Thermodynamic Swings: How Ideal Complex of Cas9–RNA/DNA Forms
Abstract
:1. Introduction
2. Results and Discussion
2.1. ITC Experiments
2.2. Computer Simulations
3. Materials and Methods
3.1. Preparation of Cas9 Enzyme
3.2. Synthesis of Single-Guide RNA
3.3. Isothermal Titration Calorimetry
3.4. Computer Simulation
3.4.1. Complexes Containing Cas9
3.4.2. Nucleic Acid Complexes
3.5. MM-PBSA Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borges, A.L.; Davidson, A.R.; Bondy-Denomy, J. The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs. Annu. Rev. Virol. 2017, 4, 37–59. [Google Scholar] [CrossRef] [PubMed]
- van der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Hruscha, A.; Krawitz, P.; Rechenberg, A.; Heinrich, V.; Hecht, J.; Haass, C.; Schmid, B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 2013, 140, 4982–4987. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Bae, S.; Park, J.; Kim, E.; Kim, S.; Yu, H.R.; Hwang, J.; Kim, J.-I.; Kim, J.-S. Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 2015, 12, 237–243. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef] [Green Version]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palermo, G.; Miao, Y.; Walker, R.C.; Jinek, M.; McCammon, J.A. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl. Acad. Sci. USA 2017, 114, 7260–7265. [Google Scholar] [CrossRef] [Green Version]
