1. Introduction
MicroRNAs (miRNAs) are endogenous non-coding RNA molecules 19–23 nucleotides in length that are important for the regulation of gene expression and various cellular processes [
1]. They are considered promising diagnostic and prognostic biomarkers for different pathological conditions, especially for various types of cancer [
2,
3]. MicroRNAs are commonly isolated from tissues, cells, and a wide variety of body fluids, such as plasma, serum, urine, saliva, or tears [
4,
5,
6]. However, their small size, high sequence homology, and extremely low concentrations in real complex samples make the analysis of miRNAs challenging [
7,
8]. It is valuable to separate miRNAs from the mixture of long RNA molecules, DNA, abundant proteins, lipids, and other contaminants present in the biological matrix to prevent their negative influence on subsequent analytical steps such as polymerase chain reaction (PCR), next-generation sequencing, or microarray [
9].
In the last few decades, many methods for miRNA isolation have been developed from conventional DNA and RNA extraction methods, including phenol/chloroform-based extraction (e.g., TRIzol Reagent) or silica-based solid-phase extraction (e.g., mirVana or miRNeasy) [
10]. Commercial sets for miRNA isolation and purification often use a helpful liquid–liquid extraction (LLE) method; however, toxic chemicals such as phenol and chloroform are utilized (e.g., all above-mentioned sets), which are hazardous to work with and can cause contamination within the final product of RNA isolation, affecting subsequent amplification or sequencing steps [
11]. In such cases, optimization of the extraction protocol is necessary and improves the quality of the isolated product; however, additional extraction and washing steps are needed [
12]. Solid-phase extraction methods, which are based on specific and/or non-specific interactions between the analyte and solid-phase material, offer a simple and efficient alternative for nucleic acid isolation, as well as a way of replacing harmful organic solvent in an assay [
13]. The most common, silicone dioxide resin packed in a column or in the form of a membrane, utilizes adsorption of negatively charged nucleic acids on the positively charged silicone dioxide resin. This is performed in the presence of chaotropic agents, and adsorbed nucleic acids are then eluted by applying a solution with a low salt concentration [
14].
Currently, new approaches using various micro- and nanomaterials are being investigated with the aim of improving the yields and purity of extracted miRNA. Such materials offer unique characteristics, e.g., a high surface-to-volume ratio and useful magnetic, optical, or biological properties. Several studies have reported advanced nanomaterials suitable for both DNA and RNA isolation, such as graphene oxide nanoplatelets, zinc oxide nanowires and nanotubes, carbon nanotubes, and magnetite nanoparticles [
15,
16,
17,
18,
19]. Another prospective material is TiO
2—a cheap and easily available material in various commercial preparations often adopted for chromatography purposes. In proteomics, TiO
2 materials are routinely used for selective enrichment of phosphorylated peptides due to the strong interactions between the TiO
2 surface and phosphate groups of phosphopeptides in harsh acidic environments. Our previous results demonstrated that the TiO
2 form, structure, and surface decoration can also significantly affect the affinity and selectivity for phosphorylated molecules [
20]. Similarly, the TiO
2 surface was used for effective interaction with DNA under strong acidic conditions (pH 2) as shown by Amano et al. [
21]. On the other hand, the TiO
2-based miRNA isolation processes described by Jimenez et al. made use of self-prepared TiO
2 nanofibers and only high concentrations of strong chaotropic salts (similar to SiO
2 isolation protocols) without a low pH buffer system. This study demonstrated miRNA isolation from serum or cell lysate spiked with cel-mir-54 standard with 18.0% miRNA recovery. However, the described protocol was not size-selective, and a broad size range of RNA molecules under 500 bases was isolated [
22]. These findings confirm that the composition of the binding buffer (molarity, pH, etc.) strongly influences the affinity of DNA or RNA of various lengths to TiO
2 material.
In our work, we aimed to develop an accessible and highly selective method for short RNA isolation (miRNA) and to avoid the extraction step with toxic phenol and chloroform.
