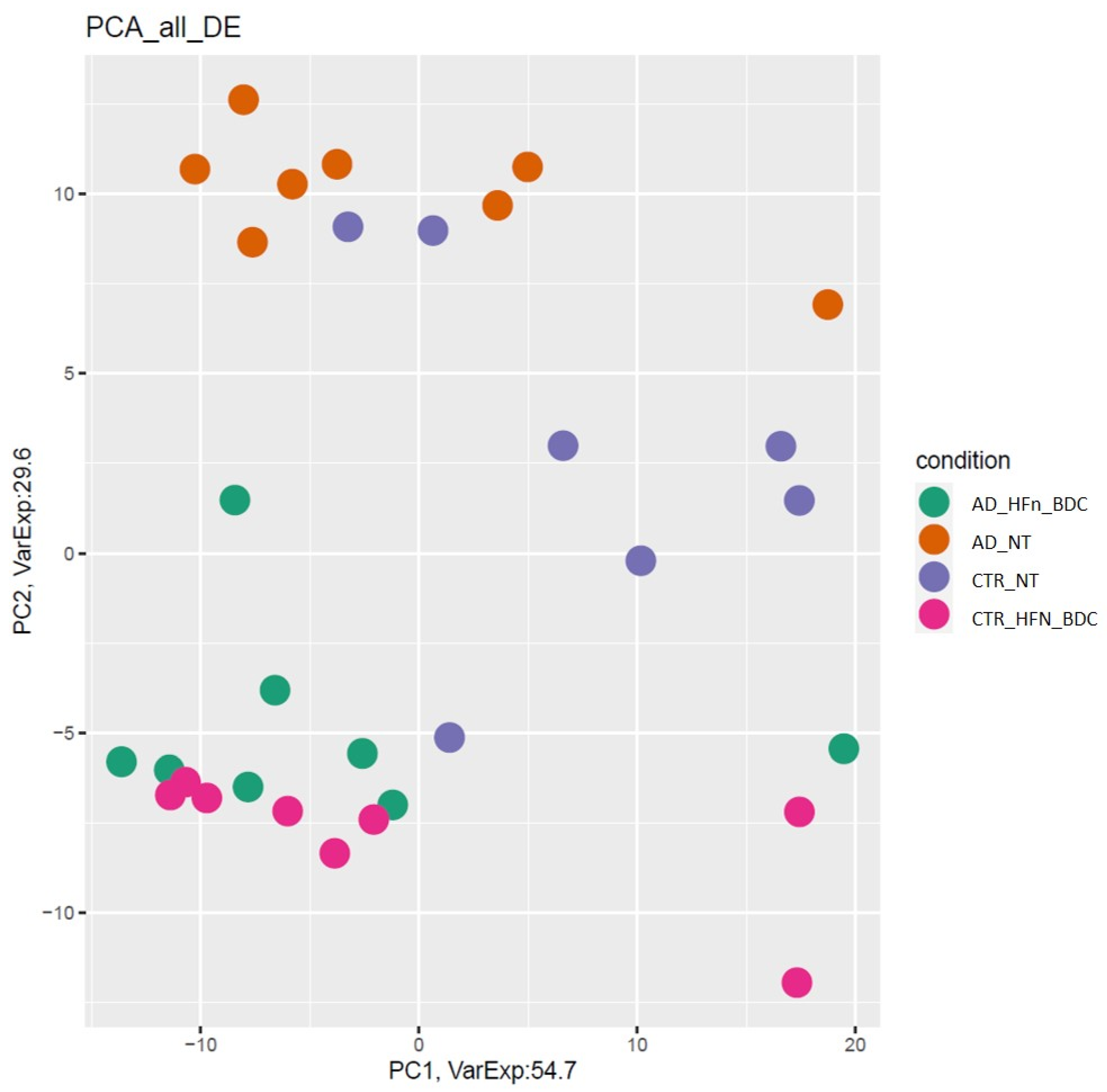
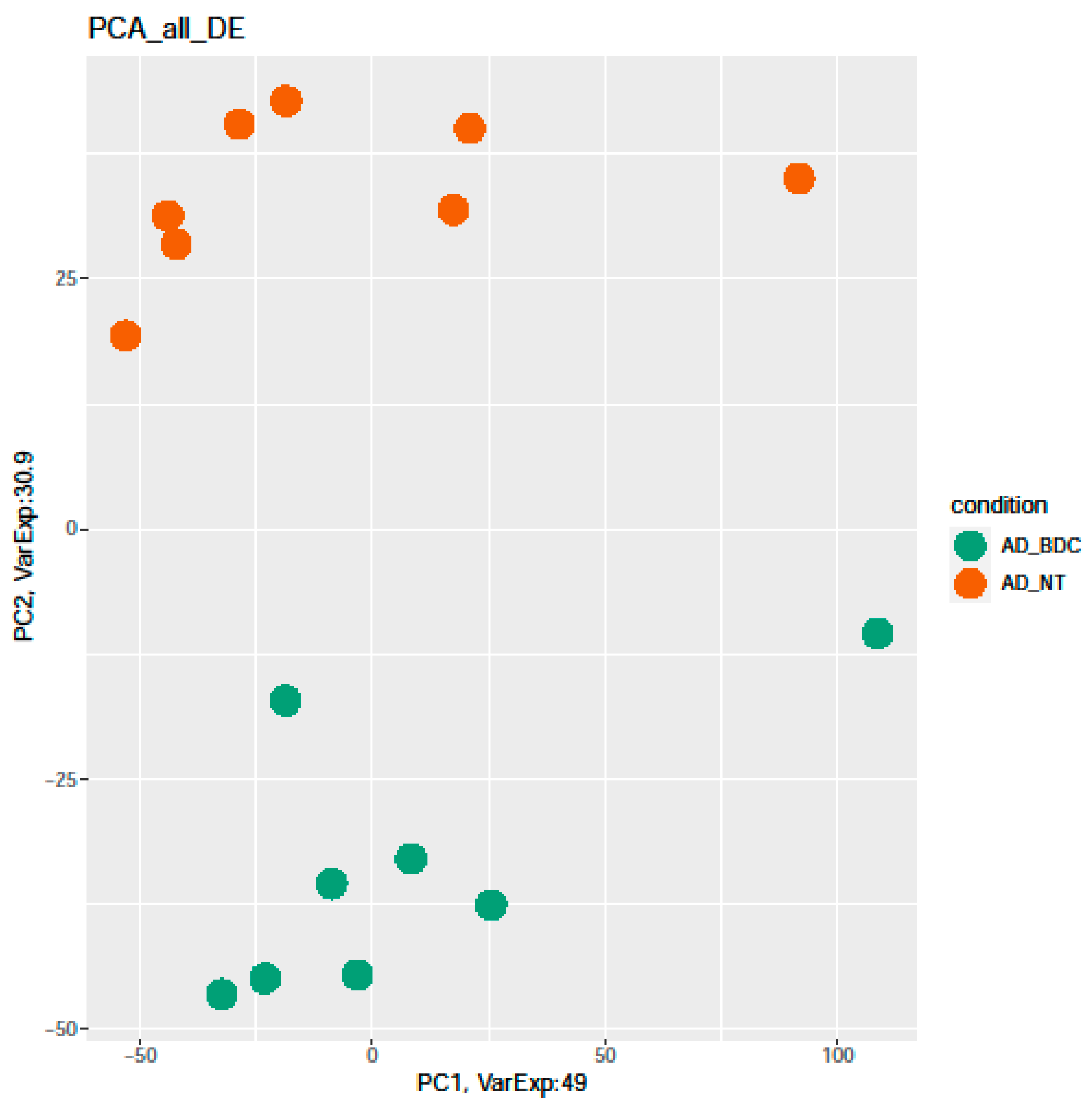
2.2. RNA-Seq Differentially Expressed mRNAs and lncRNAs
- AD NT vs. CTR NT;
- AD BDC-HFn vs. CTR BDC-HFn;
- AD NT vs. AD BDC-HFn;
- AD BDC-HFn vs. CTR NT;
- CTR NT vs. CTR BDC-HFn.
