SARS-CoV-2 and Immunity: Natural Infection Compared with Vaccination
Abstract
:1. Introduction
2. Results
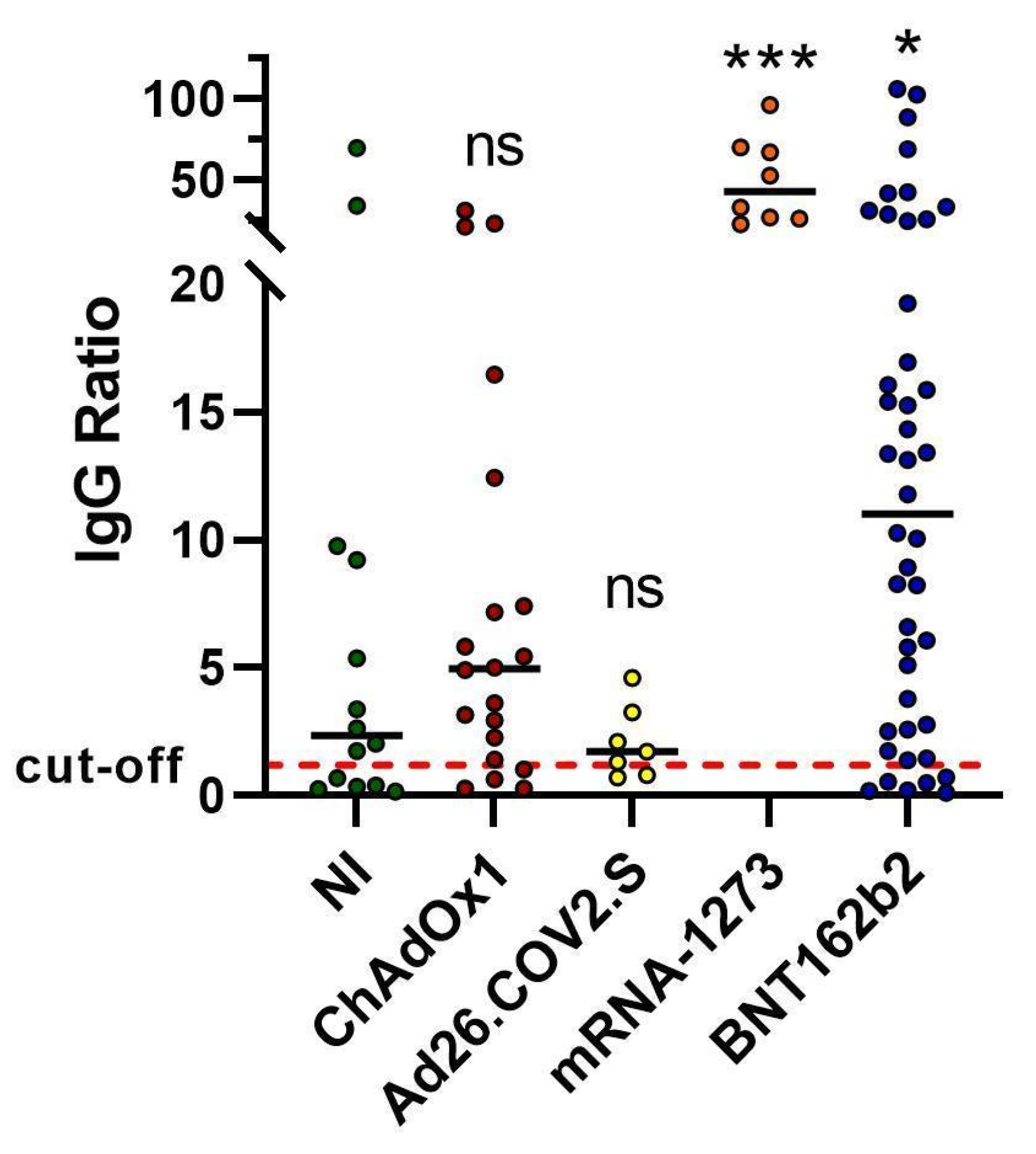
2.1. Spike Antibody Magnitude
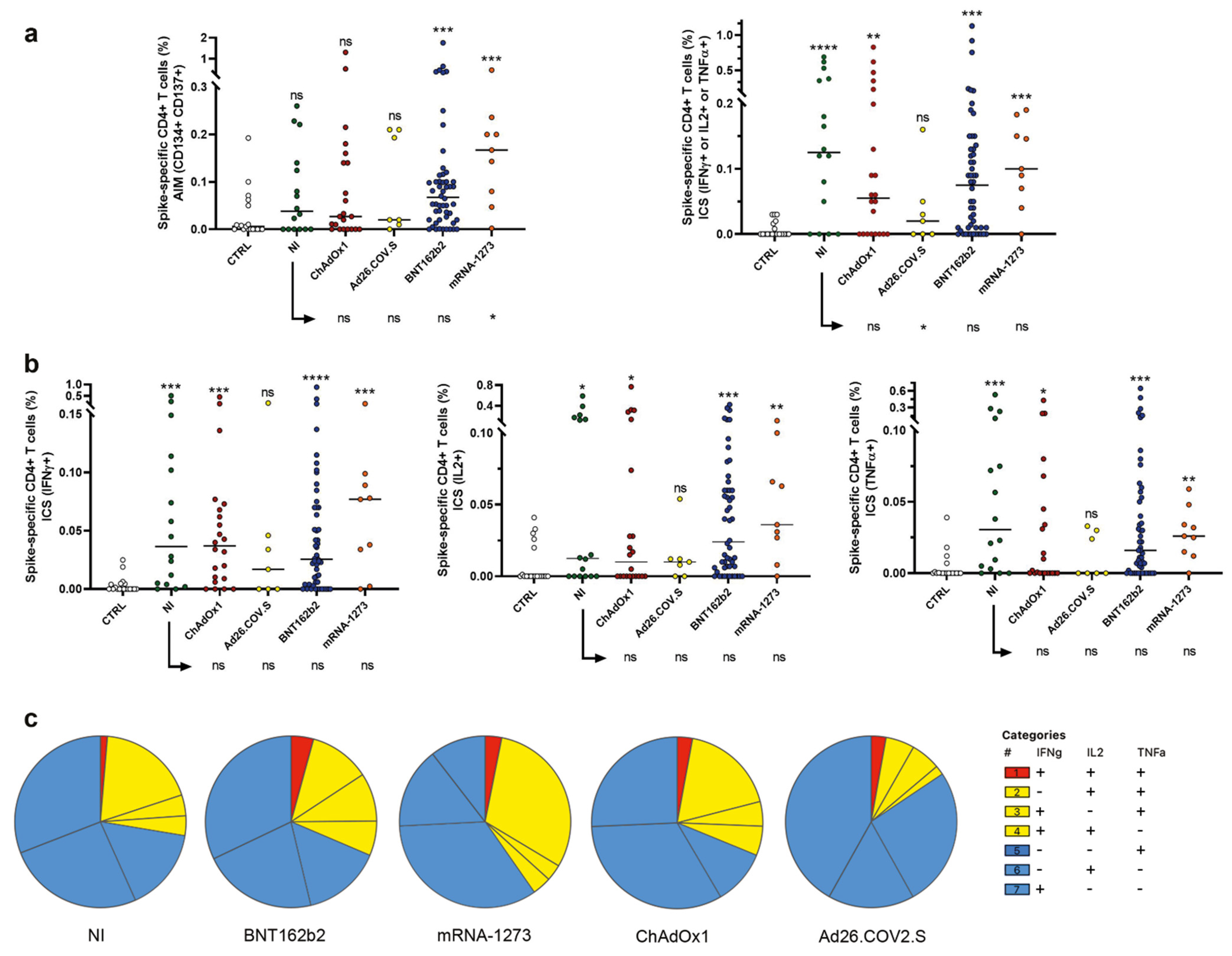
2.2. Induction of Spike Specific CD4+ T Cells
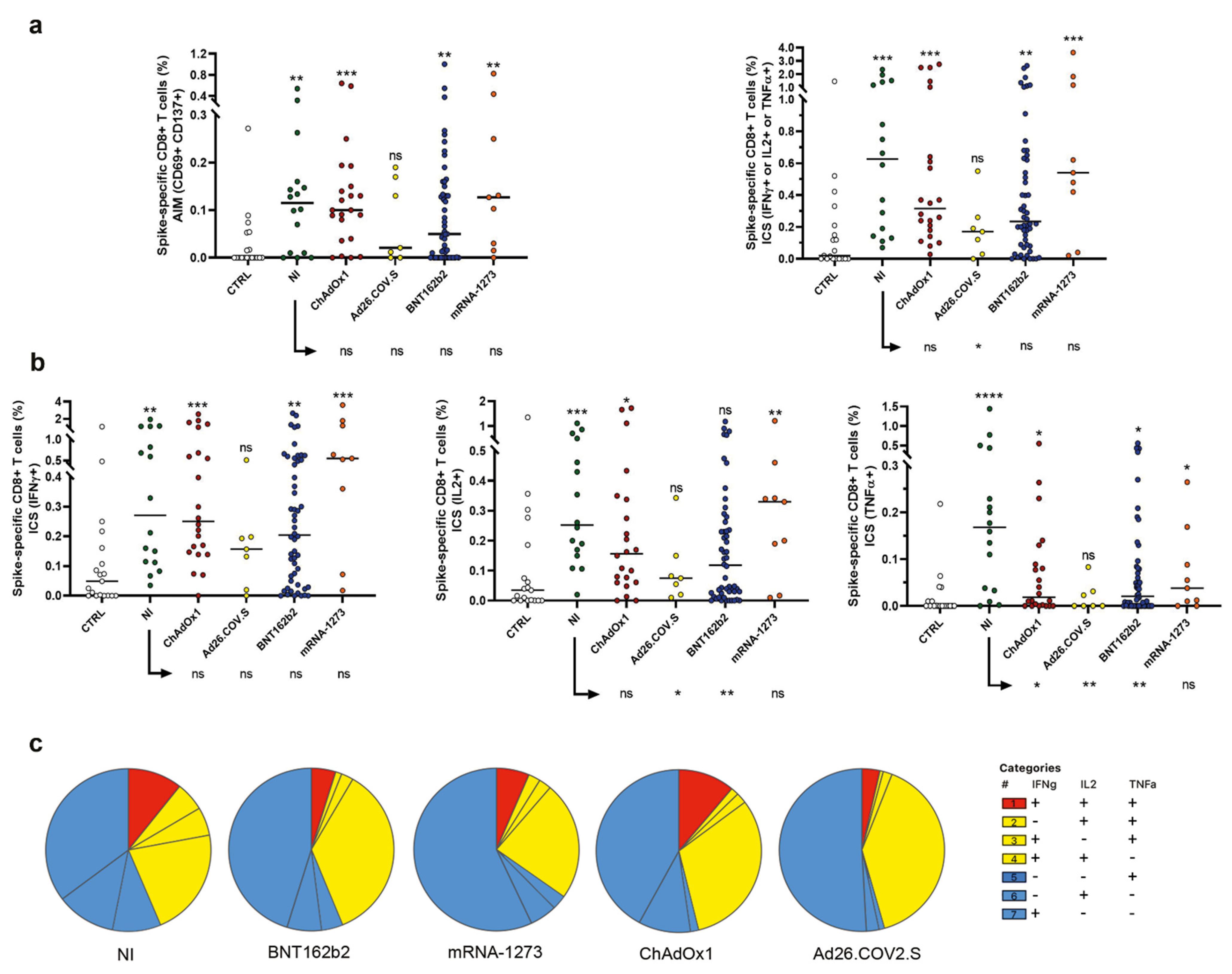
2.3. Evaluation of Spike-Specific CD8+ T Cells
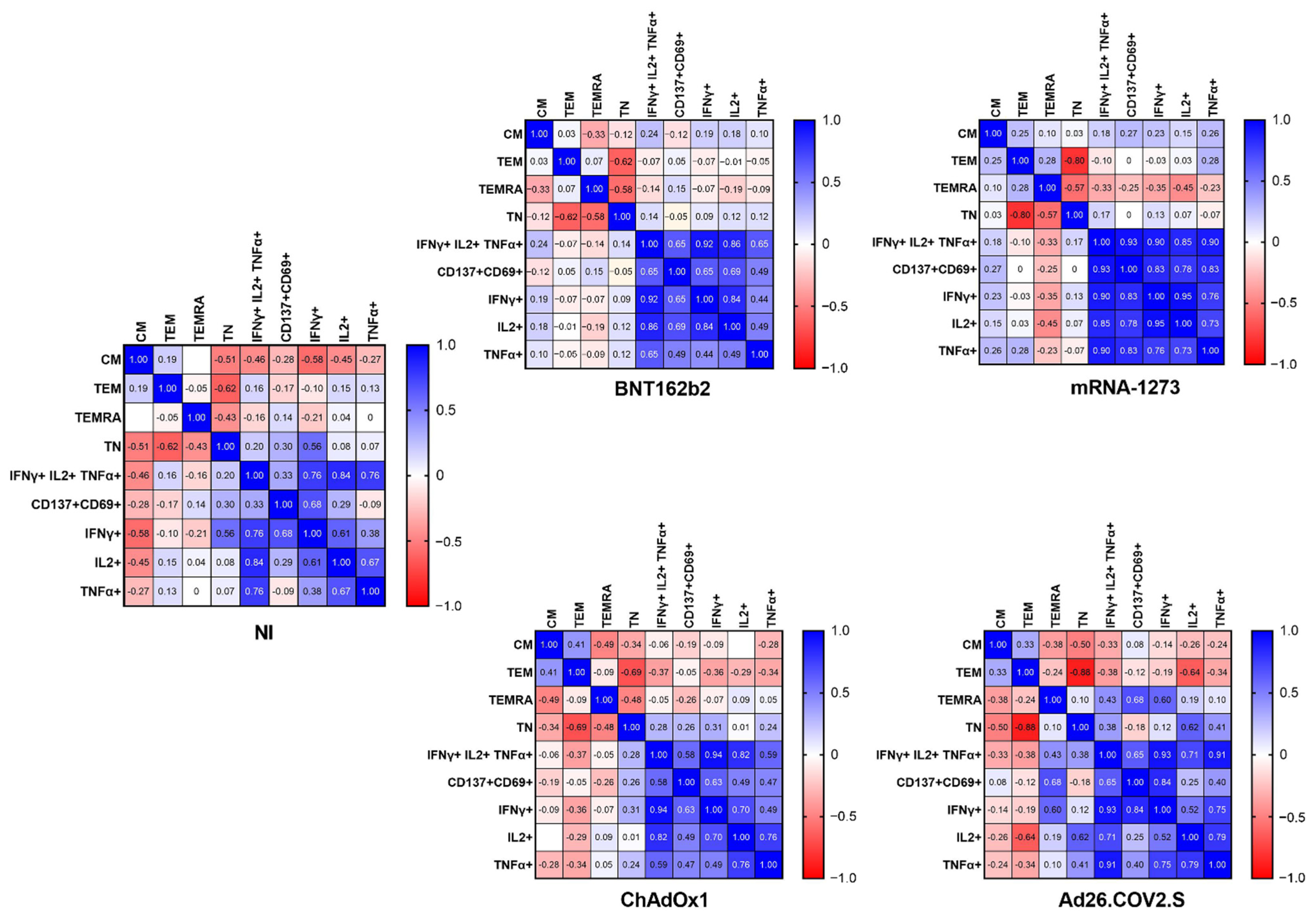
2.4. The Frequency of Naïve, Effector Memory, and Central Memory T Cells Impacts the SARS-CoV-2-Specific Responses
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Anti-S1 Spike IgG Measurement
4.3. PBMC Isolation, Stimulation, and Staining for Flow Cytometry Analysis
4.4. Gating Strategy
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heinz, F.X.; Stiasny, K. Distinguishing Features of Current COVID-19 Vaccines: Knowns and Unknowns of Antigen Presentation and Modes of Action. NPJ Vaccines 2021, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- Hasan, T.; Beardsley, J.; Marais, B.J.; Nguyen, T.A.; Fox, G.J. The Implementation of Mass-Vaccination against SARS-CoV-2: A Systematic Review of Existing Strategies and Guidelines. Vaccines 2021, 9, 326. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and Effectiveness of MRNA BNT162b2 Vaccine against SARS-CoV-2 Infections and COVID-19 Cases, Hospitalisations, and Deaths Following a Nationwide Vaccination Campaign in Israel: An Observational Study Using National Surveillance Data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Rossman, H.; Shilo, S.; Meir, T.; Gorfine, M.; Shalit, U.; Segal, E. COVID-19 Dynamics after a National Immunization Program in Israel. Nat. Med. 2021, 27, 1055–1061. [Google Scholar] [CrossRef]