2. Results
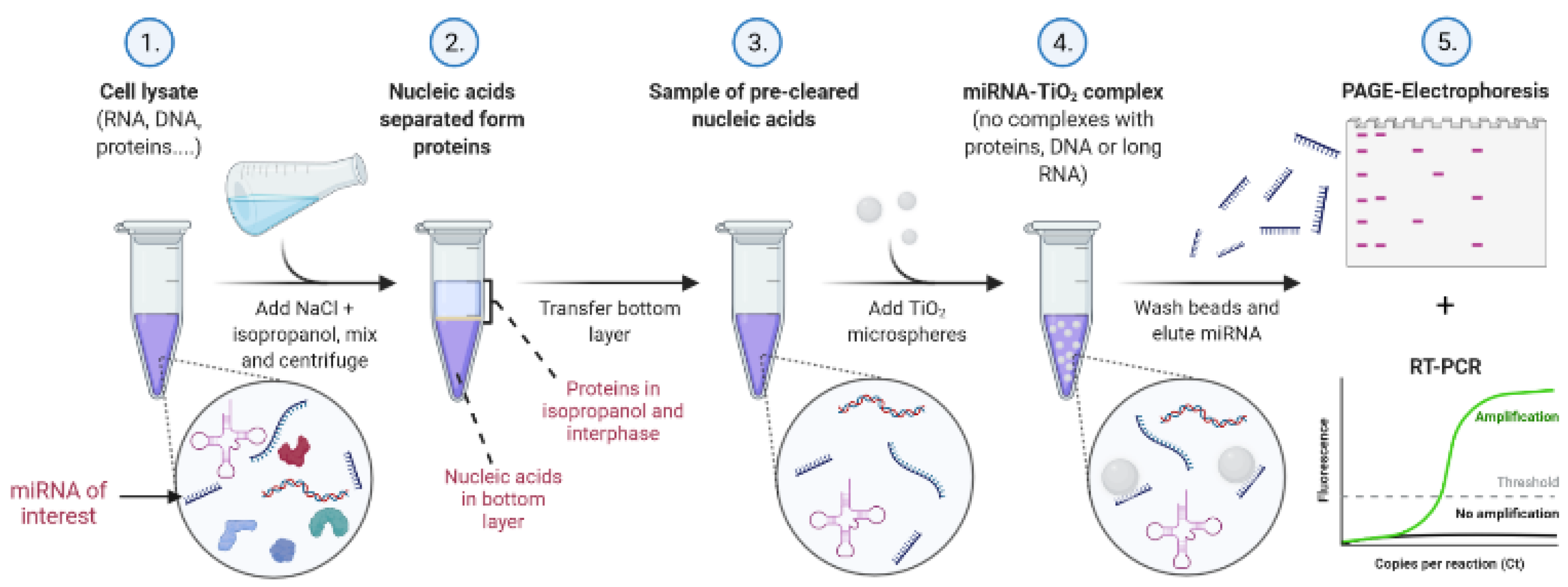
Our protocol for TiO
2-based miRNA isolation consists of a liquid–liquid extraction method using isopropanol/water/NaCl, followed by solid-phase extraction with porous 10 µm TiO
2 microspheres under low pH in acetate, MES, or glycine buffer. The final elution of isolated miRNA is performed with Na
2HPO
4, which is compatible with PCR. The scheme of the experimental procedure, including specifically optimized conditions, is illustrated in
Figure 1.
The pilot experiments showed the affinity of miRNAs towards TiO
2 material; however, the contamination with RNAs of various sizes was observed. All protocol steps were then optimized to increase the purity of the isolated miRNA. First, we prepared a model RNA mixture consisting of genomic RNA (isolated from Jurkat cells with TRIzol Reagent) and oligo RNA standard hsa-miR-18a-3p. This mixture—containing 5.28 µg of pre-isolated genomic RNA and 0.55 µg of oligo RNA (23-mer)—was then used to optimize the binding conditions. Several washing and binding buffers within the pH range of 2.4–6 were tested for this purpose.
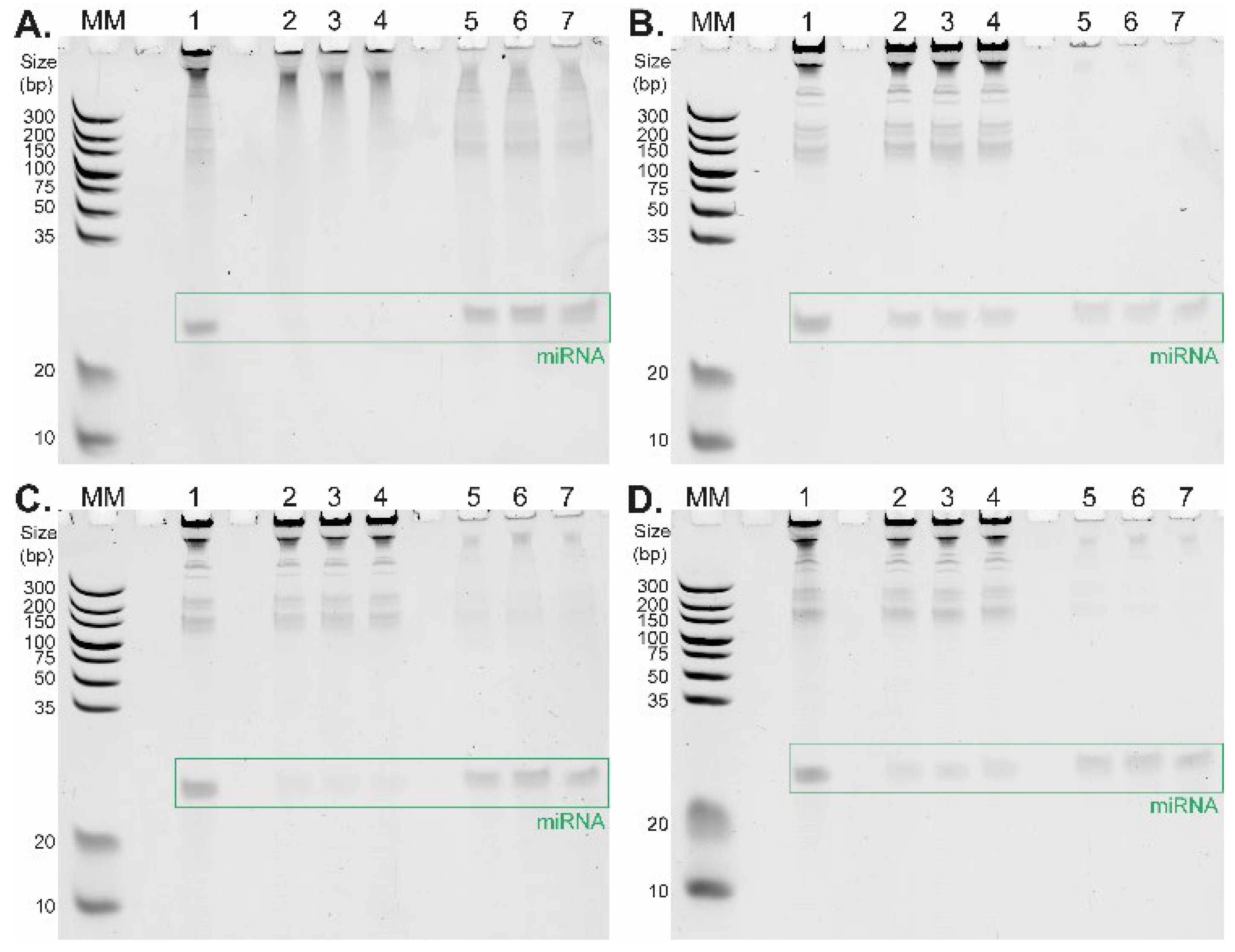
Figure 2 depicts the differences between TiO
2-based short RNA isolation procedures under four distinct binding conditions differing in washing and binding buffer composition tested in triplicate. The corresponding data are summarized in
Table 1. The selectivity of TiO
2 microspheres for short RNAs strongly depends on the pH of the selected buffer. The highest affinity of RNA molecules to TiO
2 microspheres is present under strong acidic conditions (glycine/HCl buffer, pH 2.4,
Figure 2A,
Table 1), where a large amount of RNA molecules remains in the elution fractions (lanes 5, 6, 7). This is in line with the results of the previously reported DNA–TiO
2 interaction study [
21]. Under these conditions, the relative yields of miRNA (23-mer) determined in three replicates were 57.2%, 68.9%, and 69.6%, respectively (
Table 1). Such a high yield outperformed results from the previously published study focused on TiO
2-based miRNA isolation, where only 18.0% miRNA recovery was achieved [
22]. When the pH of the washing and binding buffer was set to 4 and 4.4 (
Figure 2B: MES buffer, pH 4.0;