- Palermo, G. Structure and Dynamics of the CRISPR-Cas9 Catalytic Complex. J. Chem. Inf. Modeling 2019, 59, 2394–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Erp, P.B.G.; Patterson, A.; Kant, R.; Berry, L.; Golden, S.M.; Forsman, B.L.; Carter, J.; Jackson, R.N.; Bothner, B.; Wiedenheft, B. Conformational Dynamics of DNA Binding and Cas3 Recruitment by the CRISPR RNA-Guided Cascade Complex. ACS Chem. Biol. 2018, 13, 481–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Konermann, S.; Brideau, N.J.; Lotfy, P.; Wu, X.; Novick, S.J.; Strutzenberg, T.; Griffin, P.R.; Hsu, P.D.; Lyumkis, D. Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d. Cell 2018, 175, 212–223.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef] [Green Version]
- Dagdas, Y.S.; Chen, J.S.; Sternberg, S.H.; Doudna, J.A.; Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017, 3, eaao0027. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Sternberg, S.H.; Fei, J.; Doudna, J.A.; Ha, T. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun. 2016, 7, 12778. [Google Scholar] [CrossRef] [Green Version]
- Palermo, G.; Chen, J.S.; Ricci, C.G.; Rivalta, I.; Jinek, M.; Batista, V.S.; Doudna, J.A.; McCammon, J.A. Key role of the REC lobe during CRISPR-Cas9 activation by ‘sensing’, ‘regulating’, and ‘locking’ the catalytic HNH domain. Q. Rev. Biophys. 2018, 51, E9. [Google Scholar] [CrossRef] [Green Version]
- Zhdanova, P.V.; Chernonosov, A.A.; Prokhorova, D.V.; Stepanov, G.A.; Kanazhevskaya, L.Y.; Koval, V.V. Probing the Dynamics of Streptococcus pyogenes Cas9 Endonuclease Bound to the sgRNA Complex Using Hydrogen-Deuterium Exchange Mass Spectrometry. Int. J. Mol. Sci. 2022, 23, 1129. [Google Scholar] [CrossRef] [PubMed]