- Vasileiou, E.; Simpson, C.R.; Shi, T.; Kerr, S.; Agrawal, U.; Akbari, A.; Bedston, S.; Beggs, J.; Bradley, D.; Chuter, A.; et al. Interim Findings from First-Dose Mass COVID-19 Vaccination Roll-out and COVID-19 Hospital Admissions in Scotland: A National Prospective Cohort Study. Lancet 2021, 397, 1646–1657. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An MRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b Vaccines Protect Rhesus Macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Falsey, A.R.; Sobieszczyk, M.E.; Hirsch, I.; Sproule, S.; Robb, M.L.; Corey, L.; Neuzil, K.M.; Hahn, W.; Hunt, J.; Mulligan, M.J.; et al. Phase 3 Safety and Efficacy of AZD1222 (ChAdOx1 NCoV-19) COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 2348–2360. [Google Scholar] [CrossRef]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune Correlates Analysis of the MRNA-1273 COVID-19 Vaccine Efficacy Clinical Trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive Immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Keeton, R.; Richardson, S.I.; Moyo-Gwete, T.; Hermanus, T.; Tincho, M.B.; Benede, N.; Manamela, N.P.; Baguma, R.; Makhado, Z.; Ngomti, A.; et al. Prior Infection with SARS-CoV-2 Boosts and Broadens Ad26.COV2.S Immunogenicity in a Variant-Dependent Manner. Cell Host Microbe 2021, 29, 1611–1619.e5. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e19. [Google Scholar] [CrossRef]
