Transcriptome Analysis of Air Space-Type Variegation Formation in Trifolium pratense
Abstract
1. Introduction
2. Results
2.1. Quality Evaluation and Reliability Verification of RNA-Seq Data
2.2. Screening and Identification of Differentially Expressed Genes (DEGs) Related to Air Space Type Variegation Formation
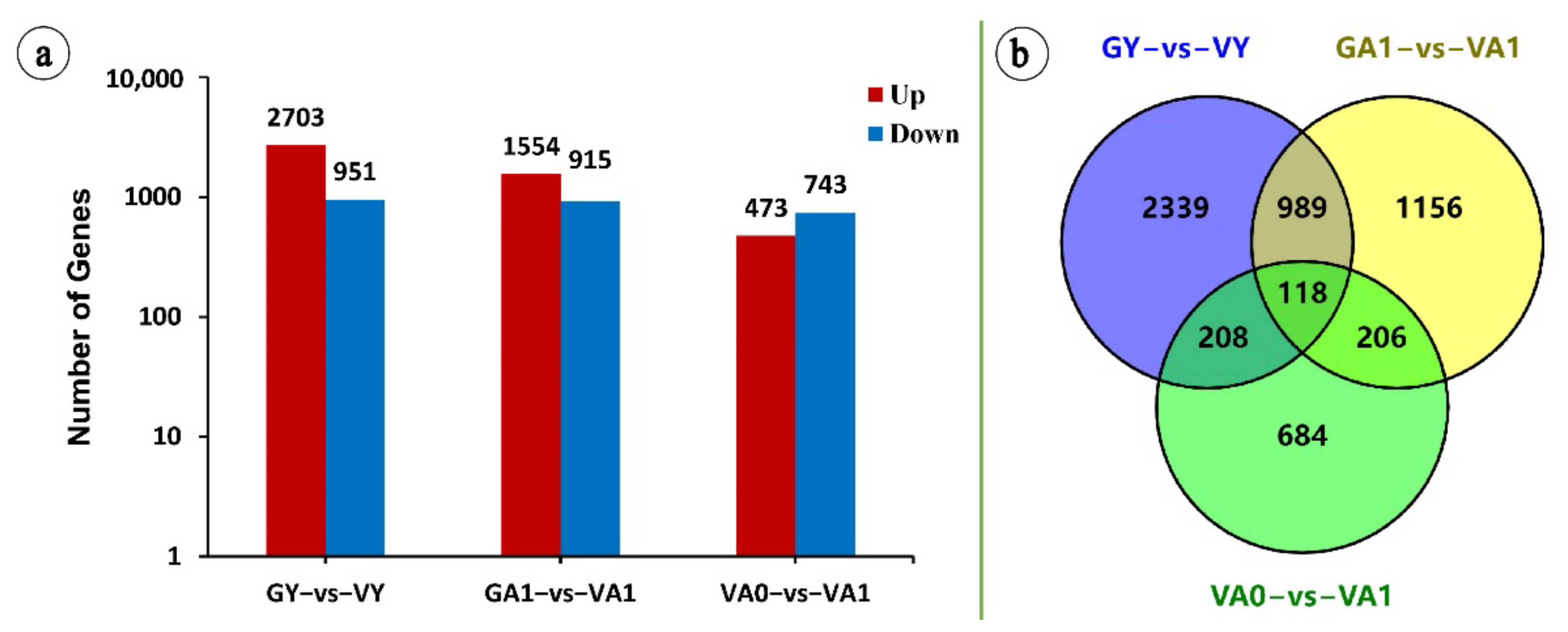
2.2.1. Differential Gene Expression Analysis
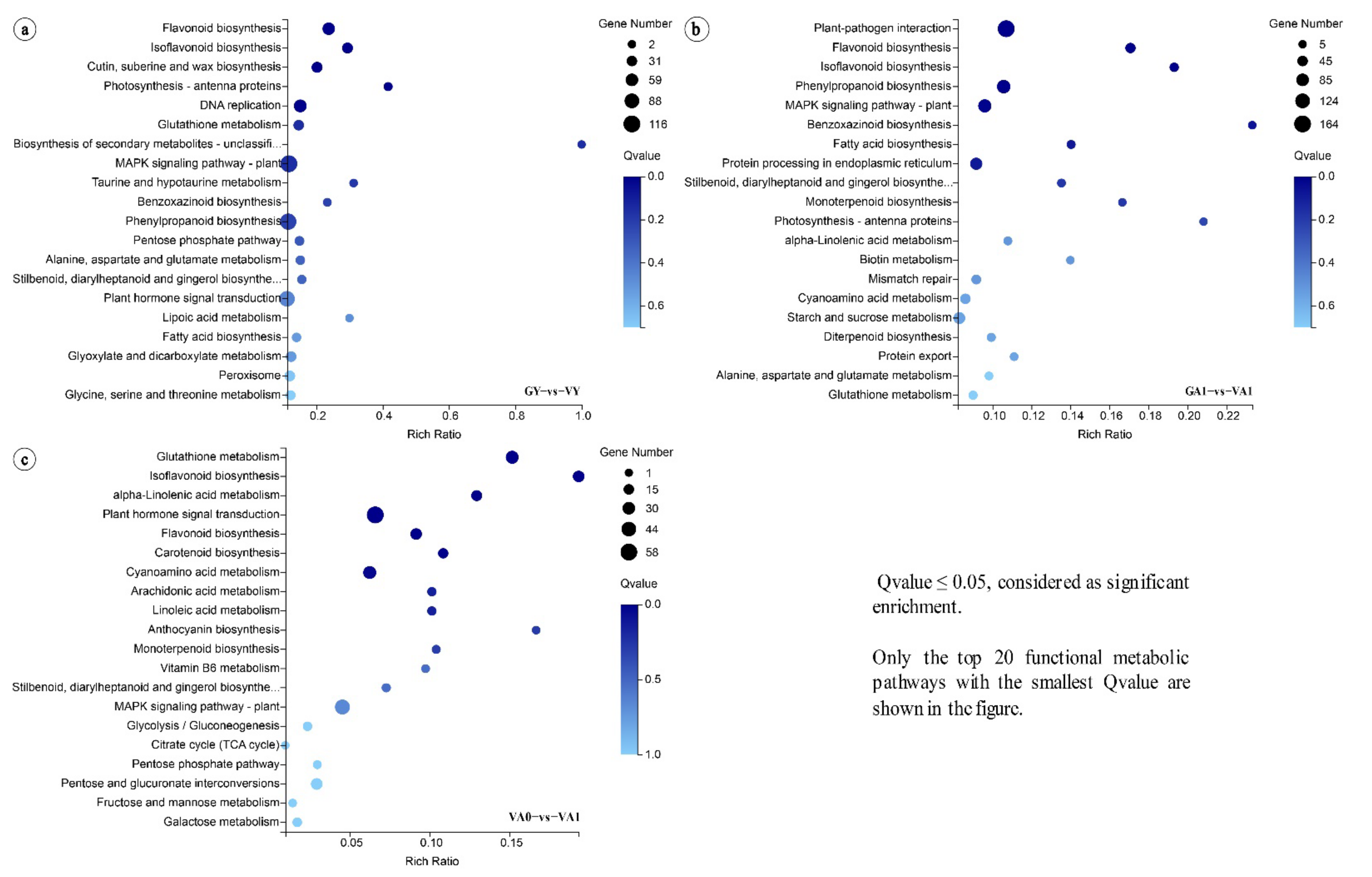
2.2.2. KEGG Enrichment Analysis
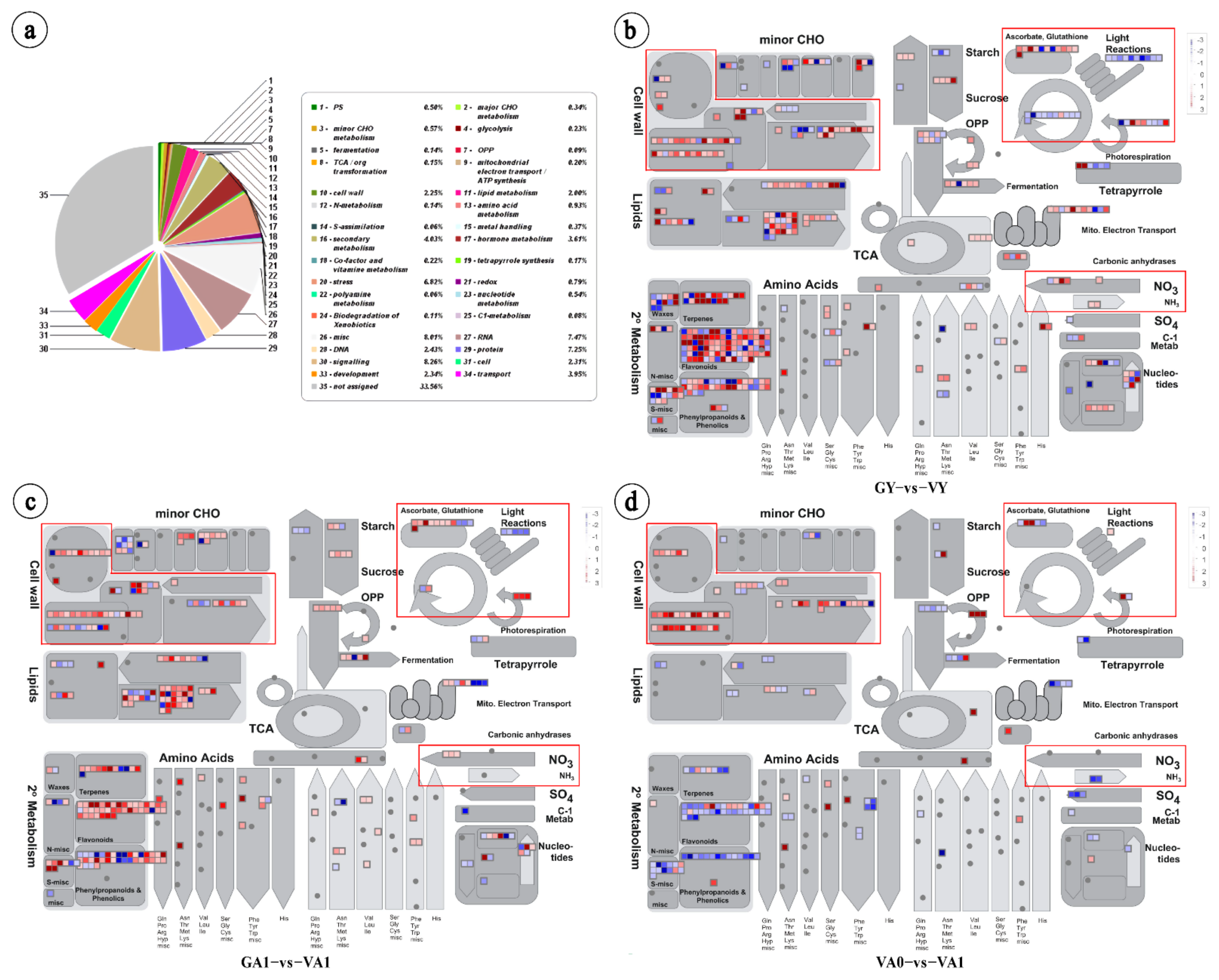
2.2.3. Identification of Genes Related to Air Space-Type Variegation Formation
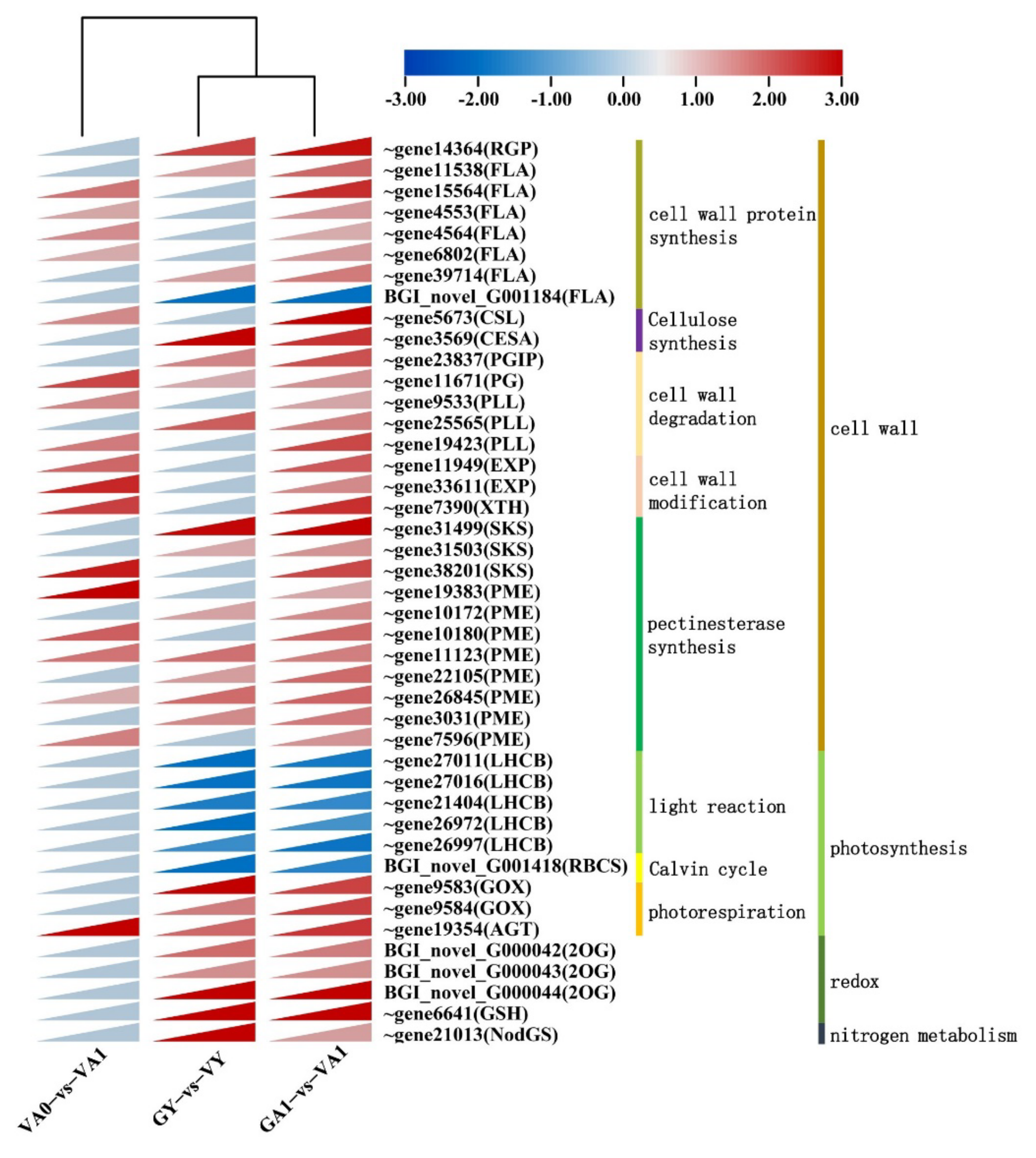
Cell Wall Development Related Pathway
Photosynthesis Pathway
Redox and Nitrogen Metabolism Pathways
Chlorophyll Metabolic Pathway
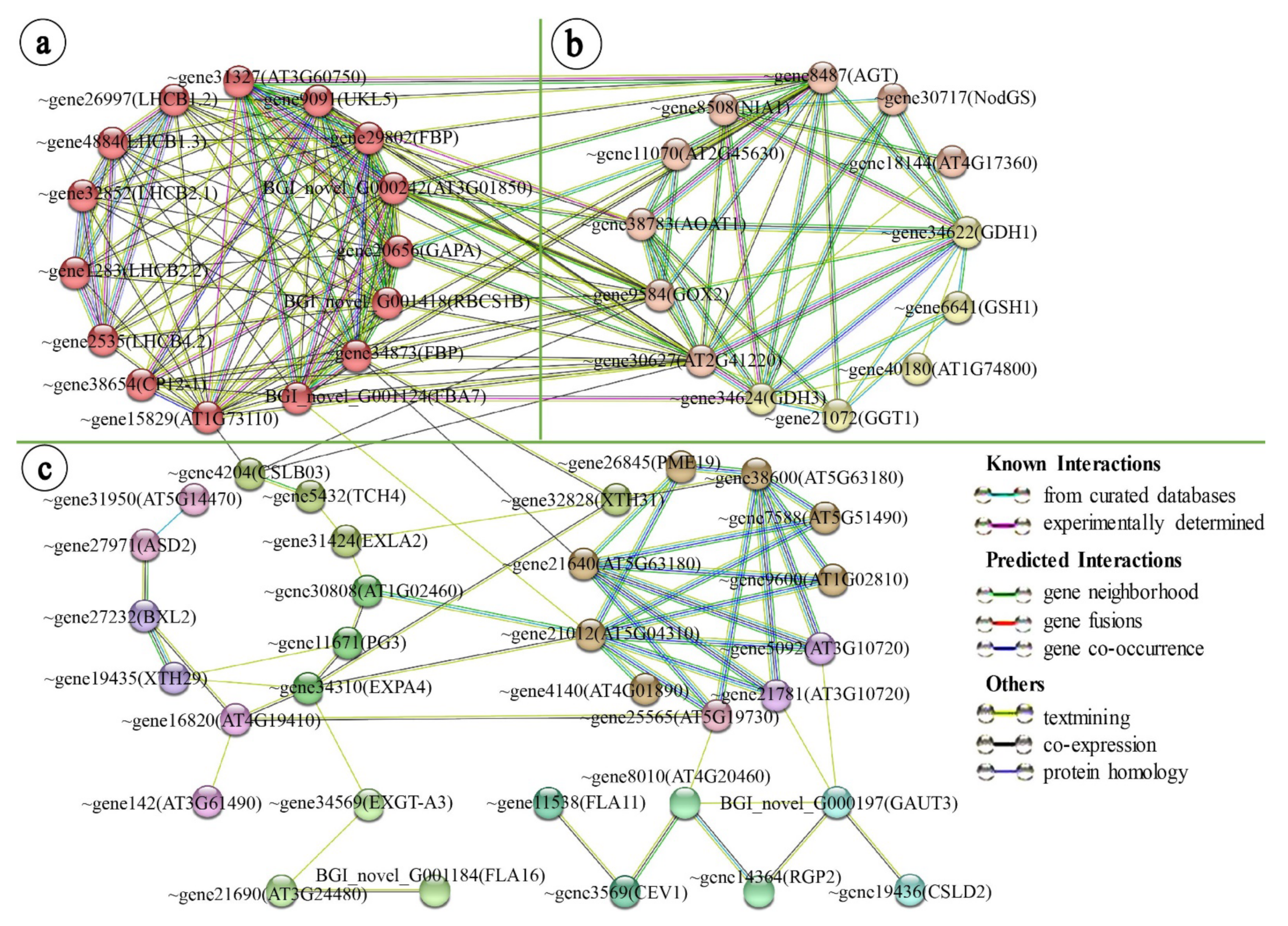
2.3. Leaf Variegation Associated Protein–Protein Network Interaction Analysis in the Young Leaflet Comparison Group
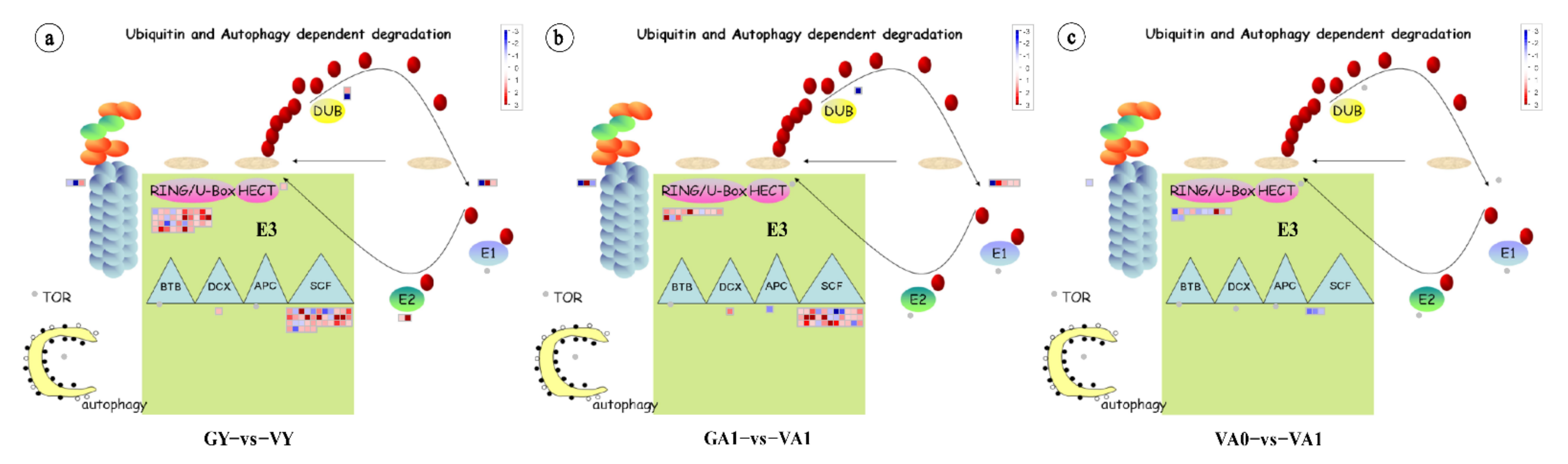
2.4. Relationship between Protease Degradation Systems and Leaf Variegation Formation
3. Discussion
3.1. Photosystem-Related Genes Play an Important Role in Air Space-Type Variegation Formation in T. pratense
3.2. Upregulated Expression of Genes Involved in Cell Wall Structure Modification Is Important for Air Space-Type Variegation Formation in T. pratense
3.3. Potential Regulatory Network of Air Space-Type Variegation in T. pratense
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA-Seq Material Collection
4.3. Transcriptome Sequencing and Preliminary Data Processing
4.3.1. mRNA Extraction, Quality Detection, and cDNA Library Construction
4.3.2. RNA-seq, Assembly, and Functional Annotation
4.4. Screening and Identification of DEGs Related to Air Space-Type Variegation Formation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.H.; Zeng, J.C.; Wang, X.M.; Chen, S.F.; Albach, D.C.; Li, H.Q. A revised classification of leaf variegation types. Flora 2020, 272, 151703. [Google Scholar] [CrossRef]
- Campitelli, B.E.; Stehlik, I.; Stinchcombe, J.R. Leaf variegation is associated with reduced herbivore damage in Hydrophyllum virginianum. Botany 2008, 86, 306–313. [Google Scholar] [CrossRef]
- Sheue, C.R.; Pao, S.H.; Chien, L.F.; Chesson, P.; Peng, C.I. Natural foliar variegation without costs? The case of Begonia. Ann. Bot. 2012, 109, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S. The proposed anti-herbivory roles of white leaf variegation. Prog. Bot. 2015, 76, 241–269. [Google Scholar] [CrossRef]
- Lev-Yadun, S.; Niemelä, P. Leaf pseudo-variegation: Definition, common types, and probably the defended models for real defensive leaf variegation mimicking them? Flora 2017, 226, 82–88. [Google Scholar] [CrossRef]
- Shelef, O.; Summerfield, L.; Lev-Yadun, S.; Villamarin-Cortez, S.; Sadeh, R.; Herrmann, I.; Rachmilevitch, S. Thermal benefits from white variegation of Silybum marianum leaves. Front. Plant Sci. 2019, 10, 688. [Google Scholar] [CrossRef]