Figure 2C,D: acetate buffer, pH 4.4), long RNA molecules remained in the supernatant fractions (lanes 2, 3, 4), whereas short RNAs were extracted from the initial samples (lanes 5, 6, 7).
An interesting effect was observed with a relatively low concentration (5 mM) of citric acid in the washing and binding buffer which significantly increased the selectivity of TiO
2 microspheres for short RNAs. Although the total miRNA yield was slightly reduced, the main advantage of this approach was the extremely high purity of miRNA in all elution fractions, as shown in
Figure 2B,D, lanes 5, 6, and 7. The positive effect on miRNA isolation selectivity is even more evident when we compare the isolation of miRNA in an acetate buffer containing citric acid (relative yields were 25.4%, 30.6%, and 29.3%,
Figure 2D,
Table 1) with miRNA isolation in an acetate buffer without citric acid (relative yields were 28.0%, 41.0%, and 34.5%,
Figure 2C,
Table 1). The combination of the acetate buffer with the citric acid addition (5 mM) provides higher relative yields for miRNA isolation compared to the application of the MES buffer with the citric acid (5 mM), with corresponding relative yields of 26.0%, 18.9%, and 24.9% (
Table 1). It was important to find an optimal buffer for cell lysis that would also be compatible with subsequent TiO
2-based miRNA isolation. For this purpose, the acetate buffer system proved to be more robust and easier to work with than the MES buffer. As demonstrated, TiO
2 microspheres successfully isolated short RNAs in the presence of all four buffer systems. In all cases, we were able to obtain high selectivity for short RNAs at higher yields compared to the previously published protocol employing neutral pH for binding conditions [
22].
To accomplish the isolation of miRNA from highly complex samples such as cells, the unique phenol/chloroform-free lysis and extraction protocol was developed. All steps prior to the TiO
2-based miRNA isolation from cells are schematically shown in
Figure 3. For the evaluation of miRNA isolation effectivity and selectivity, the initial sample of Jurkat cells was spiked with oligo RNA standard (23-mer). The cells were lysed with lysis solution (100 mM sodium acetate, 2% Triton X-100, 0.1% diethyl pyrocarbonate (DEPC)).