- Rishi, P.; Thakur, K.; Vij, S.; Rishi, L.; Singh, A.; Kaur, I.P.; Patel, S.K.S.; Lee, J.-K.; Kalia, V.C. Diet, Gut Microbiota and COVID-19. Indian J. Microbiol. 2020, 60, 420–429. [Google Scholar] [CrossRef]
- Cromer, D.; Juno, J.A.; Khoury, D.; Reynaldi, A.; Wheatley, A.K.; Kent, S.J.; Davenport, M.P. Prospects for Durable Immune Control of SARS-CoV-2 and Prevention of Reinfection. Nat. Rev. Immunol. 2021, 21, 395–404. [Google Scholar] [CrossRef]
- Feng, S.; Phillips, D.J.; White, T.; Sayal, H.; Aley, P.K.; Bibi, S.; Dold, C.; Fuskova, M.; Gilbert, S.C.; Hirsch, I.; et al. Correlates of Protection against Symptomatic and Asymptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27, 2032–2040. [Google Scholar] [CrossRef]
- Lanuti, P.; Rossi, C.; Cicalini, I.; Pierdomenico, L.; Damiani, V.; Semeraro, D.; Verrocchio, S.; Del Boccio, P.; Evangelista, A.; Sarra, A.; et al. Picture of the Favourable Immune Profile Induced by Anti-SARS-CoV-2 Vaccination. Biomedicines 2021, 9, 1035. [Google Scholar] [CrossRef]
- Rossi, C.; Lanuti, P.; Cicalini, I.; De Bellis, D.; Pierdomenico, L.; Del Boccio, P.; Zucchelli, M.; Natale, L.; Sinjari, B.; Catitti, G.; et al. BNT162b2 MRNA Vaccination Leads to Long-Term Protection from COVID-19 Disease. Vaccines 2021, 9, 1164. [Google Scholar] [CrossRef]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Capone, M.; Vanni, A.; Spinicci, M.; Mencarini, J.; Caporale, R.; Peruzzi, B.; Antonelli, A.; et al. Impaired Immune Cell Cytotoxicity in Severe COVID-19 Is IL-6 Dependent. J. Clin. Investig. 