- Li, X.; Kanakala, S.; He, Y.H.; Zhong, X.L.; Yu, S.M.; Li, R.X.; Sun, L.X.; Ma, J. Physiological characterization and comparative transcriptome analysis of white and green leaves of Ananas comosus var. bracteatus. PLoS ONE 2017, 12, e0169838. [Google Scholar] [CrossRef]
- Sun, Y.H.; Hung, C.Y.; Qiu, J.; Chen, J.J.; Kittur, F.S.; Oldham, C.E.; Henny, R.J.; Burkey, K.O.; Fan, L.J.; Xie, J.H. Accumulation of high OPDA level correlates with reduced ROS and elevated GSH benefiting white cell survival in variegated leaves. Sci. Rep. 2017, 7, 44158. [Google Scholar] [CrossRef]
- Wu, H.Y.; Shi, N.R.; An, X.Y.; Liu, C.; Fu, H.F.; Li, C.; Feng, Y.; Sun, D.J.; Zhang, L.J. Candidate genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int. J. Mol. Sci. 2018, 19, 1594. [Google Scholar] [CrossRef]
- Wang, Q.M.; Cui, J.G.; Dai, H.Y.; Zhou, Y.B.; Li, N.; Zhang, Z.H. Comparative transcriptome profiling of genes and pathways involved in leaf-patterning of Clivia miniata var. variegata. Gene 2018, 677, 280–288. [Google Scholar] [CrossRef]
- Gang, H.X.; Li, R.H.; Zhao, Y.M.; Liu, G.F.; Chen, S.; Jiang, J. Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef] [PubMed]
- Shih, T.H.; Lin, S.H.; Huang, M.Y.; Huang, W.D.; Yang, C.M. Transcriptome profile of the variegated Ficus microcarpa cv. Milky Stripe Fig Leaf. Int. J. Mol. Sci. 2019, 20, 1338. [Google Scholar] [CrossRef] [PubMed]
- Aluru, M.R.; Zola, J.; Foudree, A.; Rodermel, S.R. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 2009, 150, 904–923. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yin, H.; Zhao, C.; Lv, F. Transcriptome analysis of tessellated and green leaves in Paphiopedilum orchids using Illumina Paired-End sequencing and discovery simple sequence repeat markers. J. Plant Biochem. Physiol. 2014, 2, 2. [Google Scholar] [CrossRef]
- Wei, Z.; Vincent, M.A. Fabaceae. In Flora of China; Wu, C.Y., Raven, P.H., Eds.; Sciences Press: Beijing, China, 2010; Volume 10, pp. 548–551. [Google Scholar]
- Herbert, D.B.; Gross, T.; Rupp, O.; Becker, A. Transcriptome analysis reveals major transcriptional changes during regrowth after mowing of red clover (Trifolium pratense). BMC Plant Biol. 2021, 21, 95. [Google Scholar] [CrossRef]
- De Vega, J.J.; Ayling, S.; Hegarty, M.; Kudrna, D.; Goicoechea, J.L.; Ergon, Å.; Rognli, O.A.; Jones, C.; Swain, M.; Geurts, R.; et al. Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 2015, 5, 17394. [Google Scholar] [CrossRef]
- Chao, Y.H.; Xie, L.J.; Yuan, J.B.; Guo, T.; Li, Y.R.Z.; Liu, F.Q.; Han, L.B. Transcriptome analysis of leaf senescence in red clover (Trifolium pratense L.). Physiol. Mol. Biol. Plants 2018, 24, 753–765. [Google Scholar] [CrossRef]
- Chao, Y.H.; Yuan, J.B.; Li, S.F.; Jia, S.Q.; Han, L.B.; Xu, L.X. Analysis of transcripts and splice isoforms in red clover (Trifolium pratense L.) by single-molecule long-read sequencing. BMC Plant Biol. 2018, 18, 300. [Google Scholar] [CrossRef]
