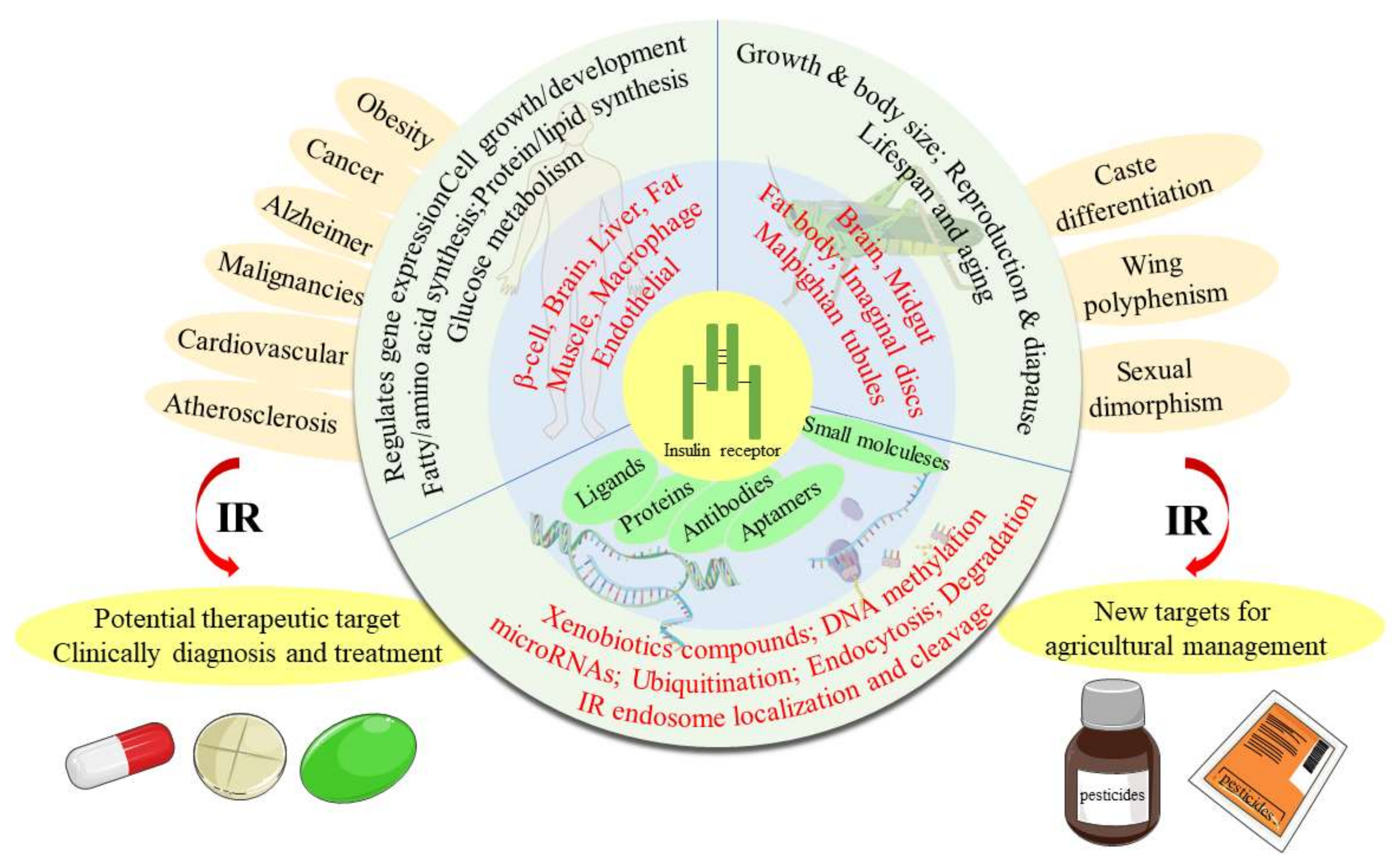
The Insulin Receptor: An Important Target for the Development of Novel Medicines and Pesticides
Abstract
1. Introduction
2. Biology Studies of the IR
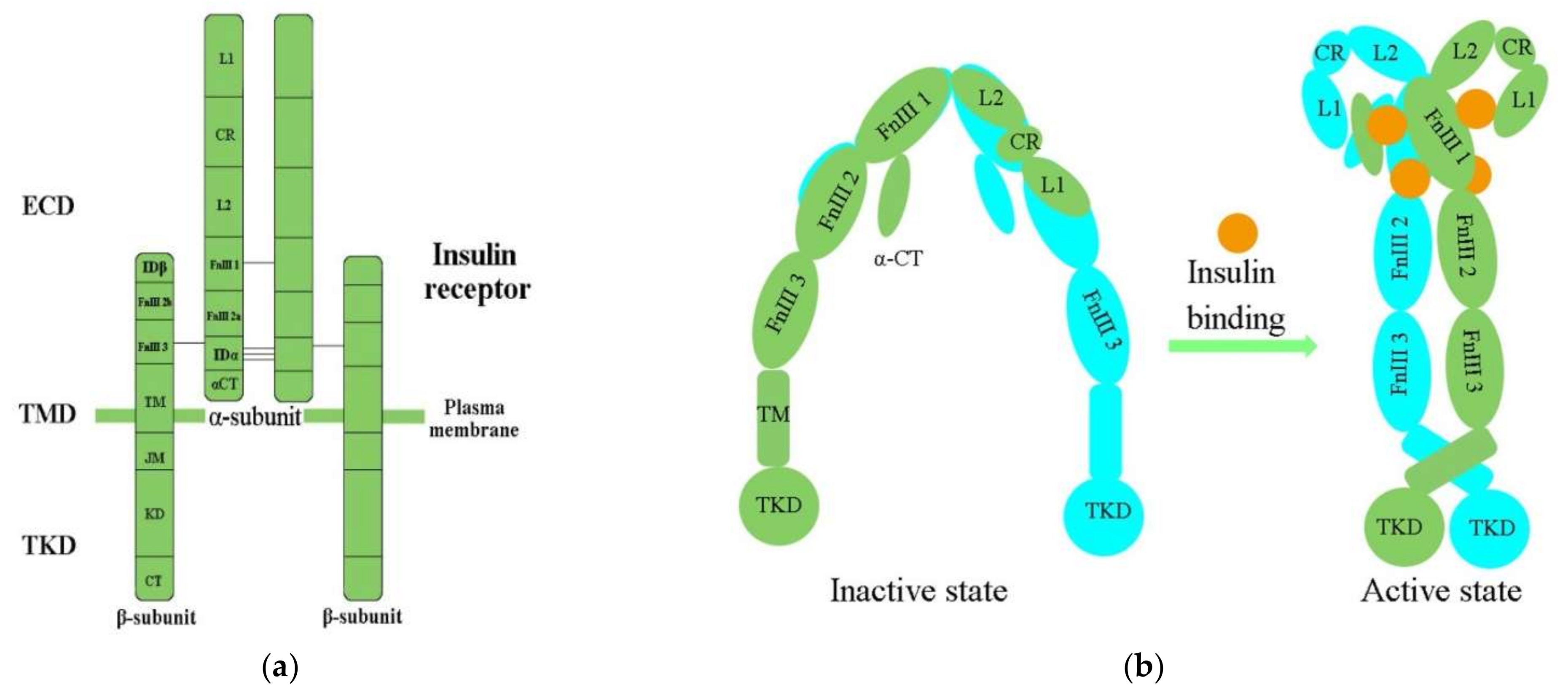
2.1. Molecular Structure of the IR
2.2. Activation of the IR
3. Functions of the IR
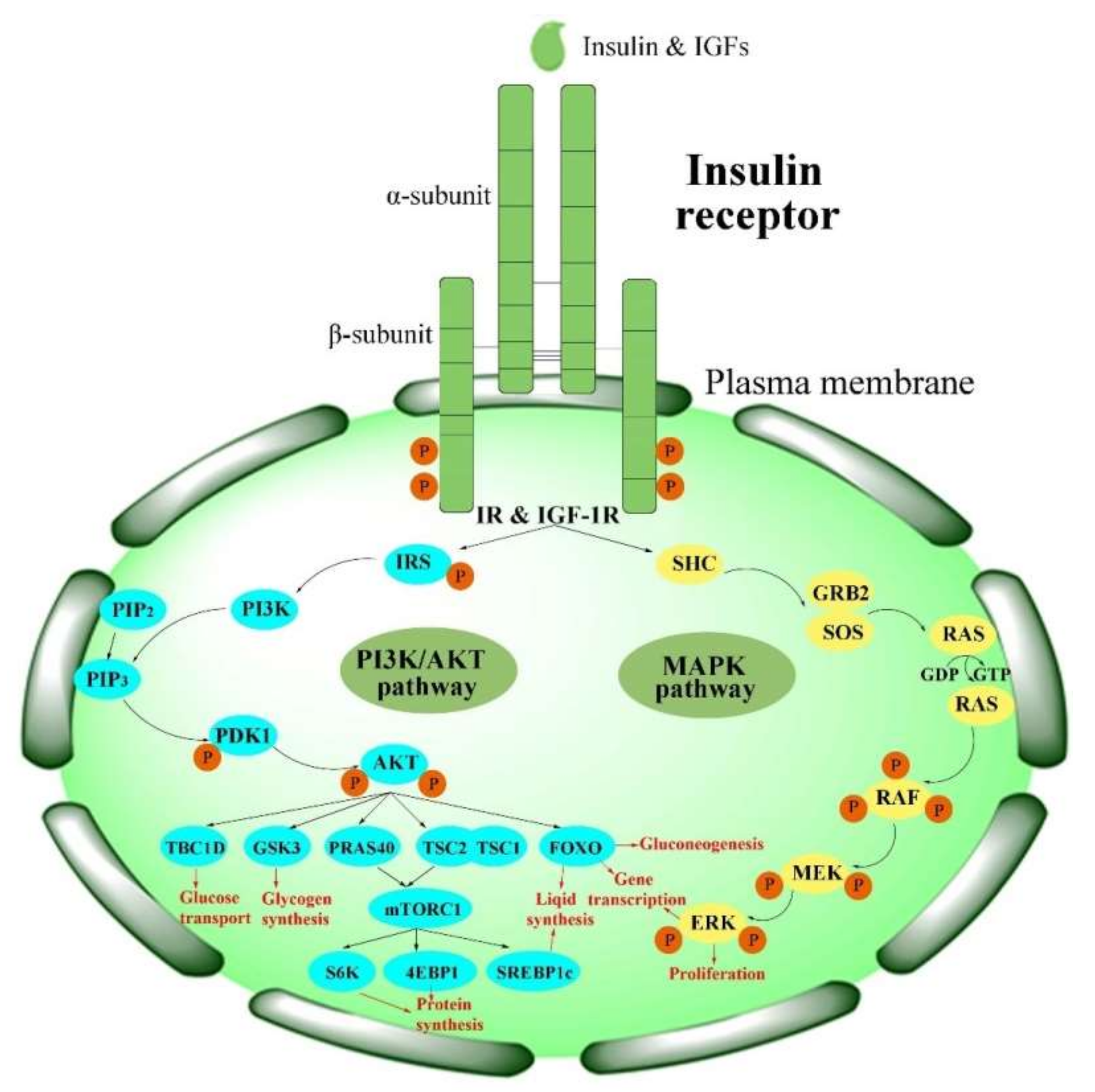
3.1. The Functions of the IR in Human Beings
3.2. The Functions of the IR in Insects
3.2.1. Gene Organization of the IR in Insects
3.2.2. The Effect of Silencing or Knockdown of IR Genes in Insects
3.2.3. Polyphenism Is Adjusted by the IR in Insects
4. Pharmacological and Physiological Modulators of IR Activation
4.1. Insulin and Its Analogs
4.2. Insulin-Mimetic Peptides
4.3. Antibodies
4.4. Allosteric Aptamers
4.5. Proteins
4.5.1. Adaptor Proteins and Regulatory Proteins
4.5.2. Membrane Proteins
4.6. Other Pharmacological and Physiological Modulators
| Classification | Modulators | IR Modulation Mechanism | Model Organisms or Cells | Side Effects | References |
|---|---|---|---|---|---|
| Insulin and insulin analogs | insulin and IGFs, | ligand-induced internalization and degradation of the IR | human | tissue irritation, abscesses, allergic edema, weight gain, risk of congestive heart failure | [3] |
| lispro, | [32,135] | ||||
| aspart, | |||||
| glulisine, | |||||
| aspb10, | |||||
| detemir, | |||||
| largine, | |||||
| degludec, | |||||
| ILPs | ligand | insects | - | [103] | |
| Insulin-mimetic peptides | S371, S446 | disrupts the primary insulin binding site of the IR | mice | - | [138] |
| - | |||||
| S519(agonist) | - | ||||
| S597 (partial agonist) | receptor activation | IR-transfected L6 myoblasts | - | [138,177] | |
| S661 | antagonist of the IR | rat adipocytes | - | [140] | |
| S961(agonist/ antagonist) | ↓IR, blocks expression of the IR without insulin | breast cancer cells | - | [141,178] | |
| Antibodies | XMetA (partial agonist) | ↑IR autophosphorylation (EC50:1.3 nmol/L); ↑Akt phosphorylation (EC50: 1.1 nmol/L) | CHO-hINSR cells (in vitro); diabetic mice (in vivo) | - | [144,145] |
| XmetS (agonist) | ↑binding affinity with IR; ↑IR autophosphorylation (insulin-dependent); ↑Akt phosphorylation | MCF-7 human breast cancer (in vitro); mouse models of insulin-resistant diabetes (in vivo) | - | [146] | |
| XmetD (X358) (antagonist) | ↓autophosphorylation of IR (interacte with IR); ↓phosphorylation of Akt and Erk | adult male CHO-hINSR cells; L6 muscle cells; COLO-205 human colon cancer cells; hyperinsulinemic hypoglycemia mice | - | [147] | |
| healthy adult | insulin resistance (3 d wherein X358-imparted) | [148] | |||
| IRAB-A (agonist/sensitizer) | ↓off-rate of insulin from the IR (stabilizes insulin binding) | diet-induced obese C57 mice | - | [132] | |
| IRAB-B (antagonist) | ↓IR phosphorylation (binds to IR) | C57BL/6N mice | - | [149] | |
| AK98 (antagonist) | competes with insulin (bind to IR) ↓IR expression levels | tumor cell (MCF-7) | - | [23] | |
| Aptamers | IR-A48 (partial agonist) (IR Tyr1150), | ↑IR autophosphorylation (allosteric binds and activates the IR, but not IGF-1R) | HEK293 and 3T3-L1 cells; Rat-1 cells overexpressing human IR (Rat-1/hIR) | - | [152] |
| IR-A43 (sensitizer) | binds to the allosteric site of IR; ↑insulin bind to IR | - | - | [153] | |
| IR-A62 (agonist and activator) | ↑insulin binding and Y1150; monophosphorylation of the IR (low concentrations); ↓insulin binding and IR phosphorylation (high concentrations) | C57BL/6 mice; Rat-1 cells overexpressing human IR (Rat-1/hIR); 3T3-L1 and MCF-7 breast cancer cells | - | [154] | |
| GL56 (inhibitor) | specifically recognizes the IR; ↓IR phosphorylation; ↓phosphorylation of AKT, ERK1/2 and IRS1 | U87MG; glioblastoma cancer cells | - | [155] | |
| Proteins | GRB10/14, | ↓activity of the IR as a pseudosubstrate of the IR-TK | mice | - | [157] |
| SOCS1/3, | mice | - | [3] | ||
| GRP78 (IGF-1R) | ↑IGF-1R phosphorylation and activation | hepatoma cells | - | [158] | |
| SH2B1 | ↑IR and IRS1 phosphorylation;↑Akt and Erk activation | CHO–IR, 3T3L1, NIH3T3, and HEK293 cells; mice | - | [157] | |
| SORLA | ↑IR surface expression (redirects internalized IR from endosomes to PM) | mouse with loss of function/tissue -specific over- expression of SORLA; obese human subjects | - | [70] | |
| Cav-2α | ↑IRS-1 recruitment and association with IR (a substrate of IR tyrosine kinase) | Hirc-B cells, HEK293T cells, 3T3L1 preadipocytes or adipocytes | - | [161] | |
| Cav-2β | desensitization of the IR; ↑IR-TK inactivation via dephospho-rylation by PTP1B and internalization via dynamin- 2-dependent endocytosis | HEK293T cells, 3T3-L1 preadipocytes (ATCC, CL-173) | - | [162] | |
| ApoE | interacts with the IR, interfering with insulin binding; ↓insulin–IR interaction and impairs IR trafficking | human ApoE -targeted replacement mice | - | [164,165] | |
| Others | Glypican-4 | interacts with the IR, causing ↑IR signaling | visceral and subcutaneous adipose tissue/3T3-L1 preadipocytes | - | [168] |
| mcIRBP-9 | ↑IR kinase activity; ↑phosphorylation of IR; ↑translocation of GLUT4; ↑uptake of glucose | 3T3-L1 preadipocytes; type 1 diabetic mice; type 2 diabetic mice (db/db mice) | - | [169] | |
| Visfatin | binds to the IR site | clonal mouse pancreatic β-cell; β-TC6 cell line (BTC) cells | - | [170] | |
| SMPDL3b | interferes with the IR isoforms binding to caveolin1 in the PM | podocytes in DKD | - | [171] | |
| PTP1B | dephosphorylates the IR, causing deactivation | mice | novel therapeutic strategy for T2DM | [172] | |
| PKCε | phosphorylates the IR, blocking IR autophosphorylation | InsrT1150A mice | improves NAFLD diagnostic screening for the early identification of patients at risk for T2D | [174] | |
| Aroclor 1254 | inhibits the expression of the IR | male C57BL/6 mice/skeletal muscle & liver | _ | [175] | |
| Subetta | increases IR β-subunit phosphorylation | human preadipocytes | _ | [176] | |
| BACE1 | cleaves the IR ECD and decreases the amount of mature IR | mouse models of diabetes (db/db) and impaired glucose tolerance (HFD mice) | _ | [21] |
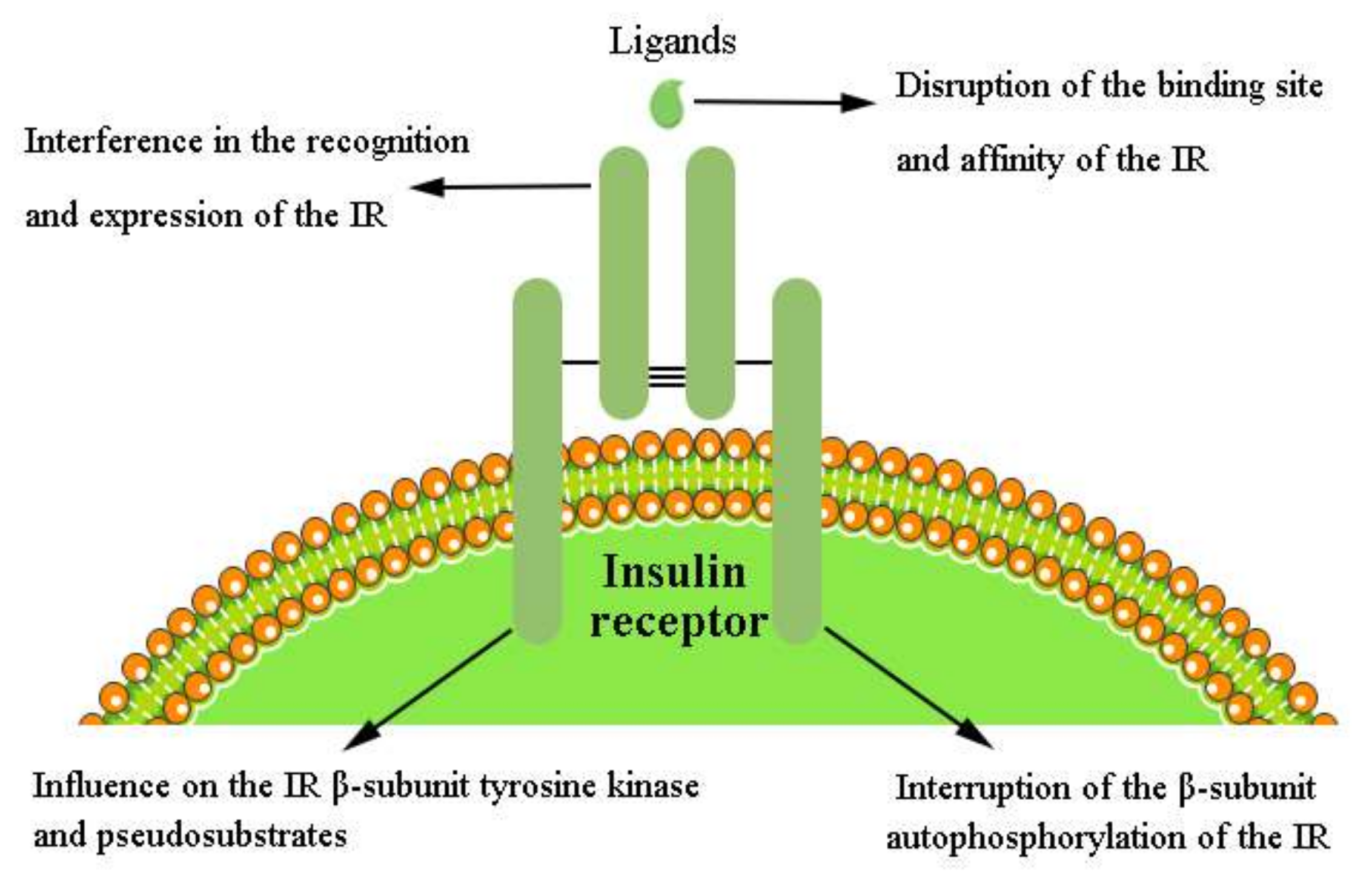
4.7. Mechanism of the Pharmacological and Physiological Modulators of IR Activation
4.7.1. Disruption of the Binding Site and Affinity of the IR
4.7.2. Interference in the Recognition and Expression of the IR in the IIS Pathway
4.7.3. Interruption of β-Subunit Autophosphorylation of the IR
4.7.4. Influence on the IR β-Subunit Tyrosine Kinase and Pseudosubstrates
5. Nonpeptide Small Molecule Modulators of the IR
5.1. Small Molecule Inducers of IR Autophosphorylation
5.2. Modulators of the Tyrosine Kinase Domain of the IR β-Subunit
5.3. Regulators of the IR and IGF-1R
6. Molecular Mechanisms of Modulators Targeting the IR
6.1. Gene Expression of the IR Changed by DNA Methylation
6.2. Gene Expression of the IR Regulated by Xenobiotic Compounds
6.3. IR Expression Regulated Posttranscriptionally via MicroRNAs
6.4. Regulation of Expression of the IR Protein
6.4.1. Ubiquitination of the IR
6.4.2. Endocytosis of the IR
6.4.3. Endosome Localization and Cleavage of the IR
7. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Ward, C.W.; Lawrence, M.C. Ligand-induced activation of the insulin receptor: A multi-step process involving structural changes in both the ligand and the receptor. Bioessays 2009, 31, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Vigneri, R.; Goldfine, I.D.; Frittitta, L. Insulin, insulin receptors, and cancer. J. Endocrinol. Investig. 2016, 39, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, R.A.; McGraw, T.E.; Accili, D. Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol. 2018, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Yunn, N.O.; Kim, J.; Kim, Y.; Leibiger, I.; Berggren, P.O.; Ryu, S.H. Mechanistic understanding of insulin receptor modulation: Implications for the development of anti-diabetic drugs. Pharmacolo. Therapeut. 2018, 185, 86–98. [Google Scholar] [CrossRef]
- Brogiolo, W.; Stocker, H.; Ikeya, T.; Rintelen, F.; Fernandez, R.; Hafen, E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001, 11, 213–221. [Google Scholar] [CrossRef]
- Lin, X.; Smagghe, G. Roles of the insulin signaling pathway in insect development and organ growth. Peptides 2019, 122, 169923. [Google Scholar] [CrossRef]
- Liu, C.Y.; Zhao, W.L.; Wang, J.X.; Zhao, X.F. Cyclin-dependent kinase regulatory subunit 1 promotes cell proliferation by insulin regulation. Cell Cycle 2015, 14, 3045–3057. [Google Scholar] [CrossRef][Green Version]
- Craft, S.; Cholerton, B.; Baker, L.D. Insulin and Alzheimer’s disease: Untangling the web. J. Alzheimer’s Dis. 2013, 33 (Suppl. S1), S263–S275. [Google Scholar] [CrossRef]
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef]
- Westermeier, F.; Saez, T.; Arroyo, P.; Toledo, F.; Gutierrez, J.; Sanhueza, C.; Pardo, F.; Leiva, A.; Sobrevia, L. Insulin receptor isoforms: An integrated view focused on gestational diabetes mellitus. Diabetes Metab. Res. Rev. 2016, 32, 350–365. [Google Scholar] [CrossRef]
- Vella, V.; Milluzzo, A.; Scalisi, N.; Vigneri, P.; Sciacca, L. Insulin receptor isoforms in cancer. Int. J. Mol. Sci. 2018, 19, 3615. [Google Scholar] [CrossRef]