- Alkan, F.; Wenzel, A.; Anthon, C.; Havgaard, J.H.; Gorodkin, J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 2018, 19, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farasat, I.; Salis, H.M. A Biophysical Model of CRISPR/Cas9 Activity for Rational Design of Genome Editing and Gene Regulation. PLoS Comput. Biol. 2016, 12, e1004724. [Google Scholar] [CrossRef]
- Prozeller, D.; Morsbach, S.; Landfester, K. Isothermal titration calorimetry as a complementary method for investigating nanoparticle-protein interactions. Nanoscale 2019, 11, 19265–19273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demarse, N.A.; Quinn, C.F.; Eggett, D.L.; Russell, D.J.; Hansen, L.D. Calibration of nanowatt isothermal titration calorimeters with overflow reaction vessels. Anal. Biochem. 2011, 417, 247–255. [Google Scholar] [CrossRef]
- Holbrook, J.A.; Capp, M.W.; Saecker, R.M.; Record, M.T. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: Interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry 1999, 38, 8409–8422. [Google Scholar] [CrossRef]
- Srivastava, V.K.; Yadav, R. Isothermal titration calorimetry. In Data Processing Handbook for Complex Biological Data Sources; Elsevier: Amsterdam, The Netherlands, 2019; pp. 125–137. ISBN 9780128165485. [Google Scholar]
- Rozners, E.; Pilch, D.S.; Egli, M. Calorimetry of Nucleic Acids. Curr. Protoc. Nucleic Acid Chem. 2015, 63, 7.4.1–7.4.12. [Google Scholar] [CrossRef]