- Mo, F.; Li, H.B.; Li, Y.H.; Chen, X.; Wang, M.S.; Li, Z.; Deng, N.C.; Yang, Y.; Huang, X.; Zhang, R.; et al. Physiological, biochemical, and transcriptional regulation in a leguminous forage Trifolium pratense L. responding to silver ions. Plant Physiol. Biochem. 2021, 162, 531–546. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Richardson, E.A.; Ye, Z.H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Cui, D.T.; Ye, Z.H. Secondary cell wall biosynthesis. New Phytol. 2018, 221, 1703–1723. [Google Scholar] [CrossRef] [PubMed]
- Ringli, C. Monitoring the outside: Cell wall-sensing mechanisms. Plant Physiol. 2010, 153, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.Y.; Tobimatsu, Y.; Takeda, Y.; Suzuki, A.; Yamamura, M.; Umezawa, T.; Loet, C. Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol. 2017, 174, 972–985. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Zhang, S.; Liu, J.Y.; Zhao, C.H.; Wu, J.; Cao, Y.P.; Fu, C.X.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Pogorelko, G.V.; Kambakam, S.; Nolan, T.; Foudree, A.; Zabotina, O.A.; Rodermel, S.R. Impaired chloroplast biogenesis in immutans, an Arabidopsis variegation mutant, modifies developmental programming, cell wall composition and resistance to Pseudomonas syringae. PLoS ONE 2016, 11, e0150983. [Google Scholar] [CrossRef]
- Rédei, G.P. Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res.-Fundam. Mol. Mech. 1973, 18, 149–162. [Google Scholar] [CrossRef]
- Li, H.S.; Culligan, K.; Dixon, R.A.; Chory, J. CUE1: A mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 1995, 7, 1599–1610. [Google Scholar] [CrossRef]
- López-Juez, E.; Jarvis, R.P.; Takeuchi, A.; Page, A.M.; Chory, J. New Arabidopsis cue mutants suggest a close connection between plastid-and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998, 118, 803–815. [Google Scholar] [CrossRef]
- Abdelnoor, R.V.; Yule, R.; Elo, A.; Christensen, A.C.; Meyer-Gauen, G.; Mackenzie, S.A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 2003, 100, 5968–5973. [Google Scholar] [CrossRef]
- Nagai, S.; Koide, M.; Takahashi, S.; Kikuta, A.; Aono, M.; Sasaki-Sekimoto, Y.; Ohta, H.; Takamiya, K.; Masuda, T. Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol. 2007, 144, 1039–1051. [Google Scholar] [CrossRef]
- Su, T.; Yang, M.Y.; Wang, P.P.; Zhao, Y.X.; Ma, C.L. Interplay between the ubiquitin proteasome system and ubiquitin-mediated autophagy in plants. Cells 2020, 9, 2219. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.W.; González-Lamothe, R.; Ewan, R.A.; Rowland, O.; Yoshioka, H.; Shenton, M.; Ye, H.; O’Donnell, E.; Jones, J.D.G.; Sadanandom, A. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 2006, 18, 1084–1098. [Google Scholar] [CrossRef] [PubMed]