- Sciacca, L.; Le Moli, R.; Vigneri, R. Insulin analogs and cancer. Front. Endocrinol. 2012, 3, 21. [Google Scholar] [CrossRef]
- Malaguarnera, R.; Belfiore, A. The insulin receptor: A new target for cancer therapy. Front. Endocrinol. 2011, 2, 93. [Google Scholar] [CrossRef]
- Stoeckel, L.E.; Arvanitakis, Z.; Gandy, S.; Small, D.; Kahn, C.R.; Pascual-Leone, A.; Pawlyk, A.; Sherwin, R.; Smith, P. Complex mechanisms linking neurocognitive dysfunction to insulin resistance and other metabolic dysfunction. F1000Res 2016, 5, 353. [Google Scholar] [CrossRef]
- Titchenell, P.M.; Lazar, M.A.; Birnbaum, M.J. Unraveling the regulation of hepatic metabolism by insulin. Trends Endocrinol Metab. 2017, 28, 497–505. [Google Scholar] [CrossRef]
- Campbell, R.K. Type 2 diabetes: Where we are today: An overview of disease burden, current treatments, and treatment strategies. J. Am. Pharm. Assoc. 2009, 49, S3–S9. [Google Scholar] [CrossRef]
- Ermakova, A.; Stauffer, M.E.; Sieradzan, R.; Taylor, S. Characterizing the clinical and economic burden of type 2 diabetes (T2DM) patients on multiple daily injections (MDI) of insulin: A systematic literature review. Value Health 2018, 21, S70. [Google Scholar] [CrossRef]
- Buyruk, B.A.; Kebapci, N.; Yorulmaz, G.; Alaguney, E.S.; Akalin, A.; Efe, B. Prevalence and risk factors of lipohypertrophy and lipoatrophy in diabetes patients receiving insulin therapy. Diabetes 2019, 68, 59. [Google Scholar] [CrossRef]
- Brietzke, S.A. Oral antihyperglycemic treatment options for type 2 diabetes mellitus. Med. Clin. North Am. 2015, 99, 87–106. [Google Scholar] [CrossRef]
- Singh, P.; Alex, J.M.; Bast, F. Insulin receptor (IR) and insulin-like growth factor receptor 1 (IGF-1R) signaling systems: Novel treatment strategies for cancer. Med. Oncol. 2014, 31, 805. [Google Scholar] [CrossRef]
- Meakin, P.J.; Mezzapesa, A.; Benabou, E.; Haas, M.E.; Bonardo, B.; Grino, M.; Brunel, J.M.; Desbois-Mouthon, C.; Biddinger, S.B.; Govers, R.; et al. The beta secretase BACE1 regulates the expression of insulin receptor in the liver. Nat. Commun. 2018, 9, 1306. [Google Scholar] [CrossRef]
- Russo, A.; Paret, C.; Alt, F.; Burhenne, J.; Fresnais, M.; Wagner, W.; Glaser, M.; Bender, H.; Huprich, S.; Harter, P.N.; et al. Ceritinib-induced regression of an insulin-like growth factor-driven neuroepithelial brain tumor. Int. J. Mol. Sci. 2019, 20, 4267. [Google Scholar] [CrossRef]
- Wenbin, K.; Xiaoqin, L.; Qiuchan, D.; Xinwen, Z.; Xiaoqin, X.; Fangyuan, S.; Dabao, H.; Shuangjiu, Z. Development of a novel insulin receptor (IR) antagonist that exhibits anti-breast tumor activity. Human Cell 2020, 33, 1204–1217. [Google Scholar] [CrossRef]
- Lin, X.; Lavine, L.C. Endocrine regulation of a dispersal polymorphism in winged insects: A short review. ScienceDirect 2018, 25, 20–24. [Google Scholar] [CrossRef]
- Guo, S.; Wang, X.; Kang, L. Special significance of non-Drosophila insects in aging. Front. Cell Dev. Biol. 2020, 8, 576571. [Google Scholar] [CrossRef]
- Alvarez-Rendon, J.P.; Salceda, R.; Riesgo-Escovar, J.R. Drosophila melanogaster as a model for diabetes type 2 progression. Biomed. Res. Int. 2018, 2018, 1417528. [Google Scholar] [CrossRef]
- Houchat, J.N.; Cartereau, A.; Le Mauff, A.; Taillebois, E.; Thany, S.H. An overview on the effect of neonicotinoid insecticides on mammalian cholinergic functions through the activation of neuronal nicotinic acetylcholine receptors. Int. J. Environ. Res. Public Health 2020, 17, 3222. [Google Scholar] [CrossRef]
- Sun, Z.; Xu, H. Ryanodine receptors for drugs and insecticides: An overview. Mini. Rev. Med. Chem. 2019, 19, 22–33. [Google Scholar] [CrossRef]
- Sun, Z.; Lv, M.; Huang, W.; Li, T.; Xu, H. Development of botanical pesticides: Exploration on the phenotype of vestigial wings of insect pests induced by plant natural products or their derivatives by blocking tyrosine phosphorylation of insulin receptor 1. J. Agric. Food Chem. 2022, 70, 2117–2126. [Google Scholar] [CrossRef]
- Belfiore, A.; Frasca, F.; Pandini, G.; Sciacca, L.; Vigneri, R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 2009, 30, 586–623. [Google Scholar] [CrossRef]
- Beneit, N.; Fernandez-Garcia, C.E.; Martin-Ventura, J.L.; Perdomo, L.; Escribano, O.; Michel, J.B.; Garcia-Gomez, G.; Fernandez, S.; Diaz-Castroverde, S.; Egido, J.; et al. Expression of insulin receptor (IR) A and B isoforms, IGF-IR, and IR/IGF-IR hybrid receptors in vascular smooth muscle cells and their role in cell migration in atherosclerosis. Cardiovasc. Diabetol. 2016, 15, 161. [Google Scholar] [CrossRef] [PubMed]
- Belfiore, A.; Malaguarnera, R.; Vella, V.; Lawrence, M.C.; Sciacca, L.; Frasca, F.; Morrione, A.; Vigneri, R. Insulin receptor isoforms in physiology and disease: An updated view. Endocr. Rev. 2017, 38, 379–431. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Morehouse, C.; Streicher, K.; Higgs, B.W.; Gao, J.; Czapiga, M.; Boutrin, A.; Zhu, W.; Brohawn, P.; Chang, Y.; et al. Altered expression of insulin receptor isoforms in breast cancer. PLoS ONE. 2011, 6, e26177. [Google Scholar] [CrossRef] [PubMed]
- Heidegger, I.; Kern, J.; Ofer, P.; Klocker, H.; Massoner, P. Oncogenic functions of IGF1R and INSR in prostate cancer include enhanced tumor growth, cell migration and angiogenesis. Oncotarget 2014, 5, 2723–2735. [Google Scholar] [CrossRef]
- Lawrence, M.C. Understanding insulin and its receptor from their three-dimensional structures. Mol Metab. 2021, 52, 101255. [Google Scholar] [CrossRef]
- De Meyts, P. Insulin/receptor binding: The last piece of the puzzle? What recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. Bioessays 2015, 37, 389–397. [Google Scholar] [CrossRef]
- Croll, T.I.; Smith, B.J.; Margetts, M.B.; Whittaker, J.; Weiss, M.A.; Ward, C.W.; Lawrence, M.C. Higher-resolution structure of the human insulin receptor ectodomain: Multi-modal inclusion of the insert domain. Structure 2016, 24, 469–476. [Google Scholar] [CrossRef]
- Ye, L.; Maji, S.; Sanghera, N.; Gopalasingam, P.; Gorbunov, E.; Tarasov, S.; Epstein, O.; Klein-Seetharaman, J. Structure and dynamics of the insulin receptor: Implications for receptor activation and drug discovery. Drug Discov. Today 2017, 22, 1092–1102. [Google Scholar] [CrossRef]
- Scapin, G.; Dandey, V.P.; Zhang, Z.; Prosise, W.; Hruza, A.; Kelly, T.; Mayhood, T.; Strickland, C.; Potter, C.S.; Carragher, B. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 2018, 556, 122–125. [Google Scholar] [CrossRef]
- McKern, N.M.; Lawrence, M.C.; Streltsov, V.A.; Lou, M.Z.; Adams, T.E.; Lovrecz, G.O.; Elleman, T.C.; Richards, K.M.; Bentley, J.D.; Pilling, P.A.; et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 2006, 443, 218–221. [Google Scholar] [CrossRef]
- Gutmann, T.; Kim, K.H.; Grzybek, M.; Walz, T.; Coskun, U. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J. Cell Biol. 2018, 217, 1643–1649. [Google Scholar] [CrossRef]
- Uchikawa, E.; Choi, E.; Shang, G.; Yu, H.; Bai, X.C. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife 2019, 8, e48630. [Google Scholar] [CrossRef]
- Gutmann, T.; Schafer, I.B.; Poojari, C.; Brankatschk, B.; Vattulainen, I.; Strauss, M.; Coskun, U. Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J. Cell Biol. 2020, 219, e201907210. [Google Scholar] [CrossRef]
- Weis, F.; Menting, J.G.; Margetts, M.B.; Chan, S.J.; Xu, Y.; Tennagels, N.; Wohlfart, P.; Langer, T.; Muller, C.W.; Dreyer, M.K.; et al. The signalling conformation of the insulin receptor ectodomain. Nat. Commun. 2018, 9, 4420. [Google Scholar] [CrossRef]
- Whittaker, J.; Whittaker, L. Characterization of the functional insulin binding epitopes of the full-length insulin receptor. J. Biol. Chem. 2005, 280, 20932–20936. [Google Scholar] [CrossRef]
- Menting, J.G.; Whittaker, J.; Margetts, M.B.; Whittaker, L.J.; Kong, G.K.; Smith, B.J.; Watson, C.J.; Zakova, L.; Kletvikova, E.; Jiracek, J.; et al. How insulin engages its primary binding site on the insulin receptor. Nature 2013, 493, 241–245. [Google Scholar] [CrossRef]
- Chrudinova, M.; Zakova, L.; Marek, A.; Socha, O.; Budesinsky, M.; Hubalek, M.; Picha, J.; Machackova, K.; Jiracek, J.; Selicharova, I. A versatile insulin analog with high potency for both insulin and insulin-like growth factor 1 receptors: Structural implications for receptor binding. J. Biol. Chem. 2018, 293, 16818–16829. [Google Scholar] [CrossRef]
- Kavran, J.M.; McCabe, J.M.; Byrne, P.O.; Connacher, M.K.; Wang, Z.; Ramek, A.; Sarabipour, S.; Shan, Y.; Shaw, D.E.; Hristova, K.; et al. How IGF-1 activates its receptor. Elife 2014, 3, e03772. [Google Scholar] [CrossRef]
- Xu, Y.; Kong, G.K.; Menting, J.G.; Margetts, M.B.; Delaine, C.A.; Jenkin, L.M.; Kiselyov, V.V.; De Meyts, P.; Forbes, B.E.; Lawrence, M.C. How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun. 2018, 9, 821. [Google Scholar] [CrossRef]
- Yunn, N.-O.; Park, M.; Noh, J.; Ryu, S.H. Stepwise autophosphorylation regulates biased agonism of the insulin receptor. Faseb. J. 2019, 34, s1.08794. [Google Scholar] [CrossRef]