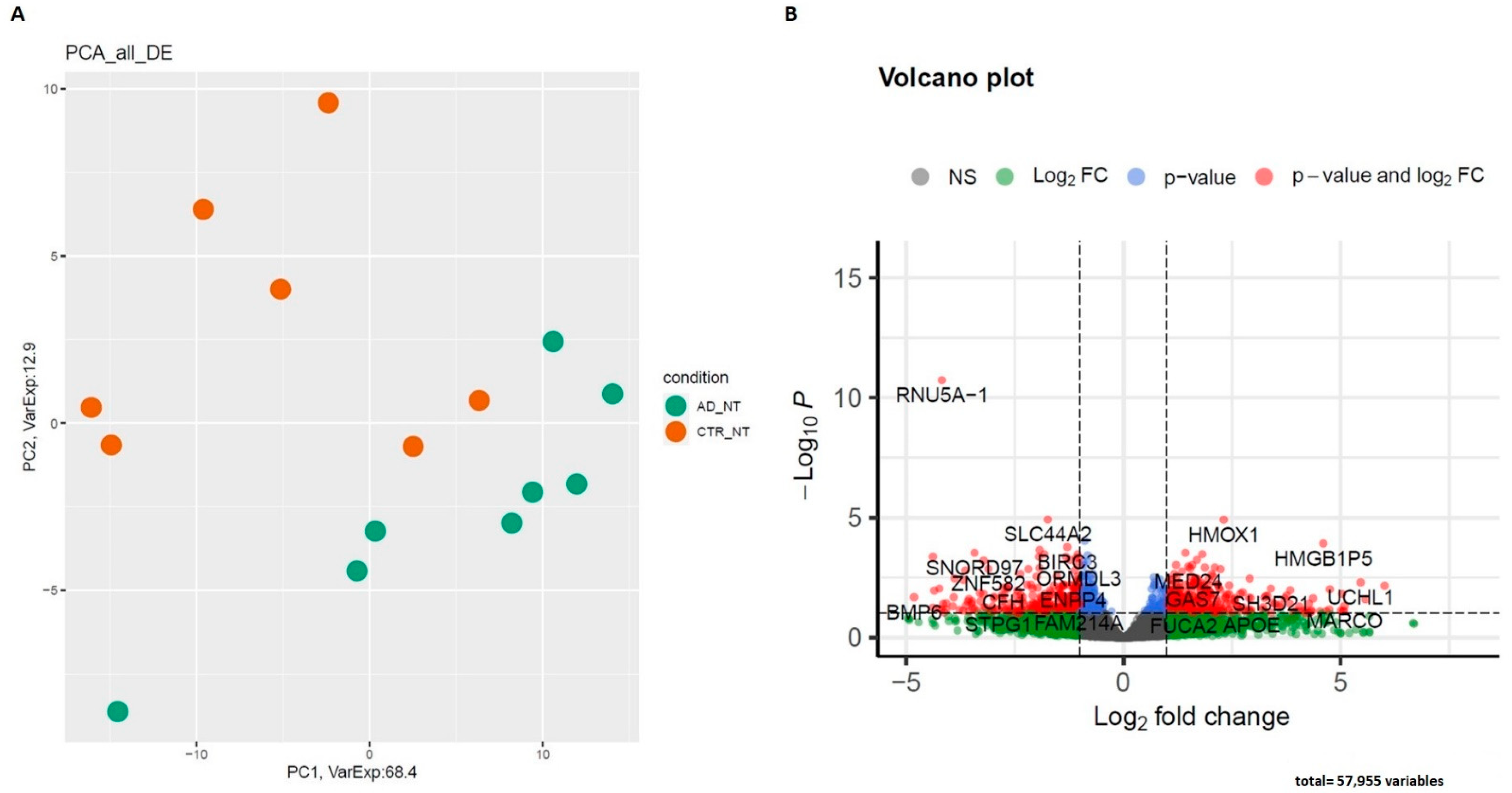
2.2.1. AD NT vs. CTR NT
2.2.2. AD NT vs. AD BDC-HFn
2.3. Validation of Deregulated Coding and Noncoding Genes
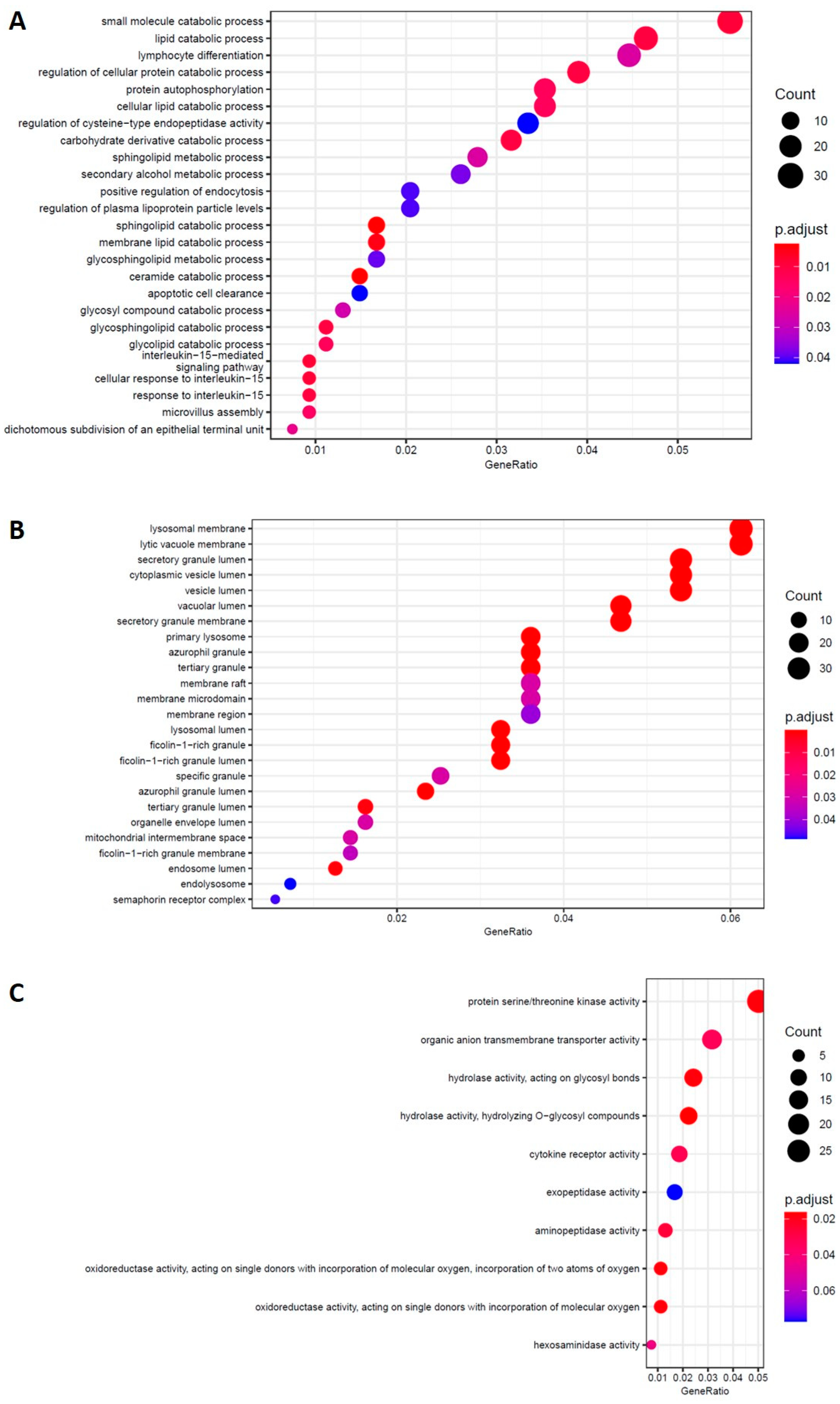
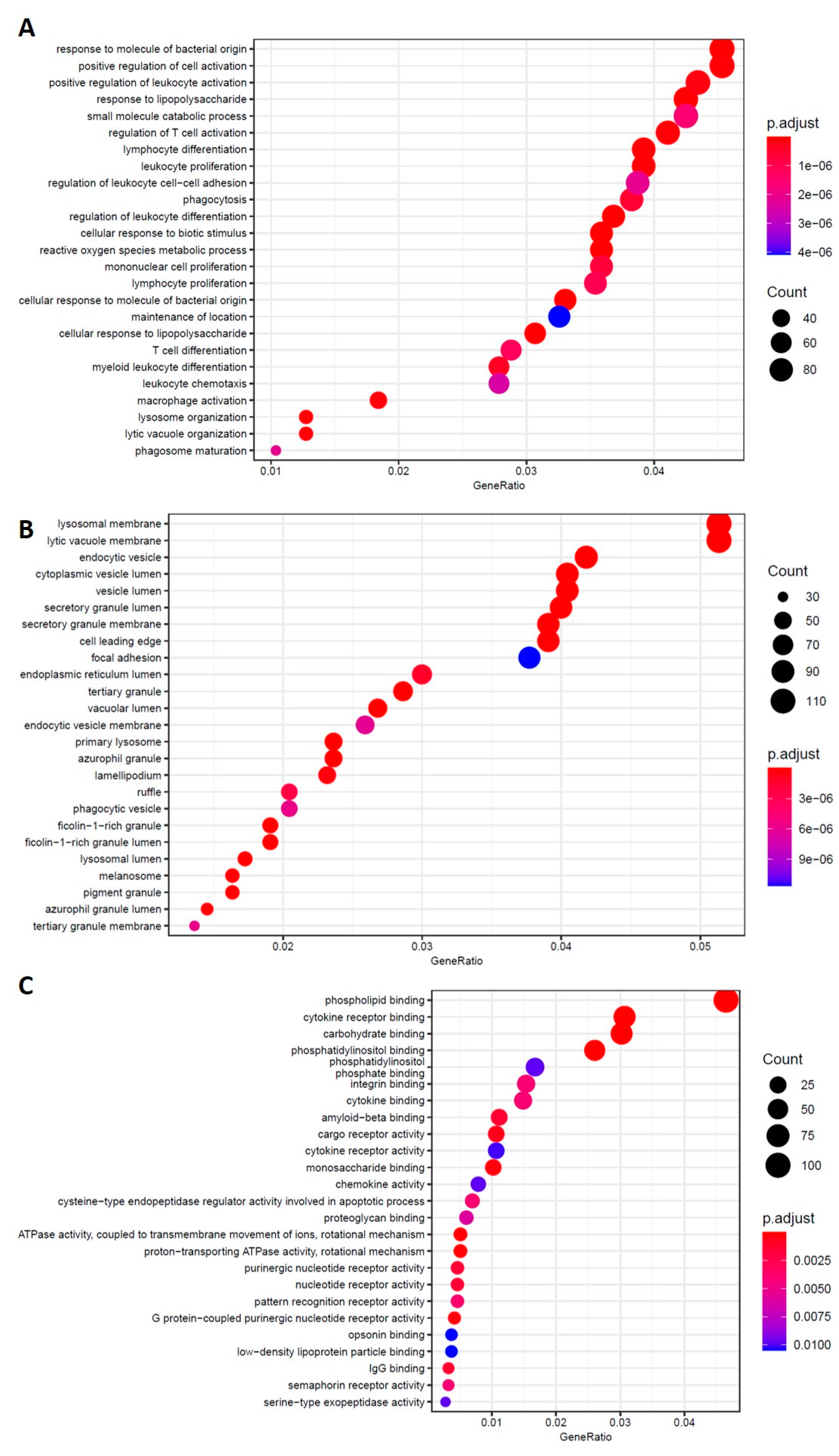
2.4. mRNA Pathway Analysis
2.4.1. AD NT vs. CTR NT
2.4.2. AD NT vs. AD Hfn-BDC
2.4.3. AD BDC-HFn vs. CTR BDC-HFn
2.4.4. AD BDC-HFn vs. CTR NT and CTR NT vs. CTR BDC-HFn
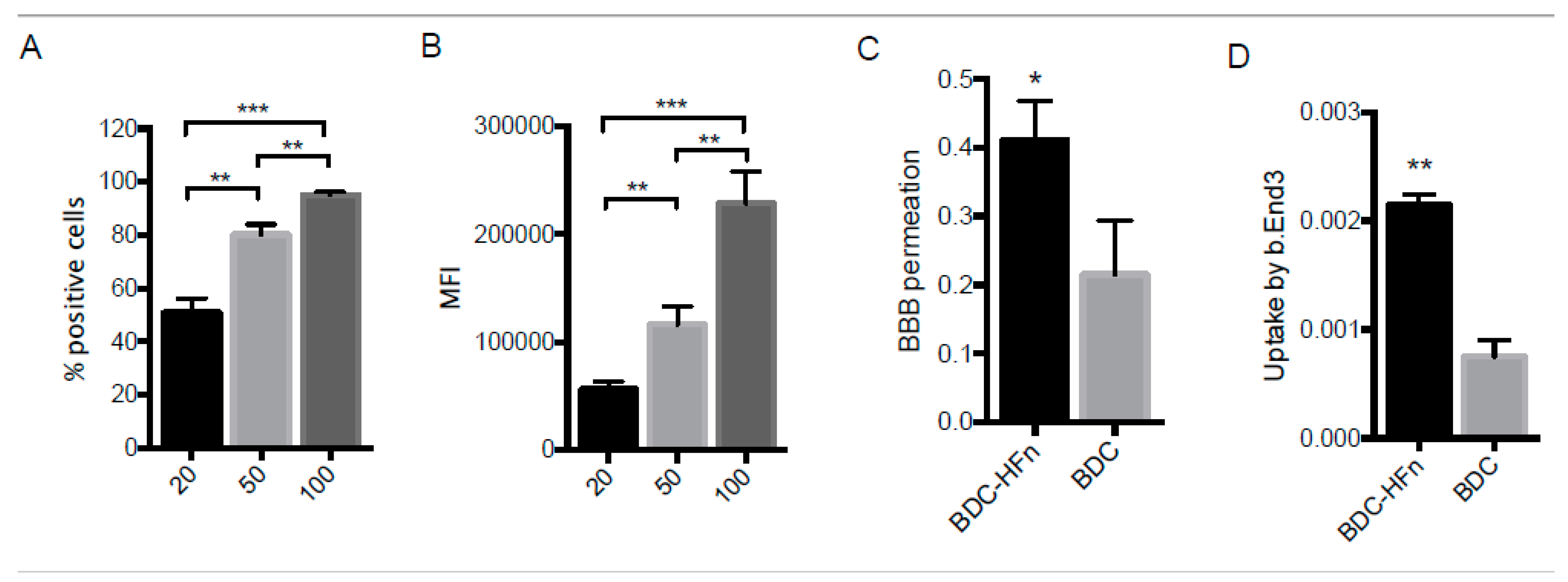
2.5. BDC-HFn Penetration across a BBB In Vitro Model
3. Discussion
4. Materials and Methods
4.1. Nanoformulation and Characterization of BDC-HFn
4.1.1. HFn Loading with Bisdemethoxycurcumin (BDC)
4.1.2. BDC-HFn Nanoparticles Characterization by Transmission Electron Microscope and Dynamic Light Scattering
4.1.3. Determination of Drug Loading Efficiency and Stability of BDC-HFn
4.1.4. Raman Spectroscopy
4.2. Blood Brain Barrier Model (BBB)
4.3. Cell Binding Assay
4.4. Endothelial Cells Viability Assay
4.5. BDC Measurement by UPLC/MS-MS
4.6. Study Subjects
4.7. PBMCs Isolation, Treatment, and RNA Extraction
- -
- PBMCs isolated from AD patients not treated (AD NT);
- -
- PBMCs isolated from AD patients treated with BDC-HFn (AD BDC-HFn);
- -
- PBMCs isolated from Controls not treated (CTR NT);
- -
- PBMCs isolated from controls treated with BDC-HFn (CTR BDC-HFn).