- Woodson, J.D.; Joens, M.S.; Sinson, A.B.; Gilkerson, J.; Salomé, P.A.; Weigel, D.; Fitzpatrick, J.A.; Chory, J. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 2015, 350, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Miao, Y. New aspects of HECT-E3 ligases in cell senescence and cell death of plants. Plants 2019, 8, 483. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.F.; Yan, H.C.; Wang, K.B.; Kuang, T.Y.; Zhang, J.P.; Gui, L.L.; An, X.M.; Chang, W.R. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 2004, 428, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.W.; Li, M.; Wan, T.; Wang, L.F.; Jia, C.J.; Hou, Z.Q.; Zhao, X.L.; Zhang, J.P.; Chang, W.R. Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat. Struct. Mol. Biol. 2011, 18, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J.D. Regulation and dynamics of the light-harvesting system. Annu. Rev. Plant Biol. 2014, 65, 287–309. [Google Scholar] [CrossRef]
- Fan, M.R.; Li, M.; Liu, Z.F.; Cao, P.; Pan, X.W.; Zhang, H.M.; Zhao, X.L.; Zhang, J.P.; Chang, W.R. Crystal structures of the PsbS protein essential for photoprotection in plants. Nat. Struct. Mol. Biol. 2015, 22, 729–735. [Google Scholar] [CrossRef]
- Fristedt, R.; Herdean, A.; Blaby-Haas, C.E.; Mamedov, F.; Merchant, S.S.; Last, R.L.; Lundin, B. PHOTOSYSTEM II PROTEIN33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiol. 2015, 167, 481–492. [Google Scholar] [CrossRef]
- Hey, D.; Rothbart, M.; Herbst, J.; Wang, P.; Müller, J.; Wittmann, D.; Gruhl, K.; Grimm, B. LIL3, a light-harvesting complex protein, links terpenoid and tetrapyr-role biosynthesis in Arabidopsis thaliana. Plant Physiol. 2017, 174, 1037–1050. [Google Scholar] [CrossRef]
- Myouga, F.; Takahashi, K.; Tanaka, R.; Nagata, N.; Kiss, A.Z.; Funk, C.; Nomura, Y.; Nakagami, H.; Jansson, S.; Shinozaki, K. Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiol. 2018, 176, 2277–2291. [Google Scholar] [CrossRef]
- Roig-Oliver, M.; Rayon, C.; Roulard, R.; Fournet, F.; Bota, J.; Flexas, J. Reduced photosynthesis in Arabidopsis thaliana atpme17.2 and atpae11.1 mutants is associated to altered cell wall composition. Physiol. Plant. 2020, 172, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y. The Study of the Mechanism of Arabidopsis Immutans Variegation. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2009. [Google Scholar]
- Mandel, M.A.; Feldmann, K.A.; Herrera-Estrella, L.; Rocha-Sosa, M.; León, P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996, 9, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Streatfield, S.J.; Weber, A.; Kinsman, E.A.; Häusler, R.E.; Li, J.M.; Post-Beittenmiller, D.; Kaiser, W.M.; Pyke, K.A.; Flügge, U.; Chory, J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 1999, 11, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Ganusova, E.E.; Reagan, B.C.; Fernandez, J.C.; Azim, M.F.; Sankoh, A.F.; Freeman, K.M.; McCray, T.N.; Patterson, K.; Kim, C.; Burch-Smith, T.M. Chloroplast-to-nucleus retrograde signalling controls intercellular trafficking via plasmodesmata formation. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190408. [Google Scholar] [CrossRef] [PubMed]