- Hernandez-Sanchez, C.; Mansilla, A.; Pablo, F.d.; Zardoya, R. Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol. Biol. Evol. 2008, 25, 1043–1053. [Google Scholar] [CrossRef]
- Li, J.; Park, J.; Mayer, J.P.; Webb, K.J.; Uchikawa, E.; Wu, J.Y.; Liu, S.; Zhang, X.W.; Stowell, M.H.B.; Choi, E.; et al. Synergistic activation of the insulin receptor via two distinct sites. Nat. Struct. Mol. Biol. 2022, 29, 357–368. [Google Scholar] [CrossRef]
- Das, D.; Arur, S. Conserved insulin signaling in the regulation of oocyte growth, development, and maturation. Mol. Reprod. Dev. 2017, 84, 444–459. [Google Scholar] [CrossRef]
- Lawrence, M.C.; McKern, N.M.; Ward, C.W. Insulin receptor structure and its implications for the IGF-1 receptor. Curr. Opin. Struct Biol. 2007, 17, 699–705. [Google Scholar] [CrossRef]
- Sim, C.; Denlinger, D.L. Insulin signaling and the regulation of insect diapause. Front. Physiol. 2013, 4, 189. [Google Scholar] [CrossRef]
- Hall, C.; Yu, H.; Choi, E. Insulin receptor endocytosis in the pathophysiology of insulin resistance. Exp. Mol. Med. 2020, 52, 911–920. [Google Scholar] [CrossRef]
- Cai, W.; Sakaguchi, M.; Kleinridders, A.; Gonzalez-Del Pino, G.; Dreyfuss, J.M.; O’Neill, B.T.; Ramirez, A.K.; Pan, H.; Winnay, J.N.; Boucher, J.; et al. Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat. Commun. 2017, 8, 14892. [Google Scholar] [CrossRef]
- Okada, Y.; Katsuki, M.; Okamoto, N.; Fujioka, H.; Okada, K. A specific type of insulin-like peptide regulates the conditional growth of a beetle weapon. PLoS Biol. 2019, 17, e3000541. [Google Scholar] [CrossRef]
- Laustsen, P.G.; Russell, S.J.; Cui, L.; Entingh-Pearsall, A.; Holzenberger, M.; Liao, R.; Kahn, C.R. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Mol. Cell Biol. 2007, 27, 1649–1664. [Google Scholar] [CrossRef]
- Nassel, D.R.; Vanden Broeck, J. Insulin/IGF signaling in Drosophila and other insects: Factors that regulate production, release and post-release action of the insulin-like peptides. Cell Mol. Life Sci. 2016, 73, 271–290. [Google Scholar] [CrossRef]
- Inoue, H. Central insulin-mediated regulation of hepatic glucose production. Endocr. J. 2016, 63, EJ15-0540. [Google Scholar] [CrossRef] [PubMed]
- Nijhout, H.F.; Smith, W.A.; Schachar, I.; Subramanian, S.; Tobler, A.; Grunert, L.W. The control of growth and differentiation of the wing imaginal disks of Manduca sexta. Dev. Biol. 2007, 302, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Pan, J.; Di, Y.Q.; Liu, W.; Hou, L.; Wang, J.X.; Zhao, X.F. Protein kinase C delta phosphorylates ecdysone receptor B1 to promote gene expression and apoptosis under 20-hydroxyecdysone regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E7121–E7130. [Google Scholar] [CrossRef] [PubMed]
- Ajaha, A.; Bouayad, N.; Aarab, A.; Rharrabe, K. Effect of 20-hydroxyecdysone, a phytoecdysteroid, on development, digestive, and detoxification enzyme activities of Tribolium castaneum (Coleoptera: Tenebrionidae). J. Insect Sci. 2019, 19, 18. [Google Scholar] [CrossRef]
- Wu, Q.; Brown, M.R. Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 2006, 51, 1–24. [Google Scholar] [CrossRef]
- Nagasawa, H.; Kataoka, H.; Isogai, A.; Tamura, S.; Suzuki, A.; Ishizaki, H.; Mizoguchi, A.; Fujiwara, Y.; Suzuki, A. Amino-terminal amino acid sequence of the silkworm prothoracicotropic hormone: Homology with insulin. Science 1984, 226, 1344–1345. [Google Scholar] [CrossRef]
- Siddle, K. Molecular basis of signaling specificity of insulin and IGF receptors: Neglected corners and recent advances. Front. Endocrinol. 2012, 3, 34. [Google Scholar] [CrossRef]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014, 6, a009191. [Google Scholar] [CrossRef]
- Versteyhe, S.; Blanquart, C.; Hampe, C.; Mahmood, S.; Christeff, N.; Meyts1, P.D.; Gray, S.G.; Issad, T. Insulin receptor substrates-5 and -6 are poor substrates for the insulin receptor. Mol. Med. Rep. 2010, 3, 189–193. [Google Scholar] [CrossRef][Green Version]
- Schmidt, V.; Schulz, N.; Yan, X.; Schurmann, A.; Kempa, S.; Kern, M.; Bluher, M.; Poy, M.N.; Olivecrona, G.; Willnow, T.E. SORLA facilitates insulin receptor signaling in adipocytes and exacerbates obesity. J. Clin. Investig. 2016, 126, 2706–2720. [Google Scholar] [CrossRef]
- Rask-Madsen, C.; Kahn, C.R. Tissue-specific insulin signaling, metabolic syndrome, and cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2052–2059. [Google Scholar] [CrossRef]
- Shi, X.; Xie, X.; Jia, Y.; Li, S. Associations of insulin receptor and insulin receptor substrates genetic polymorphisms with polycystic ovary syndrome: A systematic review and meta-analysis. J. Obstet. Gynaecol. Res. 2016, 42, 844–854. [Google Scholar] [CrossRef]
- Gralle, M.; Labrecque, S.; Salesse, C.; De Koninck, P. Spatial dynamics of the insulin receptor in living neurons. J. Neurochem. 2020, 156, 88–105. [Google Scholar] [CrossRef]
- Ling, A.V.; Gearing, M.E.; Semova, I.; Shin, D.J.; Clements, R.; Lai, Z.W.; Biddinger, S.B. FoxO1 is required for most of the metabolic and hormonal perturbations produced by hepatic insulin receptor deletion in male mice. Endocrinology 2018, 159, 1253–1263. [Google Scholar] [CrossRef]
- Titone, R.; Robertson, D.M. Insulin receptor preserves mitochondrial function by binding VDAC1 in insulin insensitive mucosal epithelial cells. FASEB J. 2019, 34, 754–775. [Google Scholar] [CrossRef]
- Oakie, A.; Zhou, L.; Rivers, S.; Cheung, C.; Li, J.; Wang, R. Postnatal knockout of beta cell insulin receptor impaired insulin secretion in male mice exposed to high-fat diet stress. Mol. Cell Endocrinol. 2020, 499, 110588. [Google Scholar] [CrossRef]
- Ardon, O.; Procter, M.; Tvrdik, T.; Longo, N.; Mao, R. Sequencing analysis of insulin receptor defects and detection of two novel mutations in INSR gene. Mol. Genet. Metab. Rep. 2014, 1, 71–84. [Google Scholar] [CrossRef]
- Greenhill, C. Insulin and the insulin receptor regulate gene expression. Nat. Rev. Endocrinol. 2019, 15, 315. [Google Scholar] [CrossRef]
- Hancock, M.L.; Meyer, R.C.; Mistry, M.; Khetani, R.S.; Wagschal, A.; Shin, T.; Ho Sui, S.J.; Naar, A.M.; Flanagan, J.G. Insulin receptor associates with promoters genome-wide and regulates gene expression. Cell 2019, 177, 722–736. [Google Scholar] [CrossRef]
- Ott, R.; Melchior, K.; Stupin, J.H.; Ziska, T.; Schellong, K.; Henrich, W.; Rancourt, R.C.; Plagemann, A. Reduced insulin receptor expression and altered DNA methylation in fat tissues and blood of women with GDM and offspring. J. Clin. Endocrinol. Metab. 2019, 104, 137–149. [Google Scholar] [CrossRef]
- Schellong, K.; Melchior, K.; Ziska, T.; Ott, R.; Henrich, W.; Rancourt, R.C.; Plagemann, A. Hypothalamic insulin receptor expression and DNA promoter methylation are sex-specifically altered in adult offspring of high-fat diet (HFD)-overfed mother rats. J. Nutr. Biochem. 2019, 67, 28–35. [Google Scholar] [CrossRef]
- Stolzenbach, F.; Valdivia, S.; Ojeda-Provoste, P.; Toledo, F.; Sobrevia, L.; Kerr, B. DNA methylation changes in genes coding for leptin and insulin receptors during metabolic-altered pregnancies. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165465. [Google Scholar] [CrossRef]
- Pan, J.; Di, Y.Q.; Li, Y.B.; Chen, C.H.; Wang, J.X.; Zhao, X.F. Insulin and 20-hydroxyecdysone oppose each other in the regulation of phosphoinositide-dependent kinase-1 expression during insect pupation. J. Biol. Chem. 2018, 293, 18613–18623. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, S.; Ma, L.; Tian, L.; Wang, S.; Sheng, Z.; Jiang, R.J.; Bendena, W.G.; Li, S. Transcriptional regulation of the insulin signaling pathway genes by starvation and 20-hydroxyecdysone in the Bombyx fat body. J. Insect Physiol. 2010, 56, 1436–1444. [Google Scholar] [CrossRef]
- Li, Y.L.; Yao, Y.X.; Zhao, Y.M.; Di, Y.Q.; Zhao, X.F. The steroid hormone 20-hydroxyecdysone counteracts insulin signaling via insulin receptor dephosphorylation. J. Biol. Chem. 2021, 296, 100318. [Google Scholar] [CrossRef]
- Emlen, D.J.; Warren, I.A.; Johns, A.; Dworkin, I.; Lavine, L.C. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 2012, 337, 860–864. [Google Scholar] [CrossRef]
- Casasa, S.; Moczek, A.P. Insulin signalling’s role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc. Biol. Sci. 2018, 285, 20181631. [Google Scholar] [CrossRef]
- Eichler, K.; Li, F.; Litwin-Kumar, A.; Park, Y.; Andrade, I.; Schneider-Mizell, C.M.; Saumweber, T.; Huser, A.; Eschbach, C.; Gerber, B.; et al. The complete connectome of a learning and memory centre in an insect brain. Nature 2017, 548, 175–182. [Google Scholar] [CrossRef]
- Thum, A.S.; Gerber, B. Connectomics and function of a memory network: The mushroom body of larval Drosophila. Curr. Opin. Neurobiol. 2019, 54, 146–154. [Google Scholar] [CrossRef]
- Eschment, M.; Franz, H.R.; Gullu, N.; Holscher, L.G.; Huh, K.E.; Widmann, A. Insulin signaling represents a gating mechanism between different memory phases in Drosophila larvae. PLoS Genet. 2020, 16, e1009064. [Google Scholar] [CrossRef]
- Toprak, U. The role of peptide hormones in insect lipid metabolism. Front. Physiol. 2020, 11, 434. [Google Scholar] [CrossRef] [PubMed]