- Bell, D.R.; Weber, J.K.; Yin, W.; Huynh, T.; Duan, W.; Zhou, R. In silico design and validation of high-affinity RNA aptamers targeting epithelial cellular adhesion molecule dimers. Proc. Natl. Acad. Sci. USA 2020, 117, 8486–8493. [Google Scholar] [CrossRef]
- Ka, D.; Jang, D.M.; Han, B.W.; Bae, E. Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2. Nucleic Acids Res. 2018, 46, 9805–9815. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Gao, A.; Zhan, Q.; Wang, Y.; Feng, H.; Liu, S.; Gao, G.; Serganov, A.; Gao, P. Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins. Mol. Cell 2019, 74, 296–309.e7. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.; Jeong, M.; Ka, D.; Han, M.; Kim, N.-K.; Bae, E.; Suh, J.-Y. Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor. Sci. Rep. 2018, 8, 3883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavaluzzi, M.J.; Borer, P.N. Revised UV extinction coefficients for nucleoside-5’-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004, 32, e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallansrud, G.; Ward, B. A comparison of measured and calculated single- and double-stranded oligodeoxynucleotide extinction coefficients. Anal. Biochem. 1996, 236, 134–138. [Google Scholar] [CrossRef]
- Lomzov, A.A.; Pyshnaya, I.A.; Ivanova, E.M.; Pyshnyi, D.V. Thermodynamic parameters for calculating the stability of complexes of bridged oligonucleotides. Dokl. Biochem. Biophys. 2006, 409, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Lomzov, A.A.; Pyshnyi, D.V. Calculating Melting Temperature of Native and Modified Oligonucleotide Complexes at Various Cation Concentrations with the Use of Enhanced Counterion Condensation Model. Chem. Biol. Med. Phys. 2008, 3, 61–75. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [Green Version]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, N.; Nakano, S.; Katoh, M.; Matsumura, A.; Nakamuta, H.; Ohmichi, T.; Yoneyama, M.; Sasaki, M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 1995, 34, 11211–11216. [Google Scholar] [CrossRef]
- SantaLucia, J.; Allawi, H.T.; Seneviratne, P.A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 1996, 35, 3555–3562. [Google Scholar] [CrossRef]
- Golyshev, V.M.; Pyshnyi, D.V.; Lomzov, A.A. Calculation of Energy for RNA/RNA and DNA/RNA Duplex Formation by Molecular Dynamics Simulation. Mol. Biol. 2021, 55, 1030–1044. [Google Scholar] [CrossRef]
- Anders, C.; Jinek, M. In Vitro Enzymology of Cas9. In The Use of CRISPR/Cas9, ZFNs, and TALENs in Generating Site-Specific Genome Alterations; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1–20. ISBN 9780128011850. [Google Scholar]
- O’Reilly, D.; Kartje, Z.J.; Ageely, E.A.; Malek-Adamian, E.; Habibian, M.; Schofield, A.; Barkau, C.L.; Rohilla, K.J.; DeRossett, L.B.; Weigle, A.T.; et al. Extensive CRISPR RNA modification reveals chemical compatibility and structure-activity relationships for Cas9 biochemical activity. Nucleic Acids Res. 2019, 47, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Šali, A. Modeller: Generation and Refinement of Homology-Based Protein Structure Models. Methods Enzymol. 2003, 374, 461–491. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Cheatham, T.E.; Case, D.A. Twenty-five years of nucleic acid simulations. Biopolymers 2013, 99, 969–977. [Google Scholar] [CrossRef]