- Pyke, K.; Zwobka, M.K.; Day, A. Marking cell layers in Brassica napus with spectinomycin provides a new tool for studying cell fate and demonstrates a requirement for chloroplasts in palisade cell differentiation. J. Exp. Bot. 2000, 51, 1713–1720. [Google Scholar] [CrossRef] [PubMed][Green Version]
- González-Bayon, R.; Kinsman, E.A.; Quesada, V.; Vera, A.; Robles, P.; Ponce, M.R.; Pyke, K.A.; Micol, J.L. Mutations in the RETICULATA gene dramatically alter internal architecture but have little effect on overall organ shape in Arabidopsis leaves. J. Exp. Bot. 2006, 57, 3019–3031. [Google Scholar] [CrossRef] [PubMed]
- Meehan, L.; Harkins, K.; Chory, J.; Rodermel, S. Lhcb transcription is coordinated with cell size and chlorophyll accumulation (studies on fluorescence-activated, cell-sorter-purified single cells from wild-type and immutans Arabidopsis thaliana). Plant Physiol. 1996, 112, 953–963. [Google Scholar] [CrossRef][Green Version]
- Gao, J.L.; Wang, H.; Yuan, Q.P.; Feng, Y. Structure and function of the photosystem supercomplexes. Front. Plant Sci. 2018, 9, 357. [Google Scholar] [CrossRef]
- Meurer, J.; Grevelding, C.; Westhoff, P.; Reiss, B. The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 1998, 258, 342–351. [Google Scholar] [CrossRef]
- Holding, D.R.; Springer, P.S.; Coomber, S.A. The chloroplast and leaf developmental mutant, pale cress, exhibits light-conditional severity and symptoms characteristic of its ABA deficiency. Ann. Bot. 2000, 86, 953–962. [Google Scholar] [CrossRef]
- Næsted, H.; Holm, A.; Jenkins, T.; Nielsen, H.B.; Harris, C.A.; Beale, M.H.; Andersen, M.; Mant, A.; Scheller, H.; Camara, B.; et al. Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. J. Cell Sci. 2004, 117, 4807–4818. [Google Scholar] [CrossRef]
- Chen, Y.S.; Chesson, P.; Wu, H.W.; Pao, S.H.; Liu, J.W.; Chien, L.F.; Yong, W.H.; Sheue, C.R. Leaf structure affects a plant’s appearance: Combined multiple mechanisms intensify remarkable foliar variegation. J. Plant Res. 2017, 130, 311–325. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, jcs207373. [Google Scholar] [CrossRef]
- Zhang, B.C.; Gao, Y.H.; Zhang, L.J.; Zhou, Y.H. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef]
- Boudart, G.; Jamet, E.; Rossignol, M.; Lafitte, C.; Borderies, G.; Jauneau, A.; Esquerré-Tugayé, M.; Pont-Lezica, R. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: Identification by mass spectrometry and bioinformatics. Proteomics 2005, 5, 212–221. [Google Scholar] [CrossRef]
- Hervé, V.; Duruflé, H.; San Clemente, H.; Albenne, C.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 2016, 16, 3183–3187. [Google Scholar] [CrossRef]
- Kimura, M.; Yamamoto, Y.Y.; Seki, M.; Sakurai, T.; Sato, M.; Abe, T.; Yoshida, S.; Manabe, K.; Shinozaki, K.; Matsui, M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003, 77, 226–233. [Google Scholar] [CrossRef]