- Smykal, V.; Pivarci, M.; Provaznik, J.; Bazalova, O.; Jedlicka, P.; Luksan, O.; Horak, A.; Vaneckova, H.; Benes, V.; Fiala, I.; et al. Complex evolution of insect insulin receptors and homologous decoy receptors, and functional significance of their multiplicity. Mol. Biol. Evol. 2020, 37, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Barbera, M.; Canas-Canas, R.; Martinez-Torres, D. Insulin-like peptides involved in photoperiodism in the aphid Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 2019, 112, 103185. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Yu, N.; Smagghe, G. Insulin receptor regulates food intake through sulfakinin signaling in the red flour beetle, Tribolium castaneum. Peptides 2016, 80, 89–95. [Google Scholar] [CrossRef]
- Sang, M.; Li, C.; Wu, W.; Li, B. Identification and evolution of two insulin receptor genes involved in Tribolium castaneum development and reproduction. Gene 2016, 585, 196–204. [Google Scholar] [CrossRef]
- Nuss, A.B.; Brown, M.R.; Murty, U.S.; Gulia-Nuss, M. Insulin receptor knockdown blocks filarial parasite development and alters egg production in the southern house mosquito, Culex quinquefasciatus. PLoS Negl. Trop. Dis. 2018, 12, e0006413. [Google Scholar] [CrossRef]
- Ihle, K.E.; Mutti, N.S.; Kaftanoglu, O.; Amdam, G.V. Insulin receptor substrate gene knockdown accelerates behavioural maturation and shortens lifespan in honeybee workers. Insects 2019, 10, 390. [Google Scholar] [CrossRef]
- Han, B.; Zhang, T.; Feng, Y.; Liu, X.; Zhang, L.; Chen, H.; Zeng, F.; Wang, M.; Liu, C.; Li, Y.; et al. Two insulin receptors coordinate oogenesis and oviposition via two pathways in the green lacewing, Chrysopa pallens. J. Insect Physiol. 2020, 123, 104049. [Google Scholar] [CrossRef]
- Petruzzelli, L.; Herrera, R.; Garcia-Arenas, R.; Rosen, O.M. Acquisition of insulin-dependent protein tyrosine kinase activity during Drosophila embryogenesis. J. Biol. Chem. 1985, 10, 351–353. [Google Scholar] [CrossRef]
- Kremer, L.P.M.; Korb, J.; Bornberg-Bauer, E. Reconstructed evolution of insulin receptors in insects reveals duplications in early insects and cockroaches. J. Exp. Zool. Part B 2018, 330, 305–311. [Google Scholar] [CrossRef]
- Chambers, D.B.; Androschuk, A.; Rosenfelt, C.; Langer, S.; Harding, M.; Bolduc, F.V. Insulin signaling is acutely required for long-term memory in Drosophila. Front. Neural. Circuits 2015, 9, 8. [Google Scholar] [CrossRef]
- Tanabe, K.; Itoh, M.; Tonoki, A. Age-related changes in insulin-like signaling lead to intermediate-term memory impairment in Drosophila. Cell Rep. 2017, 18, 1598–1605. [Google Scholar] [CrossRef]
- Teleman, A.A. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 2009, 425, 13–26. [Google Scholar] [CrossRef]
- Nassel, D.R.; Kubrak, O.I.; Liu, Y.; Luo, J.; Lushchak, O.V. Factors that regulate insulin producing cells and their output in Drosophila. Front. Physiol. 2013, 4, 252. [Google Scholar] [CrossRef]
- Erion, R.; Sehgal, A. Regulation of insect behavior via the insulin-signaling pathway. Front. Physiol. 2013, 4, 353. [Google Scholar] [CrossRef]
- Ding, B.Y.; Shang, F.; Zhang, Q.; Xiong, Y.; Yang, Q.; Niu, J.Z.; Smagghe, G.; Wang, J.J. Silencing of two insulin receptor genes disrupts nymph-adult transition of alate brown citrus aphid. Int. J. Mol. Sci. 2017, 18, 357. [Google Scholar] [CrossRef]
- Baki, A.A.; Jung, J.K.; Kim, Y. Alteration of insulin signaling to control insect pest by using transformed bacteria expressing dsRNA. Pest. Manag. Sci. 2020, 76, 1020–1030. [Google Scholar] [CrossRef]
- Gulia-Nuss, M.; Robertson, A.E.; Brown, M.R.; Strand, M.R. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS ONE 2011, 6, e20401. [Google Scholar] [CrossRef]
- Sim, C.; Denlinger, D.L. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc. Natl. Acad. Sci. USA 2008, 105, 6777–6781. [Google Scholar] [CrossRef]
- Zhao, Y.O.; Kurscheid, S.; Zhang, Y.; Liu, L.; Zhang, L.; Loeliger, K.; Fikrig, E. Enhanced survival of Plasmodium-infected mosquitoes during starvation. PLoS ONE 2012, 7, e40556. [Google Scholar] [CrossRef]
- Castillo, J.; Brown, M.R.; Strand, M.R. Blood feeding and insulin-like peptide 3 stimulate proliferation of hemocytes in the mosquito Aedes aegypti. PLoS Pathog. 2011, 7, e1002274. [Google Scholar] [CrossRef]
- Silva-Oliveira, G.; De Paula, I.F.; Medina, J.M.; Alves-Bezerra, M.; Gondim, K.C. Insulin receptor deficiency reduces lipid synthesis and reproductive function in the insect Rhodnius prolixus. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020, 1866, 158851. [Google Scholar] [CrossRef]
- Bui, L.T.; Ivers, N.A.; Ragsdale, E.J. A sulfotransferase dosage-dependently regulates mouthpart polyphenism in the nematode Pristionchus pacificus. Nat. Commun. 2018, 9, 4119. [Google Scholar] [CrossRef]
- Yang, C.H.; Andrew Pospisilik, J. Polyphenism—a window into gene-environment interactions and phenotypic plasticity. Front. Genet. 2019, 10, 132. [Google Scholar] [CrossRef]
- Hoy, M.A. Molecular genetics of insect behavior. In Insect Molecular Genetics; Elsevier Science: Amsterdam, The Netherlands, 2019; pp. 413–461. [Google Scholar]
- Wheeler, D.E.; Buck, N.; Evans, J.D. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol. Biol. 2006, 15, 597–602. [Google Scholar] [CrossRef]
- Azevedo, S.V.; Hartfelder, K. The insulin signaling pathway in honey bee (Apis mellifera) caste development-differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 2008, 54, 1064–1071. [Google Scholar] [CrossRef]
- Lu, H.L.; Pietrantonio, P.V. Insect insulin receptors: Insights from sequence and caste expression analyses of two cloned hymenopteran insulin receptor cDNAs from the fire ant. Insect Mol Biol. 2011, 20, 637–649. [Google Scholar] [CrossRef]
- Wheeler, D.E.; Buck, N.A.; Evans, J.D. Expression of insulin/insulin-like signalling and TOR pathway genes in honey bee caste determination. Insect Mol. Biol. 2014, 23, 113–121. [Google Scholar] [CrossRef]
- Corona, M.; Libbrecht, R.; Wheeler, D.E. Molecular mechanisms of phenotypic plasticity in social insects. Curr. Opin. Insect Sci. 2016, 13, 55–60. [Google Scholar] [CrossRef]
- Mutti, N.S.; Dolezal, A.G.; Wolschin, F.; Mutti, J.S.; Gill, K.S.; Amdam, G.V. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J. Exp. Biol. 2011, 214, 3977–3984. [Google Scholar] [CrossRef]
- Wolschin, F.; Mutti, N.S.; Amdam, G.V. Insulin receptor substrate influences female caste development in honeybees. Biol. Lett. 2011, 7, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.J.; Xue, J.; Lu, B.; Zhang, X.C.; Zhuo, J.C.; He, S.F.; Ma, X.F.; Jiang, Y.Q.; Fan, H.W.; Xu, J.Y.; et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 2015, 519, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Xu, L.; Li, X.; He, K.; Hua, H.; Cao, Z.; Xu, J.; Ye, W.; Zhang, J.; Yuan, Z.; et al. miR-34 modulates wing polyphenism in planthopper. PLoS Genet. 2019, 15, e1008235. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.J.; Zhang, C.X. Insulin receptors and wing dimorphism in rice planthoppers. Philos. Trans. R Soc. Lond. B Biol. Sci. 2017, 372, 20150489. [Google Scholar] [CrossRef]
- Li, X.; Liu, F.; Wu, C.; Zhao, J.; Cai, W.; Hua, H. Decapentaplegic function in wing vein development and wing morph transformation in brown planthopper, Nilaparvata lugens. Dev. Biol. 2019, 449, 143–150. [Google Scholar] [CrossRef]
- Zhang, C.X.; Brisson, J.A.; Xu, H.J. Molecular mechanisms of wing polymorphism in insects. Annu. Rev. Entomol. 2019, 64, 297–314. [Google Scholar] [CrossRef]
- Lin, X.; Gao, H.; Xu, Y.; Zhang, Y.; Li, Y.; Lavine, M.D.; Lavine, L.C. Cell cycle progression determines wing morph in the polyphenic insect Nilaparvata lugens. iScience 2020, 23, 101040. [Google Scholar] [CrossRef]
- Zhuo, J.C.; Chen, L.; Shi, J.K.; Xu, N.; Xue, W.H.; Zhang, M.Q.; Ren, Z.W.; Zhang, H.H.; Zhang, C.X. Tra-2 mediates crosstalk between sex determination and wing polyphenism in female Nilaparvata lugens. Genetics 2017, 207, 1067–1078. [Google Scholar] [CrossRef]
- Belgacem, Y.H.; Martin, J.R. Disruption of insulin pathways alters trehalose level and abolishes sexual dimorphism in locomotor activity in Drosophila. J. Neurobiol. 2006, 66, 19–32. [Google Scholar] [CrossRef]
- Belgacem, Y.H.; Martin, J.R. Hmgcr in the corpus allatum controls sexual dimorphism of locomotor activity and body size via the insulin pathway in Drosophila. PLoS ONE. 2007, 2, e187. [Google Scholar] [CrossRef]
- Hinke, S.A.; Cieniewicz, A.M.; Kirchner, T.; D’Aquino, K.; Nanjunda, R.; Aligo, J.; Perkinson, R.; Cooper, P.; Boayke, K.; Chiu, M.L.; et al. Unique pharmacology of a novel allosteric agonist/sensitizer insulin receptor monoclonal antibody. Mol. Metab. 2018, 10, 87–99. [Google Scholar] [CrossRef]
- Changeux, J.P.; Christopoulos, A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell 2016, 166, 1084–1102. [Google Scholar] [CrossRef]
- Othman, E.M.; Kreissl, M.C.; Kaiser, F.R.; Arias-Loza, P.A.; Stopper, H. Insulin-mediated oxidative stress and DNA damage in LLC-PK1 pig kidney cell line, female rat primary kidney cells, and male ZDF rat kidneys in vivo. Endocrinology 2013, 154, 1434–1443. [Google Scholar] [CrossRef][Green Version]