- Case, D.; Belfon, K.; Ben-Shalom, S.; Brozell, S.; Cerutti, D.; CheathamIII, T.; Cruzeiro, V.; Darden, T.; Duke, R.; Giambasu, G.; et al. Amber20; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
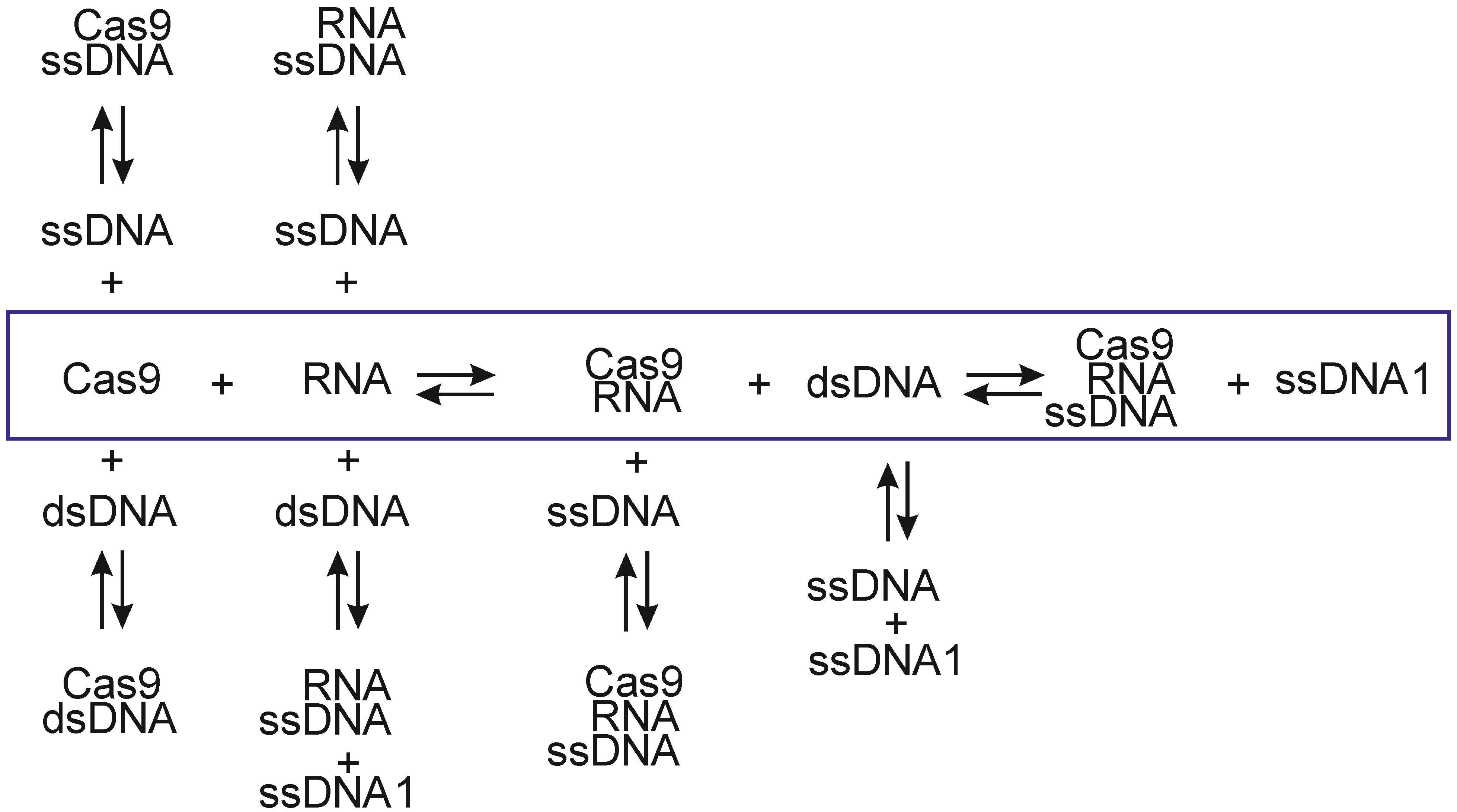
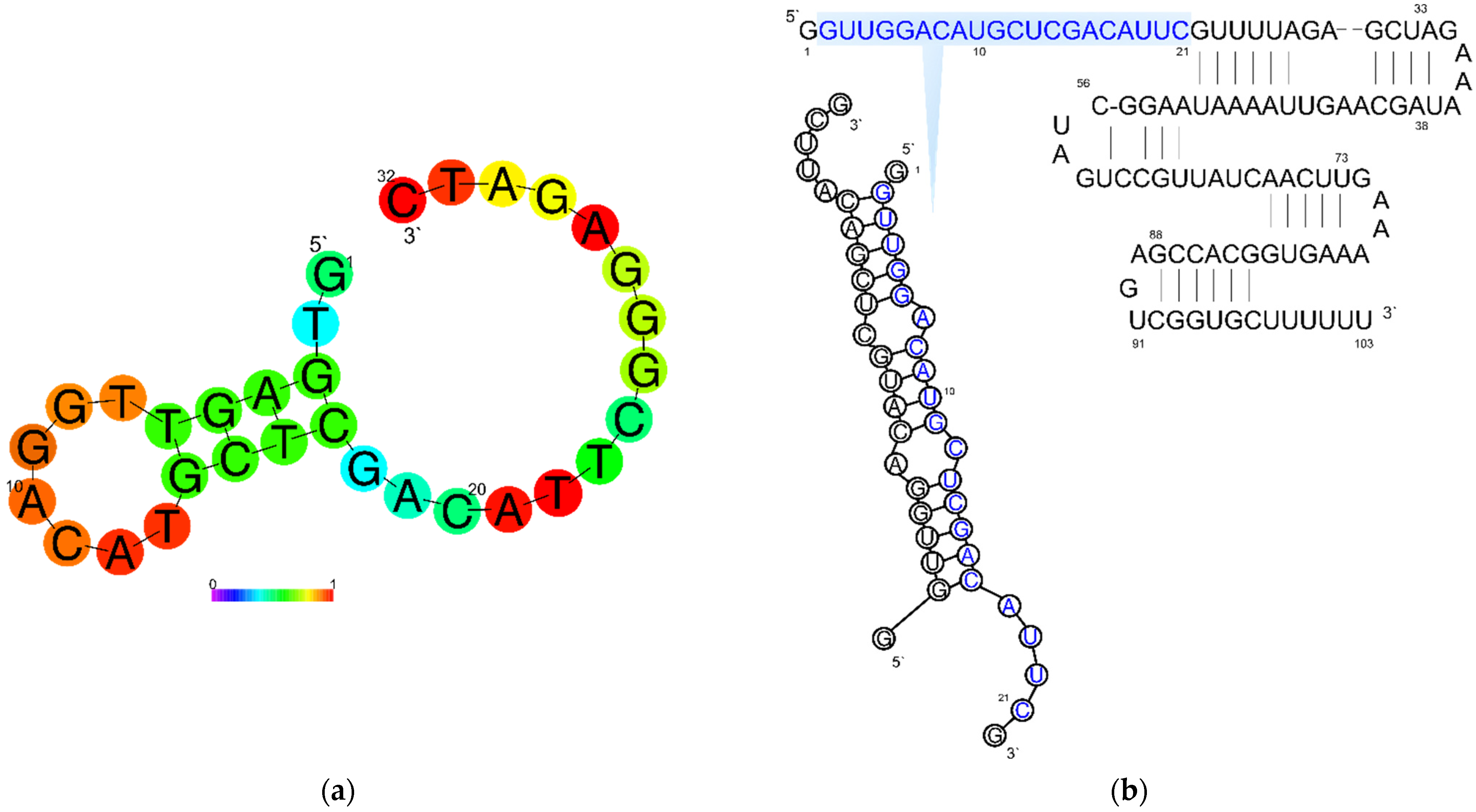

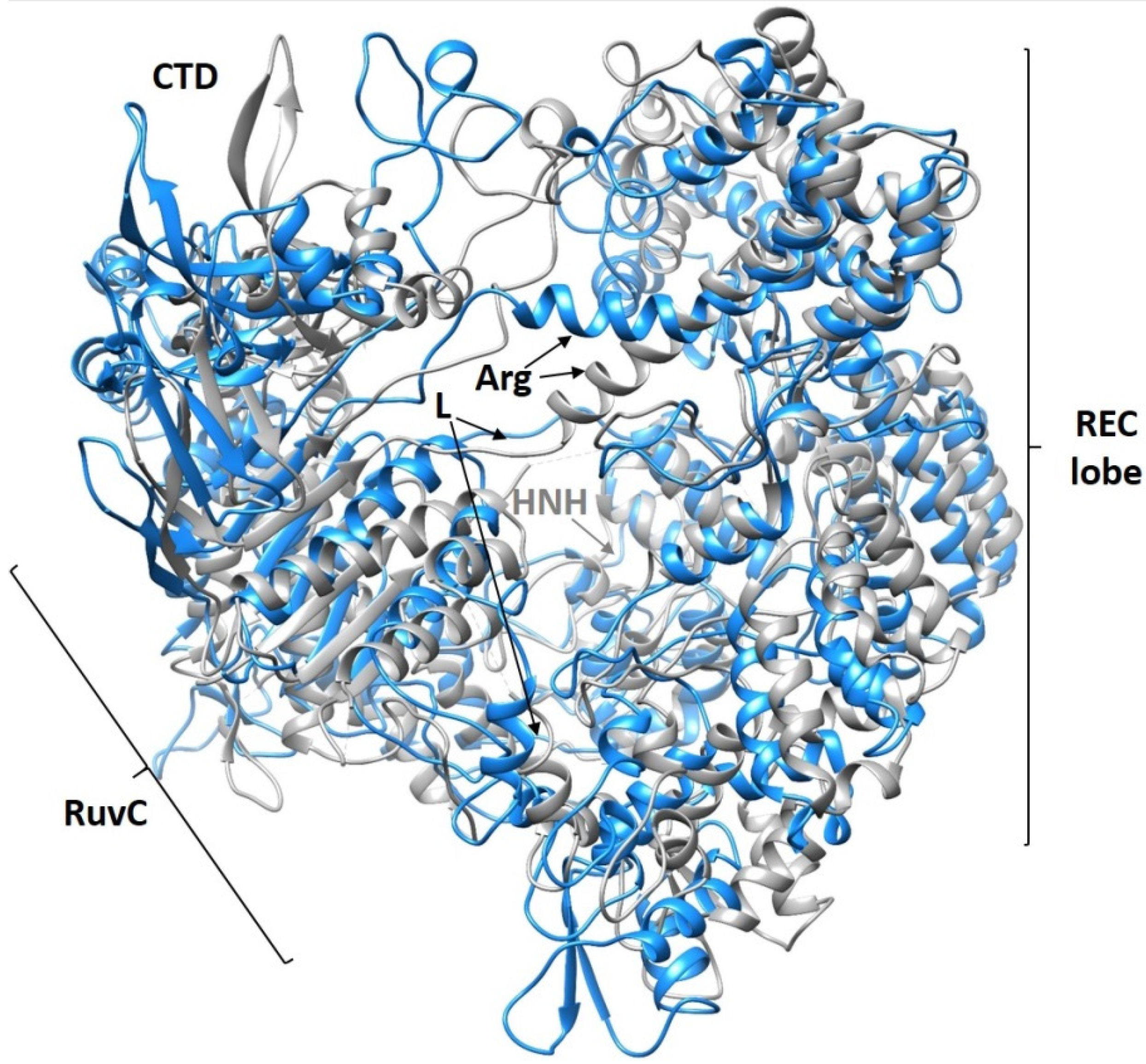
| Cell | Syringe | Kd, M | n | ΔH°, kcal/mol | ΔS°, kcal/mol·K | ΔG°(25 °C), kcal/mol |
|---|---|---|---|---|---|---|
| Substance, μM | Substance, μM | |||||
| buffer | RNA, 50 μM | 1.62 × 10−6 | - | −95.25 | −346.0 | 7.90 |
| buffer | RNA, 10 μM | ˗ | ˗ | ˗ | ˗ | ˗ |
| buffer | dsDNA, 25 μM | ˗ | ˗ | ˗ | ˗ | ˗ |
| buffer | ssDNA, 20 μM | ˗ | ˗ | ˗ | ˗ | ˗ |
| RNA,10 μM | dsDNA, 25 μM | ˗ | ˗ | ˗ | ˗ | ˗ |
| RNA, 2 μM | ssDNA, 20 μM | 6.36 × 10−7 ± 2.43 × 10−7 | 0.11 ± 0.14 | −429.2 ± 542.4 | −1411 | −8.454 |
| ssDNA, 2 μM | ssDNA1, 20 μM | 2.09 × 10−7 ± 0.67 × 10−7 | 0.83 ± 0.05 | −212.2 ± 17.7 | −681.1 | −9.112 |
| Cas9, 10 μM | ssDNA, 20 μM | ˗ | ˗ | ˗ | ˗ | ˗ |
| Cas9, 5 μM | dsDNA, 25 μM | ˗ | ˗ | ˗ | ˗ | ˗ |
| Cas9 *, 5 μM | RNA, 62.5 μM | 2.21 × 10−6 ± 0.67 × 10−6 | 2.10 ± 0.12 | −399.10 ± 39.12 | −1313 | −7.72 |
| Cas9, 5 μM RNA, 1 μM | ssDNA, 10 μM | 9.80 × 10−9 ± 1.87 × 10−8 | 0.67 ± 0.07 | −49.27 ± 8.73 | −128.60 | −10.93 |
| Cas9 **, 6 μM RNA, 1 μM | dsDNA, 25 μM | 1.02 × 10−7 ± 5.90 × 10−7 | 0.76 ± 0.68 | −45.08 ± 302.4 | −119.02 | −9.54 |
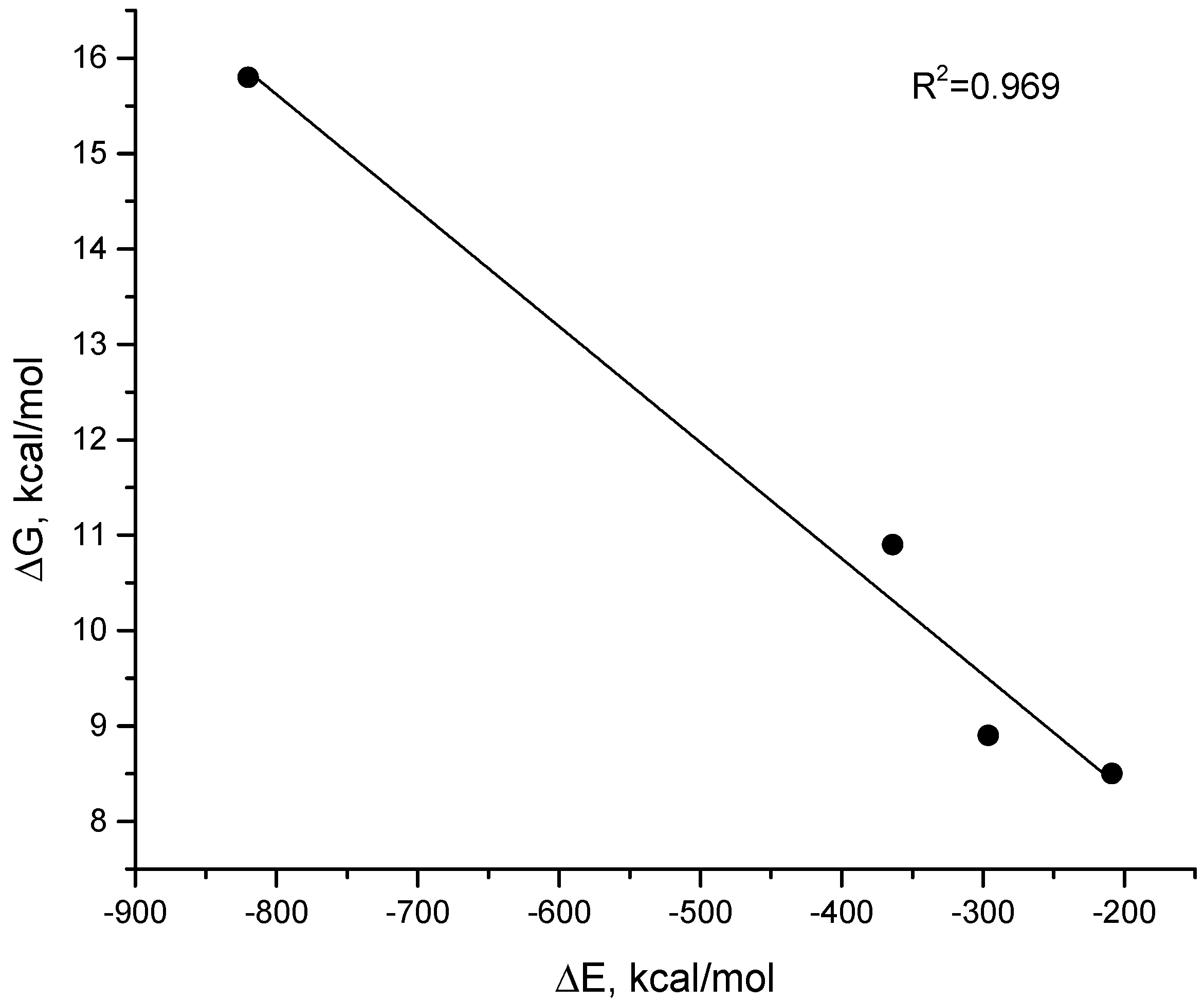