4.8. Libraries Preparation for RNA-Seq and Bioinformatic Data Analysis
4.9. Pathway Analysis
4.10. Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, S.Y.; Kim, D.S. Discovery of natural products from Curcuma longa that protect cells from beta-amyloid insult: A drug discovery effort against Alzheimer’s disease. J. Nat. Prod. 2002, 65, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- Calabrò, M.; Rinaldi, C.; Santoro, G.; Crisafulli, C. The biological pathways of Alzheimer disease: A review. AIMS Neurosci. 2020, 8, 86–132. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Cherbuin, N.; Walsh, E.I.; Leach, L.; Brüstle, A.; Burns, R.; Anstey, K.J.; Sachdev, P.S.; Baune, B.T. Systemic Inflammation Predicts Alzheimer Pathology in Community Samples without Dementia. Biomedicines 2022, 10, 1240. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Van Hoecke, L.; Vandenbroucke, R.E. The Impact of Systemic Inflammation on Alzheimer’s Disease Pathology. Front. Immunol. 2022, 12, 796867. [Google Scholar] [CrossRef]
- Holmes, C. Review: Systemic Inflammation and Alzheimer’s Disease. Neuropathol. Appl. Neurobiol. 2013, 39, 51–68. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, X.; Li, H.; Hui, S.; Zhang, Z.; Xiao, Y.; Peng, W. Non-Coding RNAs as Novel Regulators of Neuroinflammation in Alzheimer’s Disease. Front. Immunol. 2022, 13, 908076. [Google Scholar] [CrossRef]
- Arosio, B.; D’Addario, C.; Gussago, C.; Casati, M.; Tedone, E.; Ferri, E.; Nicolini, P.; Rossi, P.D.; Maccarrone, M.; Mari, D. Peripheral blood mononuclear cells as a laboratory to study dementia in the elderly. Biomed. Res. Int. 2014, 2014, 169203. [Google Scholar] [CrossRef]
- Dangond, F.; Gullans, S.R. Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem. Biophys. Res. Commun. 1998, 247, 833–837. [Google Scholar] [CrossRef]
- de Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]
- Mahapatra, G.; Gao, Z.; Bateman, J.R., 3rd; Lockhart, S.N.; Bergstrom, J.; DeWitt, A.R.; Piloso, J.E.; Kramer, P.A.; Gonzalez-Armenta, J.L.; Amick, K.A.; et al. Blood-based bioenergetic profiling reveals differences in mitochondrial function associated with cognitive performance and Alzheimer’s disease. Alzheimer’s Dement. 2022, 1–13, 12731. [Google Scholar] [CrossRef] [PubMed]
- Jörg, M.; Plehn, J.E.; Friedland, K.; Müller, W.E. Mitochondrial Dysfunction as a Causative Factor in Alzheimer’s Disease-Spectrum Disorders: Lymphocytes as a Window to the Brain. Curr. Alzheimer Res. 2021, 18, 733–752. [Google Scholar] [CrossRef] [PubMed]
- Wojsiat, J.; Prandelli, C.; Laskowska-Kaszub, K.; Martín-Requero, A.; Wojda, U. Oxidative Stress and Aberrant Cell Cycle in Alzheimer’s Disease Lymphocytes: Diagnostic Prospects. J. Alzheimer’s Dis. 2015, 46, 329–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, B. The correlation between LncRNA-17A expression in peripheral blood mononuclear cells and Wnt/β-catenin signaling pathway and cognitive function in patients with Alzheimer disease. Am. J. Transl. Res. 2021, 13, 11981–11986. [Google Scholar] [PubMed]
- Gagliardi, S.; Ghirmai, S.; Abel, K.J.; Lanier, M.; Gardai, S.J.; Lee, C.; Cashman, J.R. Evaluation in vitro of synthetic curcumins as agents promoting monocytic gene expression related to β-amyloid clearance. Chem. Res. Toxicol. 2012, 25, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Fiala, M.; Mahanian, M.; Rosenthal, M.; Mizwicki, M.T.; Tse, E.; Cho, T.; Sayre, J.; Weitzman, R.; Porter, V. MGAT3 mRNA: A biomarker for prognosis and therapy of Alzheimer’s disease by vitamin D and curcuminoids. J. Alzheimer’s Dis. 2011, 25, 135–144. [Google Scholar] [CrossRef]
- Ozben, T.; Ozben, S. Neuro-inflammation and anti-inflammatory treatment options for Alzheimer’s disease. Clin. Biochem. 2019, 72, 87–89. [Google Scholar] [CrossRef]
- Mir, R.H.; Shah, A.J.; Mohi-Ud-Din, R.; Pottoo, F.H.; Dar, M.A.; Jachak, S.M.; Masoodi, M.H. Natural Anti-inflammatory compounds as Drug candidates in Alzheimer’s disease. Curr. Med. Chem. 2020, 28, 4799–4825. [Google Scholar] [CrossRef]
- Gagliardi, S.; Morasso, C.; Stivaktakis, P.; Pandini, C.; Tinelli, V.; Tsatsakis, A.; Prosperi, D.; Hickey, M.; Corsi, F.; Cereda, C. Curcumin Formulations and Trials: What’s New in Neurological Diseases. Molecules 2020, 25, 5389. [Google Scholar] [CrossRef]
- Utomo, R.Y.; Sugie, A.; Okada, S.; Miura, K.; Nakamura, H. Detoxification of amyloid β fibrils by curcumin derivatives and their verification in a Drosophila Alzheimer’s model. Chem. Commun. 2022, 58, 2576–2579. [Google Scholar] [CrossRef]
- Gagliardi, S.; Franco, V.; Sorrentino, S.; Zucca, S.; Pandini, C.; Rota, P.; Bernuzzi, S.; Costa, A.; Sinforiani, E.; Pansarasa, O.; et al. Curcumin and Novel Synthetic Analogs in Cell-Based Studies of Alzheimer’s Disease. Front. Pharmacol. 2018, 9, 1404. [Google Scholar] [CrossRef] [PubMed]
- Pluta, R.; Furmaga-Jabłońska, W.; Januszewski, S.; Czuczwar, S.J. Post-Ischemic Brain Neurodegeneration in the Form of Alzheimer’s Disease Proteinopathy: Possible Therapeutic Role of Curcumin. Nutrients 2022, 14, 248. [Google Scholar] [CrossRef] [PubMed]
- Benameur, T.; Giacomucci, G.; Panaro, M.A.; Ruggiero, M.; Trotta, T.; Monda, V.; Pizzolorusso, I.; Lofrumento, D.D.; Porro, C.; Messina, G. New Promising Therapeutic Avenues of Curcumin in Brain Diseases. Molecules 2021, 27, 236. [Google Scholar] [CrossRef] [PubMed]