- Sasidharan, R.; Chinnappa, C.C.; Voesenek, L.A.C.J.; Pierik, R. The regulation of cell wall extensibility during shade avoidance: A study using two contrasting ecotypes of Stellaria longipes. Plant Physiol. 2008, 148, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Chinnappa, C.C.; Staal, M.; Elzenga, T.M.; Yokoyama, R.; Nishitani, K.; Voesenek, L.A.C.J.; Pierik, R. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 2010, 154, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, S.; van Orden, J.; Wolf, S.; Vissenberg, K.; Delacourt, J.; Ndong, Y.A.; Pelloux, J.; Bischoff, V.; Urbain, A.; Mouille, G.; et al. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 2010, 188, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Sénéchal, F.; Wattier, C.; Rustérucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef]
- Tabuchi, A.; Matsumoto, H. Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol. Plant. 2001, 112, 353–358. [Google Scholar] [CrossRef]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef]
- Wang, J.; Howles, P.A.; Cork, A.H.; Birch, R.J.; Williamson, R.E. Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol. 2006, 142, 685–695. [Google Scholar] [CrossRef]
- Park, J.; Cui, Y.; Kang, B.H. AtPGL3 is an Arabidopsis BURP domain protein that is localized to the cell wall and promotes cell enlargement. Front. Plant Sci. 2015, 6, 412. [Google Scholar] [CrossRef]
- Sun, C.L.; Lu, L.L.; Yu, Y.; Liu, L.J.; Hu, Y.; Ye, Y.Q.; Jin, C.W.; Lin, X.Y. Decreasing methylation of pectin caused by nitric oxide leads to higher aluminium binding in cell walls and greater aluminium sensitivity of wheat roots. J. Exp. Bot. 2016, 67, 979–989. [Google Scholar] [CrossRef]
- Zhou, K. GPI-anchored SKS proteins regulate root development through controlling cell polar expansion and cell wall synthesis. Biochem. Biophys. Res. Commun. 2019, 509, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; MacMillan, C.P.; Shafee, T.; Ma, Y.X.; Ratcliffe, J.; van de Meene, A.; Bacic, A.; Humphries, J.; Johnson, K.L. Fasciclin-like arabinogalactan-protein 16 (FLA16) is required for stem development in Arabidopsis. Front. Plant Sci. 2020, 11, 615392. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.X.; MacMillan, C.P.; de Vries, L.; Mansfield, S.D.; Hao, P.F.; Ratcliffe, J.; Bacic, A.; Johnson, K.L. FLA11 and FLA12 glycoproteins fine-tune stem secondary wall properties in response to mechanical stresses. New Phytol. 2022, 233, 1750–1767. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A.; Harpster, M.H.; Dunsmuir, P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol. Biol. 1999, 39, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.T.; Cosgrove, D.J. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 9783–9788. [Google Scholar] [CrossRef]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 2003, 15, 1591–1604. [Google Scholar] [CrossRef]
- Bosch, M.; Hepler, P.K. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 2005, 17, 3219–3226. [Google Scholar] [CrossRef]
- Jolie, R.P.; Duvetter, T.; van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef]