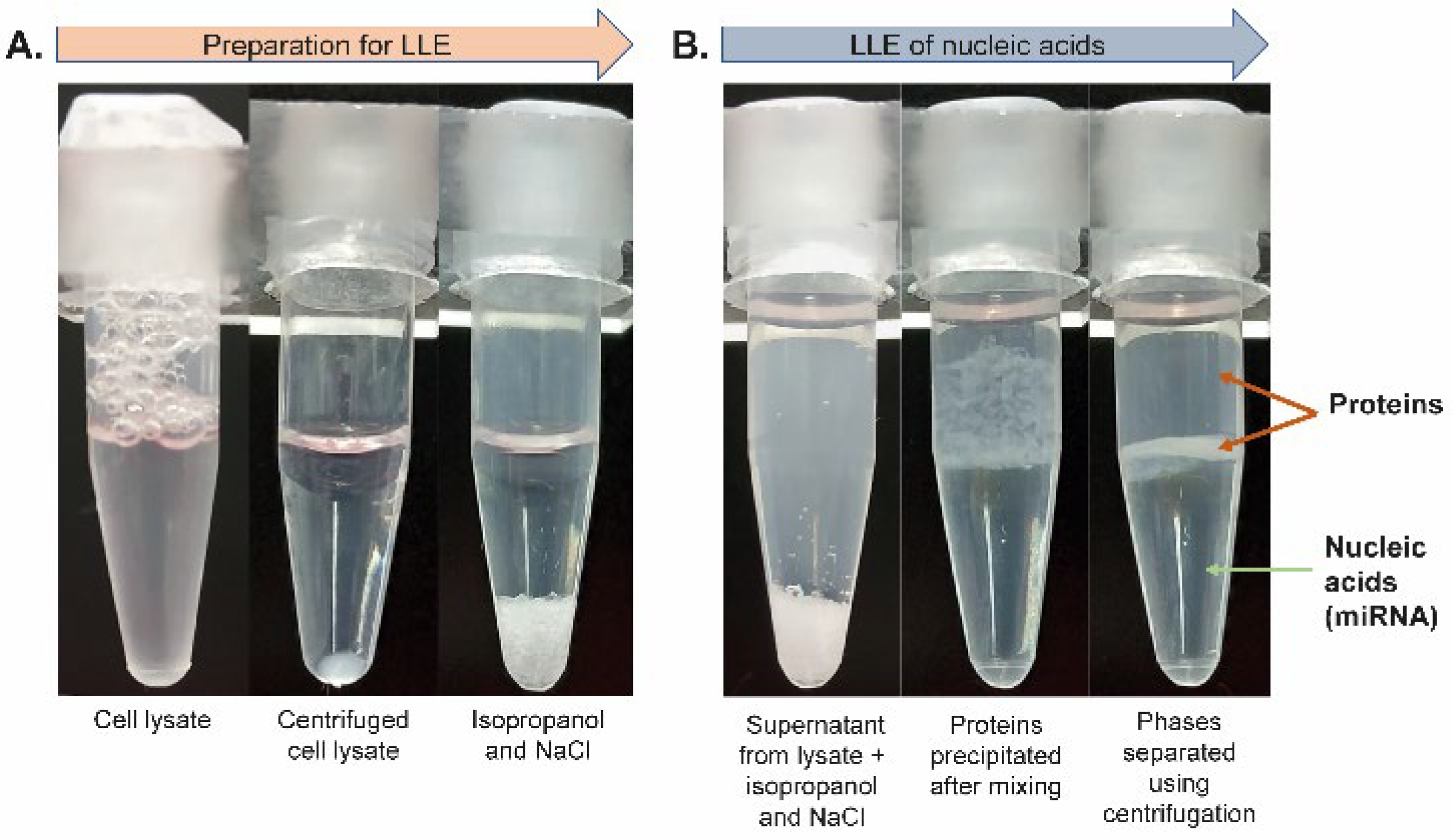
The lysate was centrifuged (see the vial before and after centrifugation in
Figure 3A) and subjected to LLE. The supernatant was transferred into a tube containing isopropanol and NaCl (see
Figure 3A). The mixture was then gently rotated to gradually dissolve NaCl in acetate buffer, which led to the separation of phases, where the top phase was organic and the lower phase was aqueous (see
Figure 3B). The proteins originating from the cell lysate were focused (concentrated) in the interphase after short centrifugation, as shown in
Figure 3B. The bottom aqueous phase containing RNA molecules was transferred into a clean microtube and acetic acid was added to adjust the pH to ~4.4 (checked with a microelectrode-equipped pH meter). Next, the aqueous phase containing RNA was added to 1 mg of TiO
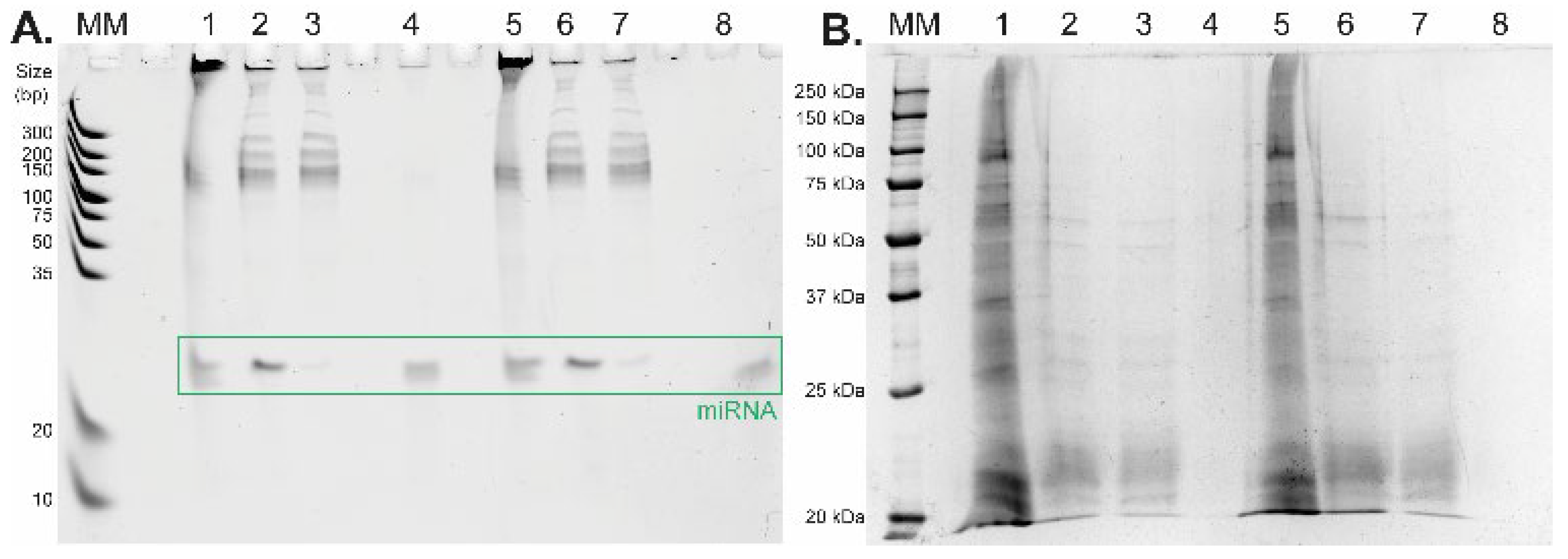
2 microspheres for further short RNA purification and isolation applying the general isolation protocol with the acetate buffer system. The amount of oligo RNA standard and sample composition during the whole isolation process was monitored by urea-PAGE. The results of the urea-PAGE analysis of all fractions collected from the whole isolation procedure are shown in
Figure 4A (the relative yields for miRNA standard were 48.1% and 68.3%,
Table 2). Besides the high yield of miRNA, the main advantage of the presented results lies in the high purity of the target short non-coding RNA after its isolation from the complex mixture—see lanes 4 and 8 (
Figure 4A). To show the purity of the eluted miRNAs, all collected fractions were also analyzed by Laemmli-SDS-PAGE electrophoresis with protein visualization by a silver staining method [
23,
24]. As shown in
Figure 4B, during the LLE, most of the proteins from the cell lysate were removed from the sample (lanes 1 and 5 containing the original cell lysate vs. lanes 2 and 6 containing the bottom aqueous phase). The rest of the proteins were then removed from the solution, and the short RNA molecules were obtained by TiO
2-based isolation during the second part of the protocol. The residual proteins are visible in the supernatant fractions after TiO
2 miRNA purification (lanes 3 and 7), whereas no proteins were detected in the elution fractions (lanes 4 and 8).