- Lefever, E.; Vliebergh, J.; Mathieu, C. Improving the treatment of patients with diabetes using insulin analogues: Current findings and future directions. Expert. Opin. Drug Saf. 2021, 20, 155–169. [Google Scholar] [CrossRef]
- Kasia, J.; Lipska, M.D. Insulin analogues for type 2 diabetes. Jama-J Am. Med. Assoc. 2019, 321, 350–351. [Google Scholar]
- Kramer, C.K.; Retnakaran, R.; Zinman, B. Insulin and insulin analogs as antidiabetic therapy: A perspective from clinical trials. Cell Metab. 2021, 33, 740–747. [Google Scholar] [CrossRef]
- Mohammadiarani, H.; Vashisth, H. Insulin mimetic peptide S371 folds into a helical structure. J. Comput Chem. 2017, 38, 1158–1166. [Google Scholar] [CrossRef]
- Jensen, M.; Palsgaard, J.; Borup, R.; de Meyts, P.; Schaffer, L. Activation of the insulin receptor (IR) by insulin and a synthetic peptide has different effects on gene expression in IR-transfected L6 myoblasts. Biochem. J. 2008, 412, 435–445. [Google Scholar] [CrossRef]
- Schaffer, L.; Brand, C.L.; Hansen, B.F.; Ribel, U.; Shaw, A.C.; Slaaby, R.; Sturis, J. A novel high-affinity peptide antagonist to the insulin receptor. Biochem. Biophys. Res. Commun. 2008, 376, 380–383. [Google Scholar] [CrossRef]
- Vikram, A.; Jena, G. S961, an insulin receptor antagonist causes hyperinsulinemia, insulin-resistance and depletion of energy stores in rats. Biochem. Biophys. Res. Commun. 2010, 398, 260–265. [Google Scholar] [CrossRef]
- Knudsen, L.; Hansen, B.F.; Jensen, P.; Pedersen, T.A.; Vestergaard, K.; Schaffer, L.; Blagoev, B.; Oleksiewicz, M.B.; Kiselyov, V.V.; Meyts, P.D. Agonism and antagonism at the insulin receptor. PLoS ONE 2012, 7, e51972. [Google Scholar] [CrossRef]
- Taylor, S.I.; Accili, D.; Cama, A.; Imano, E.; Kadowaki, H.; Kadowaki, T. Unusual forms of insulin resistance. Annu. Rev. Med. 1991, 42, 373–379. [Google Scholar] [CrossRef]
- Bhaskar, V.; Goldfine, I.D.; Bedinger, D.H.; Lau, A.; Kuan, H.F.; Gross, L.M.; Handa, M.; Maddux, B.A.; Watson, S.R.; Zhu, S.; et al. A fully human, allosteric monoclonal antibody that activates the insulin receptor and improves glycemic control. Diabetes 2012, 61, 1263–1271. [Google Scholar] [CrossRef]
- Issafras, H.; Bedinger, D.H.; Corbin, J.A.; Goldfine, I.D.; Bhaskar, V.; White, M.L.; Rubin, P.; Scannon, P.J. Selective allosteric antibodies to the insulin receptor for the treatment of hyperglycemic and hypoglycemic disorders. J. Diabetes Sci. Technol. 2014, 8, 865–873. [Google Scholar] [CrossRef]
- Corbin, J.A.; Bhaskar, V.; Goldfine, I.D.; Bedinger, D.H.; Lau, A.; Michelson, K.; Gross, L.M.; Maddux, B.A.; Kuan, H.F.; Tran, C.; et al. Improved glucose metabolism in vitro and in vivo by an allosteric monoclonal antibody that increases insulin receptor binding affinity. PLoS ONE 2014, 9, e88684. [Google Scholar] [CrossRef]
- Corbin, J.A.; Bhaskar, V.; Goldfine, I.D.; Issafras, H.; Bedinger, D.H.; Lau, A.; Michelson, K.; Gross, L.M.; Maddux, B.A.; Kuan, H.F.; et al. Inhibition of insulin receptor function by a human, allosteric monoclonal antibody: A potential new approach for the treatment of hyperinsulinemic hypoglycemia. Mabs 2014, 6, 262–272. [Google Scholar] [CrossRef]
- Johnson, K.W.; Neale, A.; Gordon, A.; Roessig, J.; Bezwada, P.; Vukelich, S.; Goldfine, I.; Rubin, P. Attenuation of insulin action by an allosteric insulin receptor antibody in healthy volunteers. J. Clin. Endocrinol. Metab. 2017, 102, 3021–3028. [Google Scholar] [CrossRef]
- Cieniewicz, A.M.; Kirchner, T.; Hinke, S.A.; Nanjunda, R.; D’Aquino, K.; Boayke, K.; Cooper, P.R.; Perkinson, R.; Chiu, M.L.; Jarantow, S.; et al. Novel monoclonal antibody is an allosteric insulin receptor antagonist that induces insulin resistance. Diabetes 2017, 66, 206–217. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Yunn, N.-O.; Koh, A.; Han, S.; Lim, J.H.; Park, S.; Lee, J.; Kim, E.; Jang, S.K.; Berggren, P.O.; Ryu, S.H. Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation. Nucleic. Acids Res. 2015, 43, 7688–7701. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Yunn, N.-O.; Ryu, S.H. Sensitizing aptamer INSR-A43 enhances insulin sensitivity and sensitizes insulin receptor signaling pathways. Faseb J. 2020, 34, S1. [Google Scholar] [CrossRef]
- Yunn, N.-O.; Lee, J.; Lee, H.S.; Oh, E.J.; Park, M.; Park, S.; Jin, S.Y.; Shin, E.; Lee, J.W.Y.; Kim, Y.; et al. An aptamer agonist of the insulin receptor acts as a positive or negative allosteric modulator, depending on its concentration. Exp. Mol. Med. 2022, 54, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Iaboni, M.; Fontanella, R.; Rienzo, A.; Capuozzo, M.; Nuzzo, S.; Santamaria, G.; Catuogno, S.; Condorelli, G.; Franciscis, V.D.; Esposito, C.L. Targeting insulin receptor with a novel internalizing aptamer. Mol. Ther-Nucl. Acids 2016, 5, e365. [Google Scholar] [CrossRef]
- Yunn, N.-O.; Park, M.; Park, S.; Lee, J.; Noh, J.; Shin, E.; Ryu, S.H. A hotspot for enhancing insulin receptor activation revealed by a conformation-specific allosteric aptamer. Nucleic Acids Res. 2021, 49, 700–712. [Google Scholar] [CrossRef]
- Desbuquois, B.; Carre, N.; Burnol, A.F. Regulation of insulin and type 1 insulin-like growth factor signaling and action by the Grb10/14 and SH2B1/B2 adaptor proteins. FEBS J. 2013, 280, 794–816. [Google Scholar] [CrossRef]
- Yin, Y.; Chen, C.; Chen, J.; Zhan, R.; Zhang, Q.; Xu, X.; Li, D.; Li, M. Cell surface GRP78 facilitates hepatoma cells proliferation and migration by activating IGF-IR. Cell Signal. 2017, 35, 154–162. [Google Scholar] [CrossRef]
- Gu, X.; Reagan, A.M.; McClellan, M.E.; Elliott, M.H. Caveolins and caveolae in ocular physiology and pathophysiology. Prog. Retin. Eye. Res. 2017, 56, 84–106. [Google Scholar] [CrossRef]
- Fernandes, I.P.G.; Oliveira-Brett, A.M. Caveolin proteins electrochemical oxidation and interaction with cholesterol. Bioelectrochemistry 2020, 133, 107451. [Google Scholar] [CrossRef]
- Kwon, H.; Lee, J.; Jeong, K.; Jang, D.; Pak, Y. Fatty acylated caveolin-2 is a substrate of insulin receptor tyrosine kinase for insulin receptor substrate-1-directed signaling activation. Biochim. Biophys. Acta 2015, 1853, 1022–1034. [Google Scholar] [CrossRef]
- Kwon, H.; Choi, M.; Ahn, Y.; Pak, Y. N-myristoylation regulates insulin-induced phosphorylation and ubiquitination of Caveolin-2 for insulin signaling. Biochem. Biophys. Res. Commun. 2020, 532, 535–540. [Google Scholar] [CrossRef]
- Kwon, H.; Jang, D.; Choi, M.; Lee, J.; Jeong, K.; Pak, Y. Alternative translation initiation of Caveolin-2 desensitizes insulin signaling through dephosphorylation of insulin receptor by PTP1B and causes insulin resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2169–2182. [Google Scholar] [CrossRef]
- Zhao, N.; Liu, C.C.; Qiao, W.; Bu, G. Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol. Psychiatry 2018, 83, 347–357. [Google Scholar] [CrossRef]
- Chan, E.S.; Shetty, M.S.; Sajikumar, S.; Chen, C.; Soong, T.W.; Wong, B.S. ApoE4 expression accelerates hippocampus-dependent cognitive deficits by enhancing Abeta impairment of insulin signaling in an Alzheimer’s disease mouse model. Sci. Rep. 2016, 6, 26119. [Google Scholar] [CrossRef]
- Zhao, N.; Liu, C.C.; Ingelgom, A.J.V.; Martens, Y.A.; Linares, C.; Knight, J.A.; Painter, M.M.; Sullivan, P.M.; Bu, G.J. Apolipoprotein E4 impairs neuronal insulin signaling by trapping insulin receptor in the endosomes. Neuron 2017, 96, 115–129. [Google Scholar] [CrossRef]
- Ong, Q.R.; Chan, E.S.; Lim, M.L.; Cole, G.M.; Wong, B.S. Reduced phosphorylation of brain insulin receptor substrate and Akt proteins in apolipoprotein-E4 targeted replacement mice. Sci. Rep. 2014, 4, 3754. [Google Scholar] [CrossRef]
- Ussar, S.; Bezy, O.; Bluher, M.; Kahn, C.R. Glypican-4 enhances insulin signaling via interaction with the insulin receptor and serves as a novel adipokine. Diabetes 2012, 61, 2289–2298. [Google Scholar] [CrossRef]
- Liao, P.Y.; Lo, H.Y.; Liu, I.C.; Lo, L.C.; Hsiang, C.Y.; Ho, T.Y. A gastro-resistant peptide from Momordica charantia improves diabetic nephropathy in db/db mice via its novel reno-protective and anti-inflammatory activities. Food Funct. 2022, 13, 1822–1833. [Google Scholar] [CrossRef]
- Brown, J.E.; Onyango, D.J.; Ramanjaneya, M.; Conner, A.C.; Patel, S.T.; Dunmore, S.J.; Randeva, H.S. Visfatin regulates insulin secretion, insulin receptor signalling and mRNA expression of diabetes-related genes in mouse pancreatic beta-cells. J. Mol. Endocrinol. 2010, 44, 171–178. [Google Scholar] [CrossRef]
- Mitrofanova, A.; Mallela, S.K.; Ducasa, G.M.; Yoo, T.H.; Rosenfeld-Gur, E.; Zelnik, I.D.; Molina, J.; Varona Santos, J.; Ge, M.; Sloan, A.; et al. SMPDL3b modulates insulin receptor signaling in diabetic kidney disease. Nat. Commun. 2019, 10, 2692. [Google Scholar] [CrossRef]
- Prabhakar, P.K.; Sivakumar, P.M. Protein tyrosine phosphatase 1B inhibitors: A novel therapeutic strategy for the management of type 2 diabetes mellitus. Curr. Pharm. Des. 2019, 25, 2526–2539. [Google Scholar] [CrossRef]
- Verma, M.; Gupta, S.J.; Chaudhary, A.; Garg, V.K. Protein tyrosine phosphatase 1B inhibitors as antidiabetic agents—A brief review. Bioorg Chem. 2017, 70, 267–283. [Google Scholar] [CrossRef]