2020, 130, 4694–4703. [Google Scholar] [CrossRef]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and Cellular Immune Memory to Four COVID-19 Vaccines. Cell 2022, 185, 2434–2451.e17. [Google Scholar] [CrossRef]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and Strong Memory CD4+ and CD8+ T Cells Induced by SARS-CoV-2 in UK Convalescent Individuals Following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Jung, J.H.; Rha, M.-S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.-H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-Specific T Cell Memory Is Sustained in COVID-19 Convalescent Patients for 10 Months with Successful Development of Stem Cell-like Memory T Cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Annunziato, F.; Cosmi, L. Hallmarks of Immune Response in COVID-19: Exploring Dysregulation and Exhaustion. Semin. Immunol. 2021, 55, 101508. [Google Scholar] [CrossRef]
- Thakur, P.; Thakur, V.; Kumar, P.; Singh Patel, S.K. Emergence of Novel Omicron Hybrid Variants: BA(x), XE, XD, XF More than Just Alphabets. Int. J. Surg. 2022, 104, 106727. [Google Scholar] [CrossRef]
- Guo, L.; Wang, G.; Wang, Y.; Zhang, Q.; Ren, L.; Gu, X.; Huang, T.; Zhong, J.; Wang, Y.; Wang, X.; et al. SARS-CoV-2-Specific Antibody and T-Cell Responses 1 Year after Infection in People Recovered from COVID-19: A Longitudinal Cohort Study. Lancet Microbe 2022, 3, e348–e356. [Google Scholar] [CrossRef]