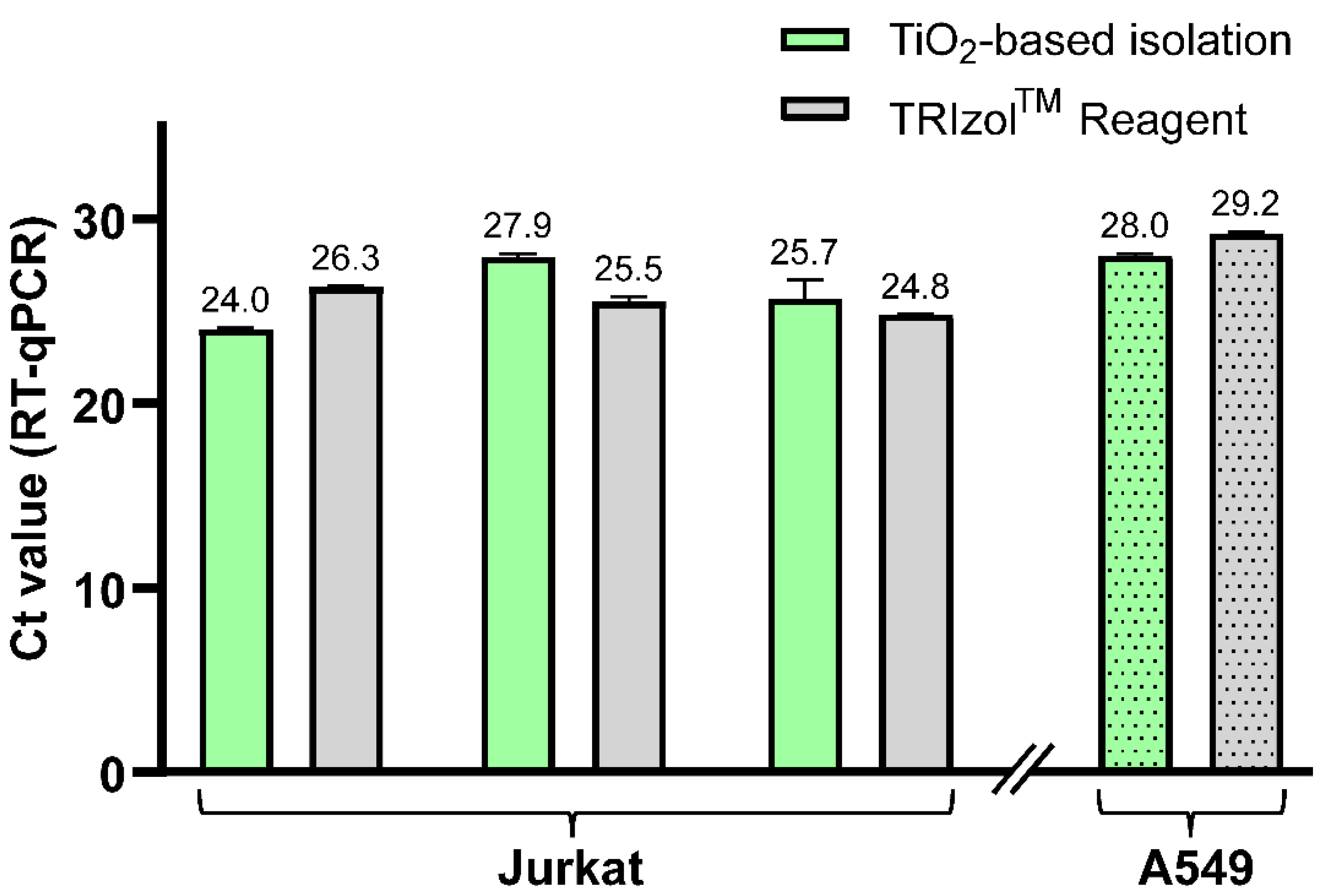
To further confirm the feasibility of this new protocol, we performed the isolation of hsa-miR-18a-3p from two types of cell lines without any added standards. Hence, we show the ability of our protocol to analyze naturally occurring miRNA in cells. The TiO
2-based method including newly developed LLE was utilized for isolation of short RNA from both Jurkat cells (three samples) and A549 cells (one sample) and compared to RNA isolation using TRIzol Reagent. Elution fractions were analyzed by RT-qPCR in triplicate, and the results are summarized in
Figure 5. The results show that we were able to isolate hsa-miR-18a-3p from both cell lines and that the newly developed TiO
2-based method provides comparable yields to the commercial TRIzol Reagent isolation method, which is demonstrated by similar Ct values of RT-qPCR analysis. This also confirms the compatibility of the whole protocol, and mainly Na
2HPO
4 used for the elution of short RNA from TiO
2 microspheres, with RT-PCR.
3. Discussion
Our results from experiments using model RNA mixture shown in
Figure 2 and
Figure 4 clearly demonstrated the benefits of using TiO
2 microspheres for short RNA isolation. They showed that the selective isolation of short RNA molecules is pH-dependent and the selectivity of miRNA isolation can be effectively modulated by changing the binding conditions. The isolation principle probably lies in the interaction of phosphate moieties of RNA with the TiO
2 surface under an acidic environment. This strong interaction in combination with the porous structure of TiO
2 particles (Titansphere, TiO) allowed selective binding of short RNA molecules, most likely due to steric hindrance of particle pores.
Moreover, the selectivity of TiO
2 toward biomolecules containing negatively charged phosphate moieties can be modulated by hydroxy acids used as excluders, which allows us to further improve selectivity. This principle was also described for the extraction of proteins containing phosphate functional groups [
25]. In our experiments, we tested several potential excluders, including citric acid, lactic acid, dihydroxybenzoic acid, glutamic acid, glycolic acid, tartaric acid, salicylic acid, 3-hydroxypicolinic acid, and sodium oxalate. However, except for citric acid (
Figure 2B,D), the excluders did not have any positive effect on isolation efficiency. This could be explained by the fact that only citric acid comprises one α-position hydroxyl group and three carboxyl groups and thus contains at least seven potential donor sites capable of coordinating metal ions and can interact with TiO
2 more strongly than other tested carboxylic acids. For applications where a high selectivity for short miRNAs is not required, the relative yields of miRNA isolation can be increased up to 41% just by omitting the citric acid from the washing and binding buffer.