| Receptor | Ligand | MM-GBSA | MM-PBSA | |
|---|---|---|---|---|
| ΔE, kcal/mol | ΔE, kcal/mol | |||
| Cas9–RNA/DNA | Cas9–RNA | ssDNA | −363.9 ± 0.2 | −149.0 ± 0.2 |
| Cas9–RNA | Cas9 | RNA | −820.0 ± 0.3 | −948.2 ± 0.4 |
| RNA/DNA | RNA | ssDNA | −208.7 ± 0.1 | 6.1 ± 0.1 |
| dsDNA, 32 nt | ssDNA | ssDNA1 | −296.3 ± 0.1 | −163.7 ± 0.1 |
| dsDNA, 24 nt | ssDNA | ssDNA1 | −221.3 ± 0.1 | −140.7 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhdanova, P.V.; Lomzov, A.A.; Prokhorova, D.V.; Stepanov, G.A.; Chernonosov, A.A.; Koval, V.V. Thermodynamic Swings: How Ideal Complex of Cas9–RNA/DNA Forms. Int. J. Mol. Sci. 2022, 23, 8891. https://doi.org/10.3390/ijms23168891
Zhdanova PV, Lomzov AA, Prokhorova DV, Stepanov GA, Chernonosov AA, Koval VV. Thermodynamic Swings: How Ideal Complex of Cas9–RNA/DNA Forms. International Journal of Molecular Sciences. 2022; 23(16):8891. https://doi.org/10.3390/ijms23168891
Chicago/Turabian StyleZhdanova, Polina V., Alexander A. Lomzov, Daria V. Prokhorova, Grigory A. Stepanov, Alexander A. Chernonosov, and Vladimir V. Koval. 2022. "Thermodynamic Swings: How Ideal Complex of Cas9–RNA/DNA Forms" International Journal of Molecular Sciences 23, no. 16: 8891. https://doi.org/10.3390/ijms23168891
APA StyleZhdanova, P. V., Lomzov, A. A., Prokhorova, D. V., Stepanov, G. A., Chernonosov, A. A., & Koval, V. V. (2022). Thermodynamic Swings: How Ideal Complex of Cas9–RNA/DNA Forms. International Journal of Molecular Sciences, 23(16), 8891. https://doi.org/10.3390/ijms23168891