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T Cell Responses to SARS-CoV-2 Spike Cross-Recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Ożarowski, M.; Seremak-Mrozikiewicz, A.; Wolski, H.; Wlodkowic, D. The 2020 Race towards SARS-CoV-2 Specific Vaccines. Theranostics 2021, 11, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- Hager, K.J.; Pérez Marc, G.; Gobeil, P.; Diaz, R.S.; Heizer, G.; Llapur, C.; Makarkov, A.I.; Vasconcellos, E.; Pillet, S.; Riera, F.; et al. Efficacy and Safety of a Recombinant Plant-Based Adjuvanted COVID-19 Vaccine. N. Engl. J. Med. 2022, 386, 2084–2096. [Google Scholar] [CrossRef] [PubMed]
- Kalimuddin, S.; Tham, C.Y.L.; Qui, M.; de Alwis, R.; Sim, J.X.Y.; Lim, J.M.E.; Tan, H.-C.; Syenina, A.; Zhang, S.L.; Le Bert, N.; et al. Early T Cell and Binding Antibody Responses Are Associated with COVID-19 RNA Vaccine Efficacy Onset. Med 2021, 2, 682–688. [Google Scholar] [CrossRef]
- Cicalini, I.; Rossi, C.; Natale, L.; Cufaro, M.C.; Catitti, G.; Vespa, S.; De Bellis, D.; Iannetti, G.; Lanuti, P.; Bucci, I.; et al. Passive Immunity to SARS-CoV-2 at Birth Induced by Vaccination in the First Trimester of Pregnancy. Int. J. Environ. Res. Public Health 2021, 18, 12789. [Google Scholar] [CrossRef] [PubMed]
- Cicalini, I.; Del Boccio, P.; Zucchelli, M.; Rossi, C.; Natale, L.; Demattia, G.; De Bellis, D.; Damiani, V.; Tommolini, M.L.; Pizzinato, E.; et al. Validation of the GSP®/DELFIA® Anti-SARS-CoV-2 IgG Kit Using Dried Blood Samples for High-Throughput Serosurveillance and Standardized Quantitative Measurement of Anti-Spike S1 IgG Antibody Responses Post-Vaccination. Vaccines 2022, 10, 514. [Google Scholar] [CrossRef] [PubMed]