For routine applications analyzing highly complex biological materials, it was necessary to adapt the protocol to extract miRNAs. Therefore, the robustness and efficiency of the protocol were tested with the whole cell lysate. For this, it was important to find an optimal buffer for cell lysis that would also be compatible with subsequent TiO
2-based miRNA isolation. For this purpose, the acetate buffer system proved to be more robust and easier to work with than the MES buffer. The protocol was also extended for the LLE method using an isopropanol/water/NaCl system (
Figure 3), followed by solid-phase extraction using TiO
2 microspheres to effectively separate miRNAs from long RNA molecules.
Different organic solvents (miscible and immiscible with water) were tested for optimal protein removal and miRNA isolation, including acetonitrile (ACN), isopropanol, acetone, ethyl acetate, and n-hexane. When using immiscible or partially miscible solvents, RNA purification from proteins was poor (e.g., n-hexane). Isopropanol was selected as the best solution because its application together with NaCl resulted in almost complete phase separation. This was in direct contrast with the use of acetone or ACN, which stayed partially mixed within the aqueous solution after centrifugation. The amount of NaCl is also important for a proper phase separation, because at lower concentrations (less than 4 M), the phase separation was incomplete.
For the isolation of a wider RNA size range with a higher yield, it is also possible to use a glycine buffer system. The pH of the lysis solution has to be adjusted to 6.4 using NaOH solution. After LLE, the pH of the acquired aqueous phase is lowered to ~2.4 by adding HCl before continuing with the general protocol of TiO2-based RNA isolation with a glycine buffer system. For effective cell lysis and LLE of RNA, it is necessary to keep the pH of the lysis solution approximately neutral, which prevents the loss of RNA molecules within the lysis procedure and exclusion from the aqueous phase into the interphase during the process of phase separation within LLE.
Our results unequivocally confirmed that this protocol combining a MES or acetate buffer containing citric acid and TiO
2 microparticles can be applied for routine miRNA extraction from eukaryotic cells with excellent purity of the final product. This protocol could also be used for the simple separation of short RNAs from a mixture of RNAs differing in chain length, e.g., from pre-isolated genomic RNA. Glycine and acetate buffers are ideal for short RNA isolation from complex mixtures such as whole cell lysates. TiO
2 microspheres in the environment of an acetate buffer with pH 4.4 possess high selectivity towards very short RNA molecules, resulting in extraordinary purity of miRNA isolated from such a highly complex mixture, which is clearly presented (miRNA bands in
Figure 2C,D or
Figure 4A). On the other hand, the glycine buffer system offers higher yields of short RNAs compared to the acetate buffer, and it allows for the isolation of a wide range of RNA lengths (
Figure 2A, lanes 5, 6, 7), meaning that it can easily be used for complex RNA isolation. Furthermore, isopropanol used for LLE procedures is a common ingredient of disinfectants and can serve as an inactivation agent for bacteria or viruses. This can be advantageous when we isolate RNA molecules, e.g., from SARS-CoV-2 samples. TiO
2-based materials are also ideal candidates for the isolation of RNA from infectious agents due to the easy decontamination of TiO
2 surfaces by exposure to UV irradiation [
26,
27].
4. Materials and Methods
4.1. Microspheres and Oligo RNA
TiO2 microspheres, Titansphere (TiO, 10 µm) by GL Sciences (Tokyo, Japan), were used as a carrier for the solid-phase extraction.
Oligo RNA standard (23 nucleotides long) was designed analogically to the sequence of hsa-miR-18a-3p (Generi Biotech, Hradec Kralove, Czech Republic). It was used for spiking in the samples and to prepare model RNA mixture containing 5.28 µg of pre-isolated genomic RNA and 0.55 µg of oligo RNA (23-mer).
4.2. Cell Cultures
Jurkat cells (T-lymphoblast cell line, clone E6.1, ATCC, Manassas, VA, USA) were used for optimization of miRNA isolation from cells as well as for the preparation of a model genomic RNA. Jurkat cells cultivated in RPMI 1640 medium were harvested and washed with PBS, and a dry pellet containing 5 × 106 cells was repeatedly frozen in liquid nitrogen and stored at −80 °C. RNA for model genomic RNA was isolated from Jurkat cells with TRIZol Reagent (Thermo Fisher Scientific, Waltham, MA, USA).
A549 cells (human lung carcinoma cell line, ATCC, Manassas, VA, USA) were cultivated in MEM medium and then washed with PBS, and a dry pellet was repeatedly frozen in liquid nitrogen and stored at −80 °C.