- Petersen, M.C.; Madiraju, A.K.; Gassaway, B.M.; Marcel, M.; Nasiri, A.R.; Butrico, G.; Marcucci, M.J.; Zhang, D.; Abulizi, A.; Zhang, X.M.; et al. Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J. Clin. Investig. 2016, 126, 4361–4371. [Google Scholar] [CrossRef]
- Xi, Z.H.; Fang, L.; Xu, J.; Li, B.S.; Zuo, Z.H.; Lv, L.J.; Wang, C.G. Exposure to Aroclor 1254 persistently suppresses the functions of pancreatic beta-cells and deteriorates glucose homeostasis in male mice. Environ. Pollut. 2019, 249, 822–830. [Google Scholar] [CrossRef]
- Gorbunov, E.A.; Nicoll, J.; Kachaeva, E.V.; Tarasov, S.A.; Epstein, O.I. Subetta increases phosphorylation of insulin receptor beta-subunit alone and in the presence of insulin. Nutr. Diabetes 2015, 5, e169. [Google Scholar] [CrossRef]
- Lawrence, C.F.; Margetts, M.B.; Menting, J.G.; Smith, N.A.; Smith, B.J.; Ward, C.W.; Lawrence, M.C. Insulin mimetic peptide disrupts the primary binding site of the insulin receptor. J. Biol. Chem. 2016, 291, 15473–15481. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, S. S961, a biosynthetic insulin receptor antagonist, downregulates insulin receptor expression & suppresses the growth of breast cancer cells. Indian J. Med. Res. 2018, 147, 545–551. [Google Scholar]
- Oakie, A.; Wang, R. Beta-cell receptor tyrosine kinases in controlling insulin secretion and exocytotic machinery: C-kit and insulin receptor. Endocrinology 2018, 159, 3813–3821. [Google Scholar] [CrossRef]
- Ward, C.W.; Menting, J.G.; Lawrence, M.C. The insulin receptor changes conformation in unforeseen ways on ligand binding: Sharpening the picture of insulin receptor activation. Bioessays 2013, 35, 945–954. [Google Scholar] [CrossRef]
- Gherzi, R.; Andraghetti, G.; Versari, G.; Cordera, R. Effect of insulin receptor autophosphorylation on insulin receptor binding. Mol. Cell Endocrinol. 1986, 45, 247–252. [Google Scholar] [CrossRef]
- Ross, J.; Miron, C.E.; Plescia, J.; Laplante, P.; McBride, K.; Moitessier, N.; Moroy, T. Targeting MYC: From understanding its biology to drug discovery. Eur. J. Med. Chem. 2020, 213, 113137. [Google Scholar] [CrossRef] [PubMed]
- Schlein, M.; Ludvigsen, S.; Olsen, H.B.; Andersen, A.S.; Danielsen, G.M.; Kaarsholm, N.C. Properties of small molecules affecting insulin receptor function. Biochemistry 2001, 40, 13520–13528. [Google Scholar] [CrossRef] [PubMed]
- Strekalova, T.; Costa-Nunes, J.P.; Veniaminova, E.; Kubatiev, A.; Lesch, K.P.; Chekhonin, V.P.; Evans, M.C.; Steinbusch, H.W. Insulin receptor sensitizer, dicholine succinate, prevents both Toll-like receptor 4 (TLR4) upregulation and affective changes induced by a high-cholesterol diet in mice. J. Affect. Disord. 2016, 196, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Cline, B.H.; Costa-Nunes, J.P.; Cespuglio, R.; Markova, N.; Santos, A.I.; Bukhman, Y.V.; Kubatiev, A.; Steinbusch, H.W.; Lesch, K.P.; Strekalova, T. Dicholine succinate, the neuronal insulin sensitizer, normalizes behavior, REM sleep, hippocampal pGSK3 beta and mRNAs of NMDA receptor subunits in mouse models of depression. Front. Behav. Neurosci. 2015, 9, 37. [Google Scholar] [CrossRef]
- Kim, W.J.; Lee, W.; Jung, Y.; Jang, H.J.; Kim, Y.K.; Kim, S.N. PPARbeta/delta agonist GW501516 inhibits TNFalpha-induced repression of adiponectin and insulin receptor in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2019, 510, 621–628. [Google Scholar] [CrossRef]
- He, K.; Chan, C.B.; Liu, X.; Jia, Y.; Luo, H.R.; France, S.A.; Liu, Y.; Wilson, W.D.; Ye, K. Identification of a molecular activator for insulin receptor with potent anti-diabetic effects. J. Biol. Chem. 2011, 286, 37379–37388. [Google Scholar] [CrossRef]
- Marsilje, T.H.; Pei, W.; Chen, B.; Lu, W.; Uno, T.; Jin, Y.; Jiang, T.; Kim, S.; Li, N.; Warmuth, M.; et al. Synthesis, structure-activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulfonyl)phenyl)pyrimidine-2,4- diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J. Med. Chem. 2013, 56, 5675–5690. [Google Scholar]
- Van Erp, A.E.M.; Hillebrandt-Roeffen, M.H.S.; van Houdt, L.; Fleuren, E.D.G.; van der Graaf, W.T.A.; Versleijen-Jonkers, Y.M.H. Targeting anaplastic lymphoma kinase (ALK) in rhabdomyosarcoma (RMS) with the second-generation ALK inhibitor ceritinib. Target. Oncol. 2017, 12, 815–826. [Google Scholar] [CrossRef]
- Vewinger, N.; Huprich, S.; Seidmann, L.; Russo, A.; Alt, F.; Bender, H.; Sommer, C.; Samuel, D.; Lehmann, N.; Backes, N.; et al. IGF1R is a potential new therapeutic target for HGNET-BCOR brain tumor patients. Int. J. Mol. Sci. 2019, 20, 3027. [Google Scholar] [CrossRef]
- Li, Y.S.; Kim, J.; Li, J.; Liu, F.; Liu, X.Q.; Himmeldirk, K.; Ren, Y.L.; Wagner, T.E.; Chen, X.Z. Natural anti-diabetic compound 1,2,3,4,6-penta-O-galloyl-D-glucopyranose binds to insulin receptor and activates insulin-mediated glucose transport signaling pathway. Biochem. Biophys. Res. Commun. 2005, 336, 430–437. [Google Scholar] [CrossRef]
- Cao, Y.; Himmeldirk, K.B.; Qian, Y.; Ren, Y.; Malki, A.; Chen, X. Biological and biomedical functions of penta-O-galloyl-D-glucose and its derivatives. J. Nat. Med. 2014, 68, 465–472. [Google Scholar] [CrossRef]
- Cao, Y.; Li, Y.; Kim, J.; Ren, Y.; Himmeldirk, K.; Liu, Y.; Qian, Y.; Liu, F.; Chen, X. Orally efficacious novel small molecule 6-chloro-6-deoxy-1,2,3,4-tetra-O-galloyl-alpha-D-glucopyranose selectively and potently stimulates insulin receptor and alleviates diabetes. J. Mol. Endocrinol. 2013, 51, 15–26. [Google Scholar] [CrossRef][Green Version]
- Lan, Z.J.; Lei, Z.; Yiannikouris, A.; Yerramreddy, T.R.; Li, X.; Kincaid, H.; Eastridge, K.; Gadberry, H.; Power, C.; Xiao, R.; et al. Non-peptidyl small molecule, adenosine, 5′-Se-methyl-5′-seleno-, 2′,3′-diacetate, activates insulin receptor and attenuates hyperglycemia in type 2 diabetic Lepr(db/db) mice. Cell Mol. Life Sci. 2020, 77, 1623–1643. [Google Scholar] [CrossRef]
- Chen, J.; Sun, J.; Prinz, R.A.; Li, Y.; Xu, X. Gingerenone A sensitizes the insulin receptor and increases glucose uptake by inhibiting the activity of p70 S6 kinase. Mol. Nutr. Food Res. 2018, 62, e1800709. [Google Scholar] [CrossRef]
- Kunkel, S.D.; Suneja, M.; Ebert, S.M.; Bongers, K.S.; Fox, D.K.; Malmberg, S.E.; Alipour, F.; Shields, R.K.; Adams, C.M. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 2011, 13, 627–638. [Google Scholar] [CrossRef]
- He, Y.; Li, W.; Li, Y.; Zhang, S.; Wang, Y.; Sun, C. Ursolic acid increases glucose uptake through the PI3K signaling pathway in adipocytes. PLoS ONE 2014, 9, e110711. [Google Scholar]
- Flory, J.; Lipska, K. Metformin in 2019. JAMA 2019, 321, 1926–1927. [Google Scholar] [CrossRef]
- Spiering, M.J. The mystery of metformin. J. Biol. Chem. 2019, 294, 6689–6691. [Google Scholar] [CrossRef]
- Xie, J.; Wang, S.; Ma, P.; Ma, F.; Li, J.; Wang, W.; Lu, F.; Xiong, H.; Gu, Y.; Zhang, S.; et al. Selection of small molecules that bind to and activate the insulin receptor from a DNA-encoded library of natural products. iScience 2020, 23, 101197. [Google Scholar] [CrossRef]
- Zhang, B.; Salituro, G.; Szalkowski, D.; Li, Z.H.; Zhang, Y.; Royo, I.; Vilella, D.; Diez, M.T.; Pelaez, F.; Ruby, C.; et al. Discovery of a small molecule insulin mimetic with antidiabetic activity in mice. Science 1999, 284, 974–977. [Google Scholar] [CrossRef]
- Diesel, B.; Kulhanek-Heinze, S.; Ho¨ltje, M.; Brandt, B.; Ho¨ltje, H.-D.; Vollmar, A.M.; Kiemer, A.K. α-Lipoic acid as a directly binding activator of the insulin receptor: Protection from hepatocyte apoptosis. Biochemistry 2007, 46, 2146–2155. [Google Scholar] [CrossRef]
- Qiang, G.; Xue, S.; Yang, J.J.; Du, G.; Pang, X.; Li, X.; Goswami, D.; Griffin, P.R.; Ortlund, E.A.; Chan, C.B.; et al. Identification of a small molecular insulin receptor agonist with potent antidiabetes activity. Diabetes 2014, 63, 1394–1409. [Google Scholar] [CrossRef]
- Manchem, V.P.; Goldfine, I.D.; Kohanski, R.A.; Cristobal, C.P.; Lum, R.T.; Schow, S.R.; Shi, S.Y.; Spevak, W.R.; Laborde, E.; Toavs, D.K.; et al. A novel small molecule that directly sensitizes the insulin receptor in vitro and in vivo. Diabetes 2001, 50, 824–830. [Google Scholar] [CrossRef][Green Version]
- Pender, C.; Goldfine, I.D.; Manchem, V.P.; Evans, J.L.; Spevak, W.R.; Shi, S.Y.; Rao, S.; Bajjalieh, S.; Maddux, B.A.; Youngren, J.F. Regulation of insulin receptor function by a small molecule insulin receptor activator. J. Biol. Chem. 2002, 277, 43565–43571. [Google Scholar] [CrossRef]
- Cheng, M.; Chen, S.; Schow, S.R.; Manchem, V.P.; Spevak, W.R.; Cristobal, C.P.; Shi, S.; Macsata, R.W.; Lum, R.T.; Goldfine, I.D.; et al. In vitro and in vivo prevention of HIV protease inhibitor-induced insulin resistance by a novel small molecule insulin receptor activator. J. Cell Biochem. 2004, 92, 1234–1245. [Google Scholar] [CrossRef]
- Wu, M.; Dai, G.; Yao, J.; Hoyt, S.; Wang, L.; Mu, J. Potentiation of insulin-mediated glucose lowering without elevated hypoglycemia risk by a small molecule insulin receptor modulator. PLoS ONE 2015, 10, e0122012. [Google Scholar] [CrossRef]