- Lanuti, P.; Ciccocioppo, F.; Bonanni, L.; Marchisio, M.; Lachmann, R.; Tabet, N.; Pierdomenico, L.; Santavenere, E.; Catinella, V.; Iacone, A.; et al. Amyloid-Specific T-Cells Differentiate Alzheimer’s Disease from Lewy Body Dementia. Neurobiol. Aging 2012, 33, 2599–2611. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vespa, S.; Simeone, P.; Catitti, G.; Buca, D.; De Bellis, D.; Pierdomenico, L.; Pieragostino, D.; Cicalini, I.; Del Boccio, P.; Natale, L.; et al. SARS-CoV-2 and Immunity: Natural Infection Compared with Vaccination. Int. J. Mol. Sci. 2022, 23, 8982. https://doi.org/10.3390/ijms23168982
Vespa S, Simeone P, Catitti G, Buca D, De Bellis D, Pierdomenico L, Pieragostino D, Cicalini I, Del Boccio P, Natale L, et al. SARS-CoV-2 and Immunity: Natural Infection Compared with Vaccination. International Journal of Molecular Sciences. 2022; 23(16):8982. https://doi.org/10.3390/ijms23168982
Chicago/Turabian StyleVespa, Simone, Pasquale Simeone, Giulia Catitti, Davide Buca, Domenico De Bellis, Laura Pierdomenico, Damiana Pieragostino, Ilaria Cicalini, Piero Del Boccio, Luca Natale, and et al. 2022. "SARS-CoV-2 and Immunity: Natural Infection Compared with Vaccination" International Journal of Molecular Sciences 23, no. 16: 8982. https://doi.org/10.3390/ijms23168982
APA StyleVespa, S., Simeone, P., Catitti, G., Buca, D., De Bellis, D., Pierdomenico, L., Pieragostino, D., Cicalini, I., Del Boccio, P., Natale, L., Owens, T., Khorooshi, R., De Laurenzi, V., Stuppia, L., & Lanuti, P. (2022). SARS-CoV-2 and Immunity: Natural Infection Compared with Vaccination. International Journal of Molecular Sciences, 23(16), 8982. https://doi.org/10.3390/ijms23168982