- Delgado, I.J.; Wang, Z.H.; de Rocher, A.; Keegstra, K.; Raikhel, N.V. Cloning and characterization of AtRGP1: A reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 1988, 116, 1339–1350. [Google Scholar] [CrossRef]
- Langeveld, S.M.J.; Vennik, M.; Kottenhagen, M.; van Wijk, R.; Buijk, R.; Kijne, J.W.; de Pater, S. Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Physiol. 2002, 129, 278–289. [Google Scholar] [CrossRef][Green Version]
- Drakakaki, G.; Zabotina, O.; Delgado, I.; Robert, S.; Keegstra, K.; Raikhel, N. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 2006, 142, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Sagi, G.; Gera, A.; Epel, B.L. The constitutive expression of Arabidopsis plasmodesmal-associated class 1 reversibly glycosylated polypeptide impairs plant development and virus spread. J. Exp. Bot. 2010, 61, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Cui, Y.; Zambryski, P.C. Reduced levels of class 1 reversibly glycosylated polypeptide increase intercellular transport via plasmodesmata. Plant Signal. Behav. 2012, 7, 62–67. [Google Scholar] [CrossRef]
- Dorokhov, Y.L.; Ershova, N.M.; Sheshukova, E.V.; Komarova, T.V. Plasmodesmata conductivity regulation: A mechanistic model. Plants 2019, 8, 595. [Google Scholar] [CrossRef]
- Antosiewicz, D.M.; Purugganan, M.M.; Polisensky, D.H.; Braam, J. Cellular localization of Arabidopsis xyloglucan endotransglycosylase-related proteins during development and after wind stimulation. Plant Physiol. 1997, 115, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.; Braam, J. Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999, 4, 361–366. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Aluru, M.R.; Bae, H.; Wu, D.Y.; Rodermel, S.R. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol. 2001, 127, 67–77. [Google Scholar] [CrossRef]
- Gotoh, E.; Suetsugu, N.; Higa, T.; Matsushita, T.; Tsukaya, H.; Wada, M. Palisade cell shape affects the light-induced chloroplast movements and leaf photosynthesis. Sci. Rep. 2018, 8, 1472. [Google Scholar] [CrossRef]
- Xu, D.R.; Dhiman, R.; Garibay, A.; Mock, H.P.; Leister, D.; Kleine, T. Cellulose defects in the Arabidopsis secondary cell wall promote early chloroplast development. Plant J. 2020, 101, 156–170. [Google Scholar] [CrossRef]
- Rosso, D.; Ivanov, A.G.; Fu, A.; Geisler-Lee, J.; Hendrickson, L.; Geisler, M.; Stewart, G.; Krol, M.; Hurry, V.; Rodermel, S.R.; et al. IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol. 2006, 142, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.J.; Huang, J.R. A review of leaf variegation mutants and mechanisms in Arabidopsis. Plant Physiol. J. 2016, 52, 1671–1680. [Google Scholar]
- Beemster, G.T.S.; de Veylder, L.; Vercruysse, S.; West, G.; Rombaut, D.; van Hummelen, P.; Galichet, A.; Gruissem, W.; Inzé, D.; Vuylsteke, M. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 2005, 138, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Kalve, S.; de Vos, D.; Beemster, G.T.S. Leaf development: A cellular perspective. Front. Plant Sci. 2014, 5, 362. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 2006, 57, 477–496. [Google Scholar] [CrossRef] [PubMed]