- Robinson, L.; Bajjalieh, S.; Cairns, N.; Lum, R.T.; Macsata, R.W.; Manchem, V.P.; Park, S.J.; Rao, S.; Schow, S.R.; Shi, S.; et al. 5-Substituted isophthalamides as insulin receptor sensitizers. Bioorg. Med. Chem. Lett. 2008, 18, 3492–3494. [Google Scholar] [CrossRef]
- Girnita, A.; Girnita, L.; Prete, F.D.; Bartolazzi, A.; Larsson, O.; Axelson, M. Cyclolignans as inhibitors of the insulin-Like growth factor-1 receptor and malignant cell growth. Cancer Res. 2004, 64, 236–242. [Google Scholar] [CrossRef]
- Rodon, J.; DeSantos, V.; Ferry, R.J.; Kurzrock, R. Early drug development of inhibitors of the insulin-like growth factor-I receptor pathway: Lessons from the first clinical trials. Mol. Cancer Ther. 2008, 7, 2575–2588. [Google Scholar] [CrossRef]
- Liu, X.; Xie, H.; Luo, C.; Tong, L.; Wang, Y.; Peng, T.; Ding, J.; Jiang, H.; Li, H. Discovery and SAR of thiazolidine-2,4-dione analogues as insulin-like growth factor-1 receptor (IGF-1R) inhibitors via hierarchical virtual screening. J. Med. Chem. 2010, 53, 2661–2665. [Google Scholar] [CrossRef]
- Ji, Q.S.; Mulvihill, M.J.; Rosenfeld-Franklin, M.; Cooke, A.; Feng, L.; Mak, G.; O’Connor, M.; Yao, Y.; Pirritt, C.; Buck, E.; et al. A novel, potent, and selective insulin-like growth factor-I receptor kinase inhibitor blocks insulin-like growth factor-I receptor signaling in vitro and inhibits insulin-like growth factor-I receptor dependent tumor growth in vivo. Mol. Cancer Ther. 2007, 6, 2158–2167. [Google Scholar] [CrossRef] [PubMed]
- Mulvihill, M.J.; Cooke, A.; Rosenfeld-Franklin, M.; Buck, E.; Foreman, K.; Landfair, D.; O’Connor, M.; Pirritt, C.; Sun, Y.C.; Yao, Y.; et al. Discovery of OSI-906: A selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Future Med. Chem. 2009, 1, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, H.; Oh, A.; Zhang, Y.; Yee, D. Enhancement of doxorubicin cytotoxicity of human cancer cells by tyrosine kinase inhibition of insulin receptor and type I IGF receptor. Breast. Cancer Res. Tr. 2012, 133, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Wittman, M.; Carboni, J.; Attar, R.; Balasubramanian, B.; Balimane, P.; Brassil, P.; Beaulieu, F.; Chang, C.; Clarke, W.; Dell, J.; et al. Discovery of a (1H-benzoimidazol-2-yl)-1H-pyridin-2-one (BMS-536924) inhibitor of insulin-like growth factor I receptor kinase with in vivo antitumor activity. J. Med. Chem. 2005, 48, 5639–5643. [Google Scholar] [CrossRef]
- Haluska, P.; Carboni, J.M.; Loegering, D.A.; Lee, F.Y.; Wittman, M.; Saulnier, M.G.; Frennesson, D.B.; Kalli, K.R.; Conover, C.A.; Attar, R.M.; et al. In vitro and in vivo antitumor effects of the dual insulin-like growth factor-i/insulin receptor inhibitor, BMS-554417. Cancer Res. 2006, 66, 362–371. [Google Scholar] [CrossRef]
- Hou, X.; Huang, F.; Macedo, L.F.; Harrington, S.C.; Reeves, K.A.; Greer, A.; Finckenstein, F.G.; Brodie, A.; Gottardis, M.M.; Carboni, J.M.; et al. Dual IGF-1R/InsR inhibitor BMS-754807 synergizes with hormonal agents in treatment of estrogen-dependent breast cancer. Cancer Res. 2011, 71, 7597–7607. [Google Scholar] [CrossRef]
- Schwartz, G.K.; Dickson, M.A.; LoRusso, P.M.; Sausville, E.A.; Maekawa, Y.; Watanabe, Y.; Kashima, N.; Nakashima, D.; Akinaga, S. Preclinical and first-in-human phase I studies of KW-2450, an oral tyrosine kinase inhibitor with insulin-like growth factor receptor-1/insulin receptor selectivity. Cancer Sci. 2016, 107, 499–506. [Google Scholar] [CrossRef]
- Umehara, H.; Maekawa, Y.; Koizumi, F.; Shimizu, M.; Ota, T.; Fouad, T.M.; Willey, J.; Kaito, H.; Shiraishi, N.; Nakashima, D.; et al. Preclinical and phase I clinical studies of KW-2450, a dual IGF-1R/IR tyrosine kinase inhibitor, in combination with lapatinib and letrozole. Ther. Adv. Med. Oncol. 2018, 10, 1–14. [Google Scholar] [CrossRef]
- Smith, D.C.; Britten, C.; Clary, D.O.; Nguyen, L.T.; Woodard, P.; Hurwitz, H.I. A phase I study of XL228, a potent IGF1R/AURORA/SRC inhibitor, in patients with solid tumors or hematologic malignancies. J. Clin. Oncol. 2009, 27, 3512. [Google Scholar] [CrossRef]
- Bell, I.M.; Stirdivant, S.M.; Ahern, J.; Culberson, J.C.; Darke, P.L.; Dinsmore, C.J.; Drakas, R.A.; Gallicchio, S.N.; Graham, S.L.; Heimbrook, D.C.; et al. Biochemical and structural characterization of a novel class of inhibitors of the type 1 insulin-like growth factor and insulin receptor kinases. Biochemistry 2005, 44, 9430–9440. [Google Scholar] [CrossRef]
- Yasujima, T.; Saito, K.; Moore, R.; Negishi, M. Phenobarbital and insulin reciprocate activation of the nuclear receptor constitutive androstane receptor through the insulin receptor. J. Pharmacol. Exp. Ther. 2016, 357, 367–374. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, Y.; Huang, T.; Hisatome, I.; Yamamoto, T.; Cheng, J. High uric acid induces insulin resistance in cardiomyocytes in vitro and in vivo. PLoS ONE 2016, 11, e0147737. [Google Scholar]
- Masi, S.; Georgiopoulos, G.; Alexopoulos, G.; Pateras, K.; Rosada, J.; Seravalle, G.; Ciuceis, C.D.; Taddei, S.; Borghi, C.; Grassi, G.; et al. The complex relationship between serum uric acid, endothelial function and small vessel remodeling in humans. J. Clin. Med. 2020, 9, 2027. [Google Scholar] [CrossRef]
- Tassone, E.J.; Cimellaro, A.; Perticone, M.; Hribal, M.L.; Sciacqua, A.; Andreozzi, F.; Sesti, G.; Perticone, F. Uric acid impairs insulin signaling by Promoting enpp1 Binding to insulin receptor in human umbilical vein endothelial cells. Front. Endocrinol. 2018, 9, 98. [Google Scholar] [CrossRef]
- Wang, Y.H.; Guan, Q.L.; Liu, G.F.; Wu, X.Y.; Wang, X.J.; Sheng, J. 2′-O-methylperlatolic acid enhances insulin-regulated blood glucose-lowering effect through insulin receptor signaling pathway. J. Diabetes Res. 2022, 2022, 2042273. [Google Scholar]
- Taochy, C.; Yu, A.; Bouche, N.; Bouteiller, N.; Elmayan, T.; Dressel, U.; Carroll, B.J.; Vaucheret, H. Post-transcriptional gene silencing triggers dispensable DNA methylation in gene body in Arabidopsis. Nucleic Acids Res. 2019, 47, 9104–9114. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenet. Chromatin. 2017, 10, 23. [Google Scholar] [CrossRef]
- Adam, S.; Anteneh, H.; Hornisch, M.; Wagner, V.; Lu, J.; Radde, N.E.; Bashtrykov, P.; Song, J.; Jeltsch, A. DNA sequence-dependent activity and base flipping mechanisms of DNMT1 regulate genome-wide DNA methylation. Nat. Commun. 2020, 11, 3723. [Google Scholar] [CrossRef]
- Nishiyama, A.; Mulholland, C.B.; Bultmann, S.; Kori, S.; Endo, A.; Saeki, Y.; Qin, W.; Trummer, C.; Chiba, Y.; Yokoyama, H.; et al. Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat. Commun. 2020, 11, 1222. [Google Scholar] [CrossRef]
- Chen, Y.T.; Lin, W.D.; Liao, W.L.; Tsai, Y.C.; Liao, J.W.; Tsai, F.J. NT5C2 methylation regulatory interplay between DNMT1 and insulin receptor in type 2 diabetes. Sci. Rep. 2020, 10, 16087. [Google Scholar] [CrossRef]
- Careddu, M.G.; Allegrini, S.; Pesi, R.; Camici, M.; Garcia-Gil, M.; Tozzi, M.G. Knockdown of cytosolic 5′-nucleotidase II (cN-II) reveals that its activity is essential for survival in astrocytoma cells. Biochim. Biophys. Acta-Mol. Cell Res. 2008, 1783, 1529–1535. [Google Scholar] [CrossRef]
- Binayi, F.; Zardooz, H.; Ghasemi, R.; Hedayati, M.; Askari, S.; Pouriran, R.; Sahraei, M. The chemical chaperon 4-phenyl butyric acid restored high-fat diet- induced hippocampal insulin content and insulin receptor level reduction along with spatial learning and memory deficits in male rats. Physiol. Behav. 2021, 231, 113312. [Google Scholar] [CrossRef]
- Che, Z.P.; Tian, Y.E.; Liu, S.M.; Jiang, J.; Hu, M.; Chen, G.Q. Stereoselective synthesis of 4beta-acyloxypodophyllotoxin derivatives as insecticidal agents. J. Asian Nat. Prod. Res. 2019, 21, 1028–1041. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, Y.; Huang, Z.; Zhou, Y.; Wang, W.; Xuan, Y.; Zhen, Y.; Ju, B.; Guo, S.; Zhang, S. Self-assembly of podophyllotoxin-loaded lipid bilayer nanoparticles for highly effective chemotherapy and immunotherapy via downregulation of programmed cell death ligand 1 production. ACS Nano. 2022, 16, 3943–3954. [Google Scholar] [CrossRef]
- Xiao, J.; Gao, M.; Sun, Z.; Diao, Q.; Wang, P.; Gao, F. Recent advances of podophyllotoxin/epipodophyllotoxin hybrids in anticancer activity, mode of action, and structure-activity relationship: An update (2010–2020). Eur. J. Med. Chem. 2020, 208, 112830. [Google Scholar] [CrossRef] [PubMed]
- Ou, K.; Kang, Y.; Chen, L.; Zhang, X.; Chen, X.; Zheng, Y.; Wu, J.; Guan, S. H2O2-responsive nano-prodrug for podophyllotoxin delivery. Biomater. Sci. 2019, 7, 2491–2498. [Google Scholar] [CrossRef] [PubMed]
- Zi, C.T.; Yang, L.; Xu, F.Q.; Dong, F.W.; Yang, D.; Li, Y.; Ding, Z.T.; Zhou, J.; Jiang, Z.H.; Hu, J.M. Synthesis and anticancer activity of dimeric podophyllotoxin derivatives. Drug Des. Devel. Ther. 2018, 12, 3393–3406. [Google Scholar] [CrossRef] [PubMed]
- Davey, S.G. Engineering etoposide. Nat. Rev. Chem. 2020, 4, 63. [Google Scholar] [CrossRef]
- Nerella, S.; Kankala, S.; Gavaji, B. Synthesis of podophyllotoxin-glycosyl triazoles via click protocol mediated by silver (I)-N-heterocyclic carbenes and their anticancer evaluation as topoisomerase-II inhibitors. Nat. Prod. Res. 2021, 35, 9–16. [Google Scholar] [CrossRef]