- Panahi, Y.; Ahmadi, Y.; Teymouri, M.; Johnston, T.P.; Sahebkar, A. Curcumin as a potential candidate for treating hyperlipidemia: A review of cellular and metabolic mechanisms. J Cell Physiol. 2018, 233, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Askarizadeh, A.; Barreto, G.E.; Henney, N.C.; Majeed, M.; Sahebkar, A. Neuroprotection by curcumin: A review on brain delivery strategies. Int. J. Pharm. 2020, 585, 119476. [Google Scholar] [CrossRef]
- Wu, S.; Guo, T.; Qi, W.; Li, Y.; Gu, J.; Liu, C.; Sha, Y.; Yang, B.; Hu, S.; Zong, X. Curcumin ameliorates ischemic stroke injury in rats by protecting the integrity of the blood-brain barrier. Exp. Ther. Med. 2021, 22, 783. [Google Scholar] [CrossRef]
- Brondino, N.; Re, S.; Boldrini, A.; Cuccomarino, A.; Lanati, N.; Barale, F.; Politi, P. Curcumin as a therapeutic agent in dementia: A mini systematic review of human studies. Sci. World J. 2014, 174282. [Google Scholar] [CrossRef]
- Tang, M.; Taghibiglou, C. The Mechanisms of Action of Curcumin in Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 58, 1003–1016. [Google Scholar] [CrossRef]
- Ravindran, J.; Subbaraju, G.V.; Ramani, M.V.; Sung, B.; Aggarwal, B.B. Bisdemethylcurcumin and structurally related hispolon analogues of curcumin exhibit enhanced prooxidant, anti-proliferative and anti-inflammatory activities in vitro. Biochem. Pharmacol. 2010, 79, 1658–1666. [Google Scholar] [CrossRef]
- Fiala, M.; Porter, V. Delineating Alzheimer’s disease progression with MGAT3, a biomarker for improved prognosis and personalized therapy. Biomark. Med. 2011, 5, 645–647. [Google Scholar] [CrossRef]
- Khanna, S.; Park, H.A.; Sen, C.K.; Golakoti, T.; Sengupta, K.; Venkateswarlu, S.; Roy, S. Neuroprotective and antiinflammatory properties of a novel demethylated curcuminoid. Antioxid. Redox Signal. 2009, 11, 449–468. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Li, Z.H.; Liu, L.; Tang, W.X.; Wang, Y.; Dong, M.R.; Xiao, C. Curcumin Attenuates Beta-Amyloid-Induced Neuroinflammation via Activation of Peroxisome Proliferator-Activated Receptor-Gamma Function in a Rat Model of Alzheimer’s Disease. Front. Pharmacol. 2016, 7, 261. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.T.; Chen, Z.T.; Hou, W.C.; Yu, L.C.; Chen, R.P. Polyhydroxycurcuminoids but not curcumin upregulate neprilysin and can be applied to the prevention of Alzheimer’s disease. Sci. Rep. 2016, 6, 29760. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.P.; Chu, T.; Yang, F.; Beech, W.; Frautschy, S.A.; Cole, G.M. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J. Neurosci. 2001, 21, 8370–8377. [Google Scholar] [CrossRef]
- Kharat, M.; Du, Z.; Zhang, G.; McClements, D.J. Physical and Chemical Stability of Curcumin in Aqueous Solutions and Emulsions: Impact of pH, Temperature, and Molecular Environment. J. Agric. Food Chem. 2017, 65, 1525–1532. [Google Scholar] [CrossRef]
- Cox, K.H.; Pipingas, A.; Scholey, A.B. Investigation of the effects of solid lipid curcumin on cognition and mood in a healthy older population. J. Psychopharmacol. 2015, 29, 642–651. [Google Scholar] [CrossRef]
- Small, G.W.; Siddarth, P.; Li, Z.; Miller, K.J.; Ercoli, L.; Emerson, N.D.; Martinez, J.; Wong, K.P.; Liu, J.; Merrill, D.A.; et al. Memory and Brain Amyloid and Tau Effects of a Bioavailable Form of Curcumin in Non-Demented Adults: A Double-Blind, Placebo-Controlled 18-Month Trial. Am. J. Geriatr. Psychiatry 2018, 26, 266–277. [Google Scholar] [CrossRef]
- Chen, L.; Bai, G.; Yang, S.; Yang, R.; Zhao, G.; Xu, C.; Leung, X. Encapsulation of curcumin in recombinant human H-chain ferritin increases its water-solubility and stability. Food Res. Int. 2014, 62, 1147–1153. [Google Scholar] [CrossRef]
- Mainini, F.; Bonizzi, A.; Sevieri, M.; Sitia, L.; Truffi, M.; Corsi, F.; Mazzucchelli, S. Protein-Based Nanoparticles for the Imaging and Treatment of Solid Tumors: The Case of Ferritin Nanocages, a Narrative Review. Pharmaceutics 2021, 13, 2000. [Google Scholar] [CrossRef]
- Fiandra, L.; Mazzucchelli, S.; Truffi, M.; Bellini, M.; Sorrentino, L.; Corsi, F. In Vitro Permeation of FITC-loaded Ferritins Across a Rat Blood-brain Barrier: A Model to Study the Delivery of Nanoformulated Molecules. J. Vis. Exp. 2016, 22, 54279. [Google Scholar] [CrossRef]
- Bellini, M.; Mazzucchelli, S.; Galbiati, E.; Sommaruga, S.; Fiandra, L.; Truffi, M.; Rizzuto, M.A.; Colombo, M.; Tortora, P.; Corsi, F.; et al. Protein nanocages for self-triggered nuclear delivery of DNA-targeted chemotherapeutics in Cancer Cells. J Control. Release 2014, 196, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Krishna Mohan, P.R.; Sreelakshmi, G.; Muraleedharan, C.V.; Joseph, R. Water soluble complexes of curcumin with cyclodextrins: Characterization by FT-Raman spectroscopy. Vib. Spectrosc. 2012, 62, 77–84. [Google Scholar] [CrossRef]
- Miloudi, L.; Bonnier, F.; Bertrand, D.; Byrne, H.J.; Perse, X.; Chourpa, I.; Munnier, E. Quantitative analysis of curcumin-loaded alginate nanocarriers in hydrogels using Raman and attenuated total reflection infrared spectroscopy. Anal. Bioanal. Chem. 2017, 409, 4593–4605. [Google Scholar] [CrossRef] [PubMed]
- Sogorb-Esteve, A.; Swift, I.J.; Woollacott, I.O.C.; Warren, J.D.; Zetterberg, H.; Rohrer, J.D. Differential chemokine alteration in the variants of primary progressive aphasia-a role for neuroinflammation. J. Neuroinflammation 2021, 18, 224. [Google Scholar] [CrossRef] [PubMed]
- Gaetani, L.; Bellomo, G.; Parnetti, L.; Blennow, K.; Zetterberg, H.; Di Filippo, M. Neuroinflammation and Alzheimer’s Disease: A Machine Learning Approach to CSF Proteomics. Cells 2021, 10, 1930. [Google Scholar] [CrossRef]