4.3. General Short RNA Isolation Protocol Using TiO2 Microspheres
The general isolation procedure described here was applied in all isolation experiments. First, 1 mg of TiO2 microspheres was dispersed in 80% ACN/0.1% trifluoroacetic acid (TFA). The suspension was centrifuged at 595× g for 1 min to separate the solid phase from the solution after each step of sample isolation. The microspheres were washed twice with suitable washing and binding buffer to remove the ACN/TFA solution. Then, 200 µL of the initial sample in the washing and binding buffer was added and the mixture was incubated for 60 min at 37 °C. The supernatant was collected for further analysis. Weakly and non-specifically bound compounds were washed out five times with 500 µL of the washing and binding buffer (repeating each step for 2 min at 37 °C). The elution of the adsorbed miRNA was performed with 100 µL of 200 mM Na2HPO4 for 10 min at 37 °C. The elution fraction was neutralized with 5% TFA to obtain an approximately neutral pH.
4.4. Short RNA Isolation from Cells
Cells (5 × 106) were lysed with 260 µL of lysis solution containing 100 mM sodium acetate, 2% Triton X-100, and 0.1% diethyl pyrocarbonate (DEPC) for 10 min. The sample was spiked with 1.10 µg of oligo RNA standard. The amount of RNA standard in the fractions was monitored by urea-PAGE throughout the protocol. The lysate was centrifuged at 10,000× g for 10 min. An aliquot of 30 µL was recovered for further analysis, and the remaining volume (230 µL) of the cell lysate was transferred into a tube containing the same volume (230 µL) of isopropanol with 53.8 mg of NaCl. The mixture was gently rotated for 10 min to gradually dissolve NaCl in acetate buffer. This led to the protein precipitation and separation of phases, where the top phase was organic and the bottom phase was aqueous. The proteins originating from the cell lysate were concentrated in the interphase after short centrifugation at 595× g for 30 s. The top phase was discarded, and the bottom aqueous phase was carefully aspirated using a Hamilton syringe and transferred into a clean microtube. Acetic acid (6 M, 3.75 µL into 230 µL of aqueous phase) was added to adjust the pH to ~4.4 (checked with a microelectrode-equipped pH meter). Next, 200 µL of the aqueous phase containing RNA was added to 1 mg of TiO2 microspheres for further short RNA purification and isolation applying the aforementioned general isolation protocol with the acetate buffer system.
For acquiring a wider range of RNA sizes, the lysis solution consisted of 50 mM glycine, 2% Triton X-100, and 0.1% DEPC with pH adjusted to 6.4 using NaOH solution. After LLE, the pH of the acquired aqueous phase was lowered to ~2.4 by adding HCl (checked with a microelectrode-equipped pH meter) before continuing with the aforementioned general isolation protocol of TiO2-based RNA isolation with a glycine buffer system.
4.5. Polyacrylamide Gel Epectrophoresis
Polyacrylamide gel electrophoresis (PAGE) was used for the subsequent analysis of the isolated miRNAs. The aliquot of separated fractions was mixed 1:1 with a sample loading buffer (89 mM tris(hydroxymethyl)aminomethane, 89 mM boric acid, 2 mM EDTA, 7 M urea, 12% Ficoll, 0.01% xylene cyanole FF/bromphenol blue) and incubated for 4 min at 70 °C. All collected fractions were then loaded onto a 0.75 mm urea-polyacrylamide gel (20% gel with 8 M urea), and electrophoresis was performed with the Mini-Protean System (Bio-Rad, Hercules, CA, USA) at a constant voltage of 180 V in a Tris-Borate-EDTA running buffer. The gels were stained with SYBR Green II RNA Gel Stain (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Stained gels were analyzed using the ChemiDoc XRS + System (Bio-Rad, Hercules, CA, USA), with subsequent data processing in ImageLab software (Bio-Rad, Hercules, CA, USA).
Laemmli-SDS-PAGE electrophoresis with protein visualization by a silver staining method was used for the detection of protein residue in the samples of purified RNA [
23,
24]. Stained gels were analyzed using the ChemiDoc XRS + System (Bio-Rad), with subsequent data processing in ImageLab software (Bio-Rad).
4.6. Real-Time Polymerase Chain Reaction
Elution fractions were analyzed by RT-qPCR (TaqMan miRNA assay, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. First, miRNA was reverse transcribed into cDNA using hsa-miR-18a-3p-specific RT primer. A TProfessional Basic Thermocycler (Biometra Ltd., Göttingen, Germany) was used for cDNA synthesis with the following program: 30 min at 16 °C, 30 min at 42 °C, and 5 min at 85 °C. The cDNA transcript was then used for detection by real-time quantitative PCR with TaqMan miRNA assay. The reaction was carried out on RotorGene RG-3000A (Corbett Research, Cambridge, United Kingdom) using a real-time quantitative program: 10 min at 95 °C, then 15 s at 95 °C and 60 s at 60 °C for a total of 40 cycles.