- Routier-Kierzkowska, A.L.; Kierzkowski, D. Coping with stress: Mechanics of the expanding leaf. J. Exp. Bot. 2016, 67, 5288–5290. [Google Scholar] [CrossRef][Green Version]
- Runions, A.; Tsiantis, M.; Prusinkiewicz, P.A. common developmental program can produce diverse leaf shapes. New Phytol. 2017, 216, 401–418. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Fleming, A.J. The control of leaf development. New Phytol. 2005, 166, 9–20. [Google Scholar] [CrossRef]
- Chen, Y.X.; Chen, Y.S.; Shi, C.M.; Huang, Z.B.; Zhang, Y.; Li, S.K.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.T. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]

| Sample | Raw Reads (M) | Clean Reads (M) | Clean Bases (Gb) | Q20 | Q30 | Mapped Genome | Mapped Genes |
|---|---|---|---|---|---|---|---|
| GY_1 | 47.43 | 43.17 | 6.48 | 96.69% | 88.28% | 78.99% | 69.76% |
| GY_2 | 47.43 | 42.90 | 6.43 | 96.58% | 88.02% | 79.16% | 70.29% |
| GY_3 | 47.43 | 43.22 | 6.48 | 96.74% | 88.37% | 79.70% | 71.19% |
| VY_1 | 49.19 | 44.12 | 6.62 | 96.69% | 88.35% | 80.04% | 67.25% |
| VY_2 | 47.43 | 42.55 | 6.38 | 96.84% | 88.70% | 79.78% | 67.26% |
| VY_3 | 49.19 | 44.66 | 6.70 | 96.77% | 88.53% | 80.14% | 69.17% |
| GA1_1 | 47.43 | 43.62 | 6.54 | 96.67% | 88.18% | 77.55% | 70.51% |
| GA1_2 | 47.43 | 43.97 | 6.60 | 96.78% | 88.47% | 77.79% | 71.49% |
| GA1_3 | 49.19 | 44.40 | 6.66 | 96.81% | 88.59% | 78.43% | 70.59% |
| VA0_1 | 51.96 | 47.11 | 7.07 | 96.55% | 88.44% | 77.54% | 69.49% |
| VA0_2 | 52.70 | 47.92 | 7.19 | 96.69% | 88.75% | 78.37% | 68.71% |
| VA0_3 | 52.70 | 47.72 | 7.16 | 96.70% | 88.66% | 79.03% | 69.88% |
| VA1_1 | 52.47 | 47.49 | 7.12 | 96.78% | 88.94% | 78.22% | 69.17% |
| VA1_2 | 51.19 | 46.95 | 7.04 | 97.50% | 90.33% | 79.91% | 70.93% |
| VA1_3 | 47.39 | 43.02 | 6.45 | 95.85% | 87.14% | 76.59% | 69.41% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Li, J.; Zou, L.; Li, H. Transcriptome Analysis of Air Space-Type Variegation Formation in Trifolium pratense. Int. J. Mol. Sci. 2022, 23, 7794. https://doi.org/10.3390/ijms23147794
Zhang J, Li J, Zou L, Li H. Transcriptome Analysis of Air Space-Type Variegation Formation in Trifolium pratense. International Journal of Molecular Sciences. 2022; 23(14):7794. https://doi.org/10.3390/ijms23147794
Chicago/Turabian StyleZhang, Jianhang, Jiecheng Li, Lu Zou, and Hongqing Li. 2022. "Transcriptome Analysis of Air Space-Type Variegation Formation in Trifolium pratense" International Journal of Molecular Sciences 23, no. 14: 7794. https://doi.org/10.3390/ijms23147794
APA StyleZhang, J., Li, J., Zou, L., & Li, H. (2022). Transcriptome Analysis of Air Space-Type Variegation Formation in Trifolium pratense. International Journal of Molecular Sciences, 23(14), 7794. https://doi.org/10.3390/ijms23147794