- Bogdan, S.; Puścion-Jakubik, A.; Klimiuk, K.; Socha, K.; Kochanowicz, J.; Gorodkiewicz, E. UCHL1 and Proteasome in Blood Serum in Relation to Dietary Habits, Concentration of Selected Antioxidant Minerals and Total Antioxidant Status among Patients with Alzheimer’s Disease. J. Clin. Med. 2022, 11, 412. [Google Scholar] [CrossRef]
- Hochstrasser, T.; Weiss, E.; Marksteiner, J.; Humpel, C. Soluble cell adhesion molecules in monocytes of Alzheimer’s disease and mild cognitive impairment. Exp. Gerontol. 2010, 45, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.T.; Chen, S.D.; Xu, W.; Chen, K.L.; Wang, H.F.; Tan, C.C.; Cui, M.; Dong, Q.; Tan, L.; Yu, J.T. Alzheimer’s Disease Neuroimaging Initiative. Genome-wide association study identifies CD1A associated with rate of increase in plasma neurofilament light in non-demented elders. Aging 2019, 11, 4521–4535. [Google Scholar] [CrossRef]
- Zhang, K.; Tian, L.; Liu, L.; Feng, Y.; Dong, Y.B.; Li, B.; Shang, D.S.; Fang, W.G.; Cao, Y.P.; Chen, Y.H. CXCL1 contributes to β-amyloid-induced transendothelial migration of monocytes in Alzheimer’s disease. PLoS ONE 2013, 8, e72744. [Google Scholar] [CrossRef]
- Nitsch, L.; Schneider, L.; Zimmermann, J.; Müller, M. Microglia-Derived Interleukin 23: A Crucial Cytokine in Alzheimer’s Disease? Front. Neurol. 2021, 12, 639353. [Google Scholar] [CrossRef]
- Zettergren, A.; Höglund, K.; Kern, S.; Thorvaldsson, V.; Johan Skoog, M.; Hansson, O.; Andreasen, N.; Bogdanovic, N.; Blennow, K.; Skoog, I.; et al. Association of IL1RAP-related genetic variation with cerebrospinal fluid concentration of Alzheimer-associated tau protein. Sci. Rep. 2019, 9, 2460. [Google Scholar] [CrossRef] [PubMed]
- Hirayasu, K.; Arase, H. Leukocyte Immunoglobulin-Like Receptor (LILR). In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Chen, D.; Wang, X.; Huang, T.; Jia, J. Sleep and Late-Onset Alzheimer’s Disease: Shared Genetic Risk Factors, Drug Targets, Molecular Mechanisms, and Causal Effects. Front Genet. 2022, 13, 794202. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, P.; Li, A.; Ladd, T.B.; Strickland, M.R.; Koller, E.J.; Burgess, J.D.; Funk, C.C.; Cruz, P.E.; Allen, M.; Yaroshenko, M.; et al. TLR5 decoy receptor as a novel anti-amyloid therapeutic for Alzheimer’s disease. J. Exp. Med. 2018, 215, 2247–2264. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression pro-files. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Rajaei, S.; Karima, S.; Tehrani, H.S.; Shateri, S.; Baram, S.M.; Mahdavi, M.; Mokhtari, F.; Alimohammadi, A.; Tafakhori, A.; Amiri, A.; et al. Conformational change and GTPase activity of human tubulin: A comparative study on Alzheimer’s disease and healthy brain. J. Neurochem. 2020, 155, 207–224. [Google Scholar] [CrossRef]
- Fan, K.; Jia, X.; Zhou, M.; Wang, K.; Conde, J.; He, J.; Tian, J.; Yan, X. Ferritin Nanocarrier Traverses the Blood Brain Barrier and Kills Glioma. ACS Nano 2018, 12, 4105–4115. [Google Scholar] [CrossRef]
- Rizzuto, M.A.; Dal Magro, R.; Barbieri, L.; Pandolfi, L.; Sguazzini-Viscontini, A.; Truffi, M.; Salvioni, L.; Corsi, F.; Colombo, M.; Re, F.; et al. H-Ferritin nanoparticle-mediated delivery of antibodies across a BBB in vitro model for treatment of brain malignancies. Biomater. Sci. 2021, 9, 2032. [Google Scholar] [CrossRef]
- Omidi, Y.; Campbell, L.; Barar, J.; Connell, D.; Akhtar, S.; Gumbleton, M. Evaluation of the immortalised mouse brain capillary endothelial cell line, b.End3, as an in vitro blood–brain barrier model for drug uptake and transport studies. Brain Res. 2003, 990, 95–112. [Google Scholar] [CrossRef]
- Weksler, B.; Romero, I.A.; Couraud, P.O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS 2013, 10, 16. [Google Scholar] [CrossRef]
- Paudel, A.; Raijada, D.; Rantanen, J. Raman spectroscopy in pharmaceutical product design. Adv. Drug Deliv. Rev. 2015, 89, 3–20. [Google Scholar] [CrossRef]
- Sanphui, P.; Goud, N.R.; Khandavilli, U.B.; Bhanoth, S.; Nangia, A. New polymorphs of curcumin. Chem. Commun. 2011, 47, 5013–5015. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019, 332, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wise, L.; Fukuchi, K.I. TLR4 Cross-Talk with NLRP3 Inflammasome and Complement Signaling Pathways in Alzheimer’s Disease. Front. Immunol. 2020, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Hermans, S.J.; Nero, T.L.; Morton, C.J.; Gooi, J.H.; Crespi, G.A.N.; Hancock, N.C.; Gao, C.; Ishii, K.; Markulić, J.; Parker, M.W. Structural biology of cell surface receptors implicated in Alzheimer’s disease. Biophys. Rev. 2021, 14, 233–255. [Google Scholar] [CrossRef]
- Hur, J.Y.; Frost, G.R.; Wu, X.; Crump, C.; Pan, S.J.; Wong, E.; Barros, M.; Li, T.; Nie, P.; Zhai, Y.; et al. The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer’s disease. Nature 2020, 586, 735–740. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Bawa, K.K.; Ouk, M.; Leung, N.; Yu, D.; Lanctôt, K.L.; Herrmann, N.; Pakosh, M.; Swardfager, W. Neutrophil activation in Alzheimer’s disease and mild cognitive impairment: A systematic review and meta-analysis of protein markers in blood and cerebrospinal fluid. Ageing Res. Rev. 2020, 62, 101130. [Google Scholar] [CrossRef]
- Wang, H.; Huang, L.; Wu, L.; Lan, J.; Feng, X.; Li, P.; Peng, Y. The MMP-2/TIMP-2 System in Alzheimer Disease. CNS Neurol Disord Drug Targets 2020, 19, 402–416. [Google Scholar] [CrossRef]
- Chen, M.; Du, Z.Y.; Zheng, X.; Li, D.L.; Zhou, R.P.; Zhang, K. Use of curcumin in diagnosis, prevention, and treatment of Alzheimer’s disease. Neural Regen. Res. 2018, 13, 742–752. [Google Scholar]
- Williams-Medina, A.; Deblock, M.; Janigro, D. In vitro Models of the Blood-Brain Barrier: Tools in Translational Medicine. Front. Med Technol. 2021, 2, 623950. [Google Scholar] [CrossRef]
- Yang, S.; Mei, S.; Jin, H.; Zhu, B.; Tian, Y.; Huo, J.; Cui, X.; Guo, A.; Zhao, Z. Identification of two immortalized cell lines, ECV304 and bEnd3, for in vitro permeability studies of blood-brain barrier. PLoS ONE 2017, 12, e0187017. [Google Scholar] [CrossRef]
- García-Salvador, A.; Domínguez-Monedero, A.; Gómez-Fernández, P.; García-Bilbao, A.; Carregal-Romero, S.; Castilla, J.; Goñi-de-Cerio, F. Evaluation of the Influence of Astrocytes on In Vitro Blood-Brain Barrier Models. Altern. Lab. Anim. 2020, 48, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Janigro, D.; Hossain, M.; Oby, E.; Rapp, E.; Cucullo, L. Side by side comparison between dynamic versus static models of blood-brain barrier in vitro: A permeability study. Brain Res. 2006, 1109, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Puscas, I.; Bernard-Patrzynski, F.; Jutras, M.; Lécuyer, M.A.; Bourbonnière, L.; Prat, A.; Leclair, G.; Roullin, V.G. Assessment of Two Mouse Brain Endothelial Cell Models for Drug Screening. Pharmaceutics 2019, 11, 587. [Google Scholar] [CrossRef]
- Jagtiani, E.; Yeolekar, M.; Naik, S.; Patravale, V.J. In vitro blood brain barrier models: An overview. J. Control. Release 2022, 343, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Sitia, L.; Catelani, T.; Guarnieri, D.; Pompa, P.P. In Vitro Blood-Brain Barrier Models for Nanomedicine: Particle-Specific Effects and Methodological Drawbacks. ACS Appl. Bio Mater. 2019, 2, 3279–3289. [Google Scholar] [CrossRef]
- Pandolfi, L.; Bellini, M.; Vanna, R.; Morasso, C.; Zago, A.; Carcano, S.; Avvakumova, S.; Bertolini, J.A.; Rizzuto, M.A.; Colombo, M.; et al. H-Ferritin Enriches the Curcumin Uptake and Improves the Therapeutic Efficacy in Triple Negative Breast Cancer Cells. Biomacromolecules 2017, 18, 3318–3330. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001, Appendix 3, Appendix 3B. [Google Scholar]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for cell viability assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 2073. [Google Scholar] [CrossRef]
- Carrara, M.; Lum, J.; Cordero, F.; Beccuti, M.; Poidinger, M.; Donatelli, S.; Calogero, R.A.; Zolezzi, F. Alternative splicing detection workflow needs a careful combination of sample prep and bioinformatics analysis. BMC Bioinform. 2015, 16 (Suppl. S9), S2. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, 90–97. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]

| Upregulated | Downregulated | Total RNAs | |||
|---|---|---|---|---|---|
| mRNA | lncRNA | mRNA | lncRNA | ||
| AD NT vs. CTR NT | 333 | 11 | 234 | 52 | 630 |
| AD NT vs. AD BDC-HFn | 1419 | 88 | 952 | 58 | 2517 |
| AD BDC-HFn vs. CTR BDC-HFn | 29 | 7 | 52 | 10 | 98 |
| AD Hfn-BDC vs. CTR NT | 151 | 30 | 566 | 24 | 771 |
| CTR NT vs. CTR BDC-HFn | 164 | 26 | 724 | 22 | 936 |
| Most Deregulated Genes in AD-NT Group Compared to CTR-NT | ||
|---|---|---|
| Gene | Fold Change | Role in AD |
| CXCL5 | −5.28042797835226 | monocytes migrating from blood to brain in AD patients [48] |
| CD1E | −3.76621256916995 | CD1A is involved in longitudinal changes AD phenotypes [49] |
| IL12RB2 | −2.36344776700486 | associated with cognitive aging [50] |
| IL18RAP | −1.994385464 | associated to Tau concentration in CSF of AD patients [51] |
| LILRA6 | 1.930851051 | expressed in monocyte, function unclear [52] |
| LILRB5 | 4.048756658 | expressed in monocyte, function unclear [52] |
| HLA-DRB6 | 3.84178295 | associated to late onset AD [53] |
| TLR5 | 3.589139449 | may regulate Aβ clearance [54] |
| CTRs | AD | |
| Recruited subjects | 15 | 15 |
| Age (mean ± SD) | 56.1 ± 5.2 | 74.4 ± 8.8 |
| Males % | 43% | 47% |
| Females % | 57% | 53% |
| MMSE | 29.625 ± 0.74 | 18.5 ± 3.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gagliardi, S.; Truffi, M.; Tinelli, V.; Garofalo, M.; Pandini, C.; Cotta Ramusino, M.; Perini, G.; Costa, A.; Negri, S.; Mazzucchelli, S.; et al. Bisdemethoxycurcumin (BDC)-Loaded H-Ferritin-Nanocages Mediate the Regulation of Inflammation in Alzheimer’s Disease Patients. Int. J. Mol. Sci. 2022, 23, 9237. https://doi.org/10.3390/ijms23169237
Gagliardi S, Truffi M, Tinelli V, Garofalo M, Pandini C, Cotta Ramusino M, Perini G, Costa A, Negri S, Mazzucchelli S, et al. Bisdemethoxycurcumin (BDC)-Loaded H-Ferritin-Nanocages Mediate the Regulation of Inflammation in Alzheimer’s Disease Patients. International Journal of Molecular Sciences. 2022; 23(16):9237. https://doi.org/10.3390/ijms23169237
Chicago/Turabian StyleGagliardi, Stella, Marta Truffi, Veronica Tinelli, Maria Garofalo, Cecilia Pandini, Matteo Cotta Ramusino, Giulia Perini, Alfredo Costa, Sara Negri, Serena Mazzucchelli, and et al. 2022. "Bisdemethoxycurcumin (BDC)-Loaded H-Ferritin-Nanocages Mediate the Regulation of Inflammation in Alzheimer’s Disease Patients" International Journal of Molecular Sciences 23, no. 16: 9237. https://doi.org/10.3390/ijms23169237
APA StyleGagliardi, S., Truffi, M., Tinelli, V., Garofalo, M., Pandini, C., Cotta Ramusino, M., Perini, G., Costa, A., Negri, S., Mazzucchelli, S., Bonizzi, A., Sitia, L., Busacca, M., Sevieri, M., Mocchi, M., Ricciardi, A., Prosperi, D., Corsi, F., Cereda, C., & Morasso, C. (2022). Bisdemethoxycurcumin (BDC)-Loaded H-Ferritin-Nanocages Mediate the Regulation of Inflammation in Alzheimer’s Disease Patients. International Journal of Molecular Sciences, 23(16), 9237. https://doi.org/10.3390/ijms23169237