- Lv, M.; Zhang, Y.; Wang, F.; Zhang, S.; Xu, H. Non-food renewable and bioactive forest products for pest management: Valuation of agricultural properties of podophyllotoxin analogs derived from Podophyllum hexandrum as botanical pesticides. Ind. Crop. Prod. 2020, 153, 112608. [Google Scholar] [CrossRef]
- Yu, X.; Che, Z.; Xu, H. Recent advances in the chemistry and biology of podophyllotoxins. Chemistry 2017, 23, 4467–4526. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, Y.; Wang, F.; Zhang, S.; Lv, M.; Hui, X. Renewable forest bioresources for pest management: Semisynthesis of esters containing ferrocene scaffolds of podophyllotoxin from Juniperus sabina L. as botanical pesticides. Ind. Crop. Prod. 2020, 152, 112510. [Google Scholar] [CrossRef]
- Che, Z.P.; Tian, Y.; Yang, J.M.; Liu, S.M.; Jiang, J.; Hu, M.; Chen, G.Q. Screening of insecticidal activity of podophyllotoxin analogues against Athetis dissimilis. Nat. Prod. Commun. 2019, 14, 117–120. [Google Scholar] [CrossRef]
- Sun, Z.; Lv, M.; Yu, X.; Xu, H. Application of sustainable natural bioesources in crop protection: Insight into a podophyllotoxin-derived botanical pesticide for regulating insect vestigial wing of Mythimna separata Walker. ACS Sustain. Chem. Eng. 2017, 5, 3945–3954. [Google Scholar] [CrossRef]
- Wang, R.; Qu, C.; Wang, Z.; Yang, G. Cross-resistance, biochemical mechanism and fitness costs of laboratory-selected resistance to pyridalyl in diamondback moth, Plutella xylostella. Pestic. Biochem. Phys. 2020, 163, 8–13. [Google Scholar] [CrossRef]
- Abbasi-Mojdehia, M.R.; Hajizadeha, J.; Zibaeea, A.; Keyhanianc, A.A.; Rajaei, F. Alterations in some physiological processes of Bactrocera oleae Rossi (Diptera: Tephritidae) following pyridalyl treatment. Pestic. Biochem. Phys. 2019, 164, 85–90. [Google Scholar] [CrossRef]
- Abbasi-Mojdehi, M.R.; Hajizadeh, J.; Zibaee, A.; Keyhanian, A.A. Effect of pyridalyl on mortality, fecundity and physiological performance of olive fruit fly, Bactrocera oleae Rossi (Diptera: Tephritidae). J. Asia-Pac Entomol. 2019, 22, 506–512. [Google Scholar] [CrossRef]
- Powell, G.F.; Ward, D.A.; Prescott, M.C.; Spiller, D.G.; White, M.R.; Turner, P.C.; Earley, F.G.; Phillips, J.; Rees, H.H. The molecular action of the novel insecticide, Pyridalyl. Insect Biochem. Mol. Biol. 2011, 41, 459–469. [Google Scholar] [CrossRef]
- Shao, X.H.; Lai, D.; Zhang, L.; Xu, H. Induction of autophagy and apoptosis via PI3K/AKT/TOR pathways by azadirachtin A in Spodoptera litura Cells. Sci. Rep. 2016, 6, 35482. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, biogenesis, their Web-based tools, and databases. MicroRNA 2019, 8, 4–27. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. MicroRNAs regulating MicroRNAs in cancer. Trends Cancer. 2018, 4, 465–468. [Google Scholar] [CrossRef]
- Asgari, S. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 2013, 43, 388–397. [Google Scholar] [CrossRef]
- Sun, X.; Liu, J.; Xu, C.; Tang, S.C.; Ren, H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J. Cell Mol. Med. 2016, 20, 1779–1788. [Google Scholar] [CrossRef]
- Nweke, E.E.; Brand, M. Downregulation of the let-7 family of microRNAs may promote insulin receptor/insulin-like growth factor signalling pathways in pancreatic ductal adenocarcinoma. Oncol Lett. 2020, 20, 2613–2620. [Google Scholar] [CrossRef]
- Frutos, M.F.; Galan-Chilet, I.; Goedeke, L.; Kim, B.; Pardo-Marques, V.; Perez-Garcia, A.; Herrero, J.I.; Fernandez-Hernando, C.; Kim, J.; Ramirez, C.M. MicroRNA 7 impairs insulin signaling and regulates A beta levels through posttranscriptional regulation of the insulin receptor substrate 2, insulin receptor, insulin-degrading enzyme, and liver X receptor pathway. Mol. Cell Biol. 2019, 39, e00170-19. [Google Scholar]
- Ling, L.; Kokoza, V.A.; Zhang, C.; Aksoy, E.; Raikhel, A.S. MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. USA 2017, 114, E8017–E8024. [Google Scholar] [CrossRef]
- Shang, F.; Niu, J.; Ding, B.Y.; Zhang, W.; Wei, D.D.; Wei, D.; Jiang, H.B.; Wang, J.J. The miR-9b microRNA mediates dimorphism and development of wing in aphids. Proc. Natl. Acad. Sci. USA 2020, 117, 8404–8409. [Google Scholar] [CrossRef]
- Mansour, M.A. Ubiquitination: Friend and foe in cancer. Int. J. Biochem. Cell Biol. 2018, 101, 80–93. [Google Scholar] [CrossRef]
- Foot, N.; Henshall, T.; Kumar, S. Ubiquitination and the regulation of membrane proteins. Physiol. Rev. 2017, 97, 253–281. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Z.; Yang, Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer. 2020, 19, 146. [Google Scholar] [CrossRef] [PubMed]
- Rui, L.Y.; Yuan, M.S.; Frantz, D.; Shoelson, S.; White, M.F. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 2002, 277, 42394–42398. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Sarikas, A.; Dias-Santagata, D.C.; Dolios, G.; Lafontant, P.J.; Tsai, S.-C.; Zhu, W.; Nakajima, H.; Nakajima, H.O.; Field, L.J.; et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell. 2008, 30, 403–414. [Google Scholar] [CrossRef]
- Yi, J.S.; Park, J.S.; Ham, Y.M.; Nguyen, N.; Lee, N.R.; Hong, J.; Kim, B.W.; Lee, H.; Lee, C.S.; Jeong, B.C.; et al. MG53-induced IRS-1 ubiquitination negatively regulates skeletal myogenesis and insulin signalling. Nat. Commun. 2013, 4, 2354. [Google Scholar] [CrossRef]
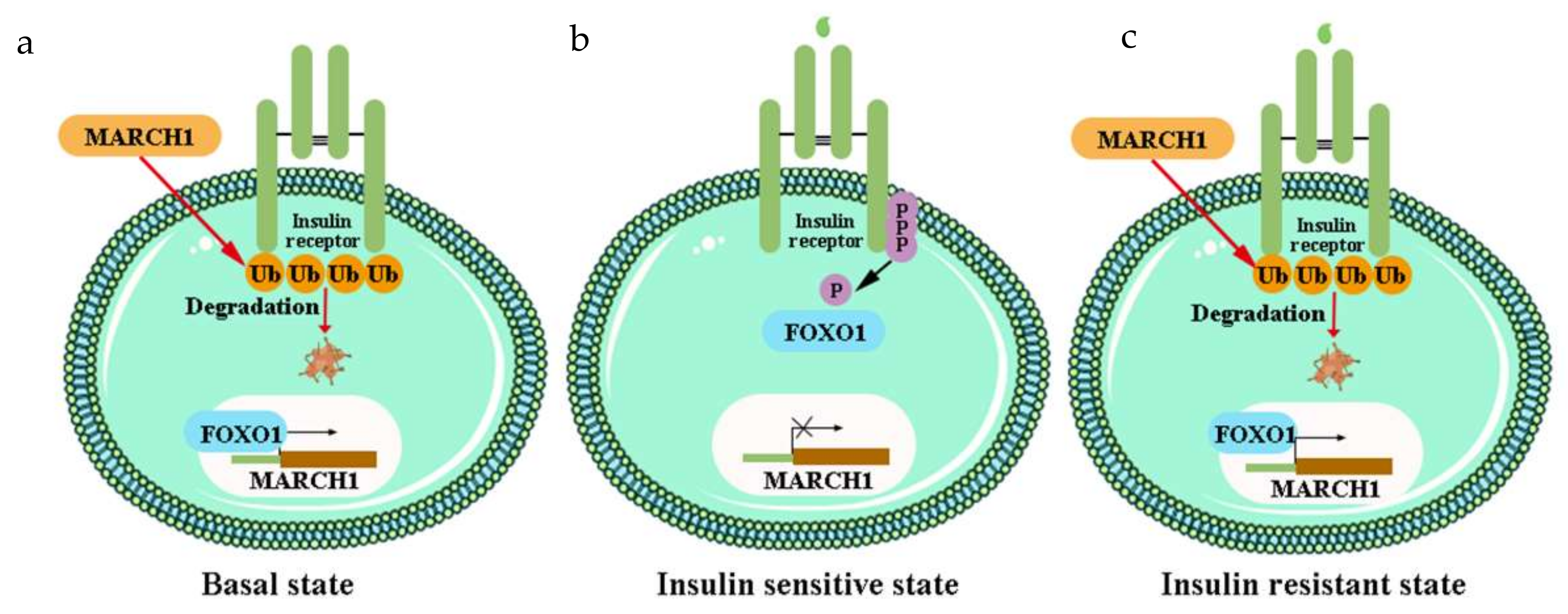
- Nagarajan, A.; Petersen, M.C.; Nasiri, A.R.; Butrico, G.; Fung, A.; Ruan, H.B.; Kursawe, R.; Caprio, S.; Thibodeau, J.; Bourgeois-Daigneault, M.C.; et al. MARCH1 regulates insulin sensitivity by controlling cell surface insulin receptor levels. Nat. Commun. 2016, 7, 12639. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
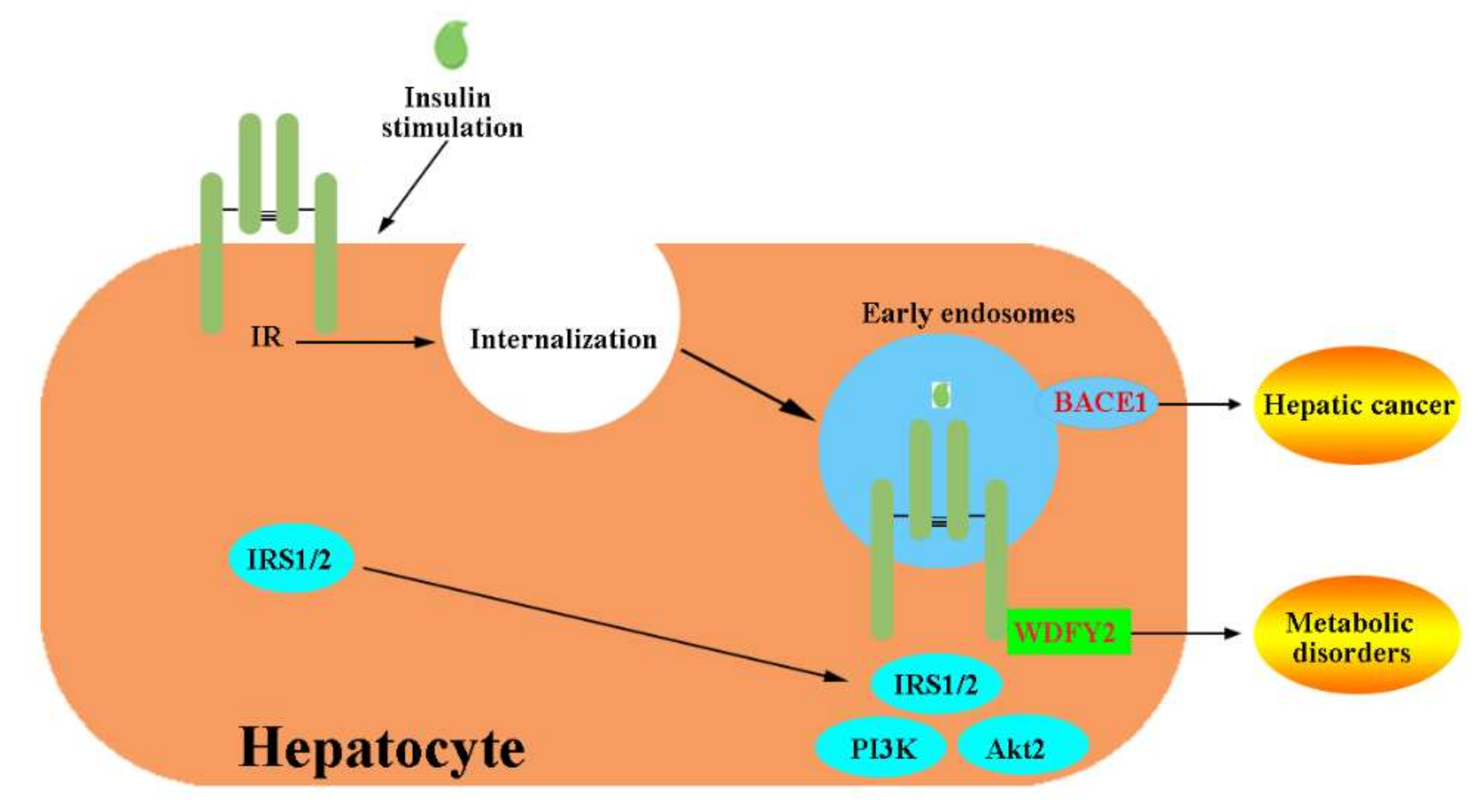
- Choi, E.; Kikuchi, S.; Gao, H.; Brodzik, K.; Nassour, I.; Yopp, A.; Singal, A.G.; Zhu, H.; Yu, H. Mitotic regulators and the SHP2-MAPK pathway promote IR endocytosis and feedback regulation of insulin signaling. Nat. Commun. 2019, 10, 1473. [Google Scholar] [CrossRef]
- Choi, E.; Yu, H. Spindle checkpoint regulators in insulin signaling. Front. Cell Dev. Biol. 2018, 6, 161. [Google Scholar] [CrossRef]
- Hagan, R.S.; Manak, M.S.; Buch, H.K.; Meier, M.G.; Meraldi, P.; Shah, J.V.; Sorger, P.K. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol. Biol. Cell. 2011, 22, 4236–4246. [Google Scholar] [CrossRef]
- Yonei, Y.; Takabe, W. Liver training: Yes or no? Glycative Stress Res. 2015, 2, 121–128. [Google Scholar]
- Murphy, J.E.; Padilla, B.E.; Hasdemir, B.; Cottrell, G.S.; Bunnett, N.W. Endosomes: A legitimate platform for the signaling train. Proc. Natl. Acad. Sci. USA 2009, 106, 17615–17622. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Tong, S.; Zhou, H.; Sun, J.; Gou, Y.; Wu, F.; Hu, J.; Xu, J.; Ding, G. Overexpression of WDFY2 inhibits prostate cancer cell growth and migration via inactivation of Akt pathway. Tumor. Biol. 2017, 39, 1010428317704821. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Zhang, N.; Yang, X.; Hou, T.; Fu, W.; Yuan, F.; Wang, L.; Wen, H.; Tian, Y.; et al. WDFY2 potentiates hepatic insulin sensitivity and controls endosomal localization of the insulin receptor and IRS1/2. Diabetes 2020, 69, 1887–1902. [Google Scholar] [CrossRef]
- Hayakawa, A.; Leonard, D.; Murphy, S.; Hayes, S.; Soto, M.; Fogarty, K.; Standley, C.; Bellve, K.; Lambright, D.; Mello, C.; et al. The WD40 and FYVE domain containing protein 2 defines a class of early endosomes necessary for endocytosis. Proc. Natl. Acad. Sci. USA 2006, 103, 11928–11933. [Google Scholar] [CrossRef]
- Evin, G.; Hince, C. BACE1 as a therapeutic target in Alzheimer’s disease: Rationale and current status. Drugs Aging. 2013, 30, 755–764. [Google Scholar] [CrossRef]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef]
- Obata, T.; Yokota, I.; Yokoyama, K.; Okamoto, E.; Kanezaki, Y.; Tanaka, Y.; Maegawa, H.; Teshigawara, K.; Hirota, F.; Yuasa, T.; et al. Soluble insulin receptor ectodomain is elevated in the plasma of patients with diabetes. Diabetes 2007, 56, 2028–2035. [Google Scholar]
- Mesallamy, H.O.E.; Hamdy, N.M.; Mostafa, D.M.; Amin, A.I. The serine protease granzyme B as an inflammatory marker, in relation to the insulin receptor cleavage in human obesity and type 2 diabetes mellitus. J. Interf. Cytok. Res. 2014, 34, 179–186. [Google Scholar] [CrossRef]
- Dassano, A.; Loretelli, C.; Fiorina, P. Idebenone and T2D: A new insulin-sensitizing drug for personalized therapy. Pharmacol. Res. 2019, 139, 469–470. [Google Scholar] [CrossRef]
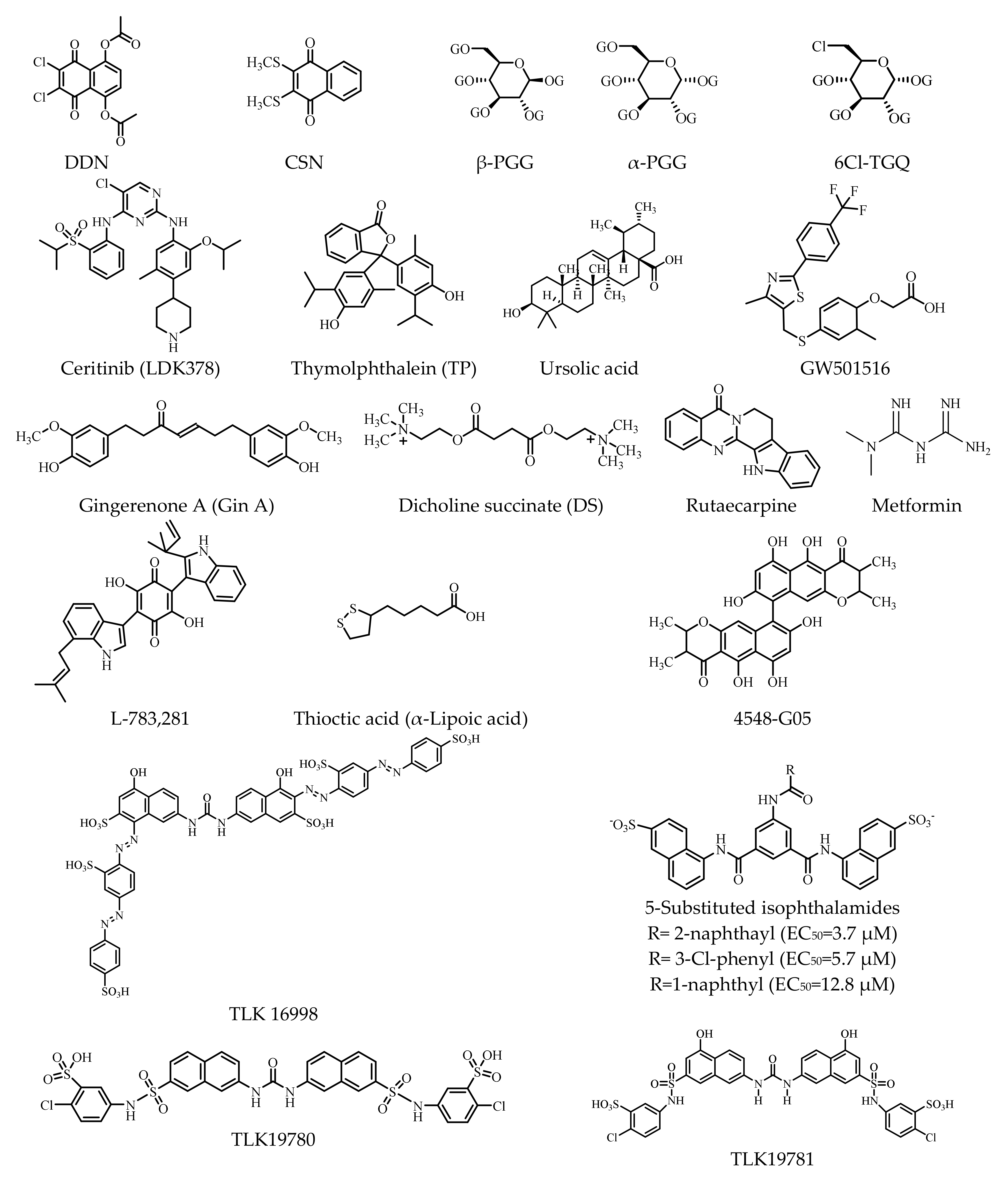
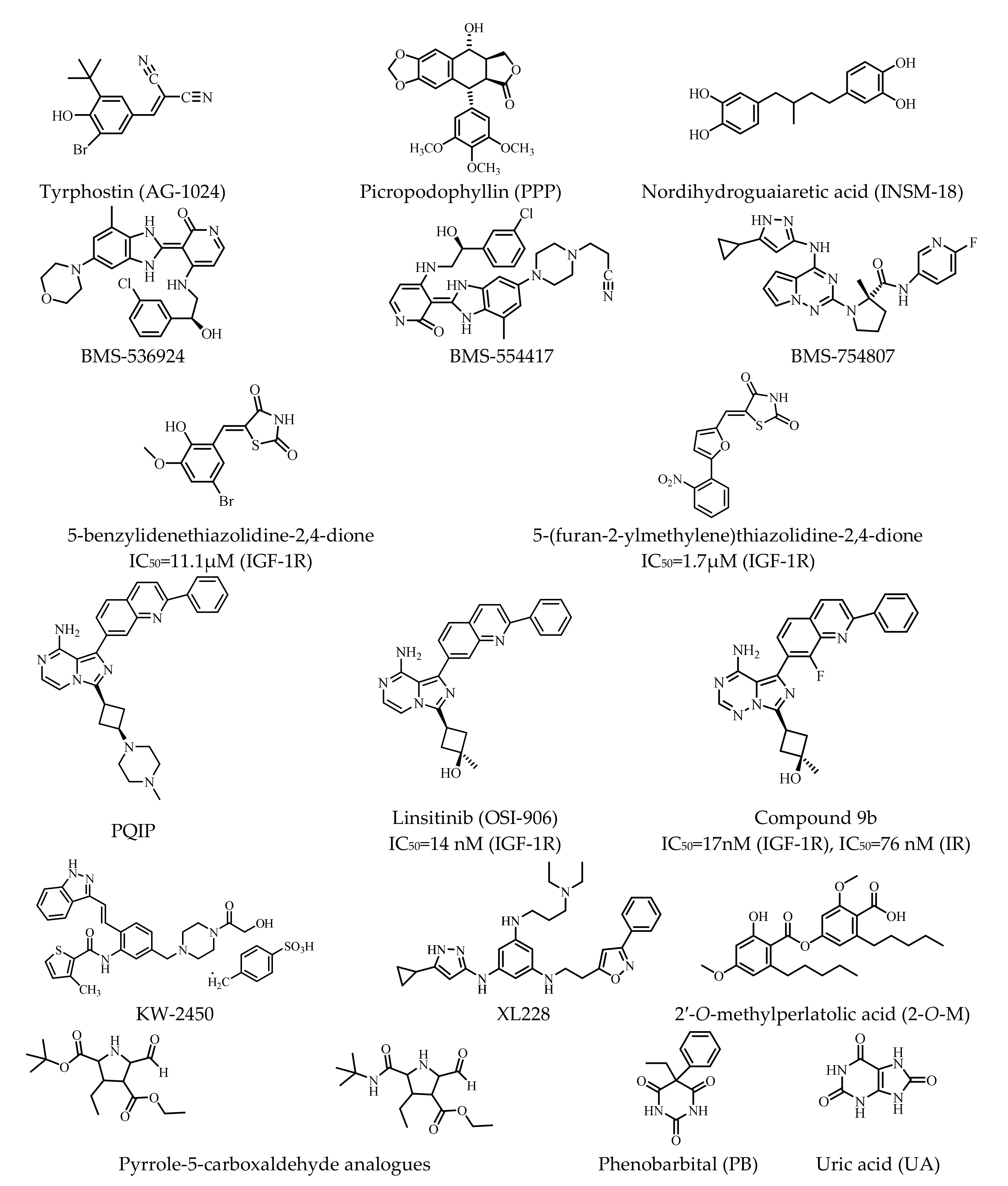

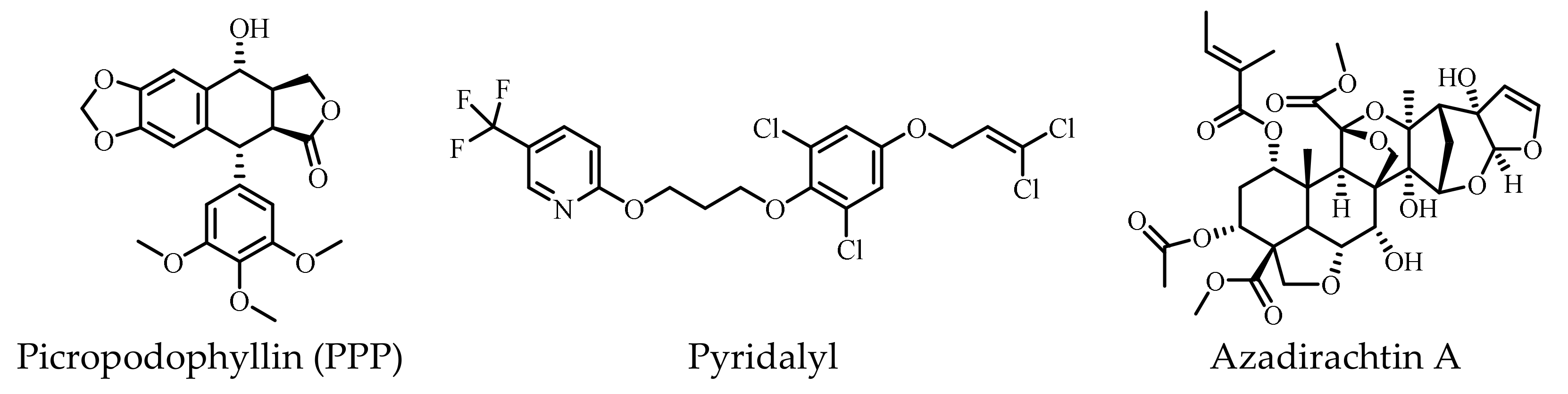

| Classification | Structure of IR | References |
|---|---|---|
| Domain layout | an (αβ)2 disulfide-linked homodimer | [35] |
| cDNA sequenced | α chain lies on the N-terminal of the β chain | |
| 3D structure of human apo IR ectodomain | intracellular unphosphorylated from TKD (2.1 Å resolution, PDB 1IRK) | |
| receptor’s isolated L1-CR-L2 module (2.32 Å resolution, PDB 2HR7) | ||
| intact receptor ectodomain in apo form (3.8 Å resolution, PDB 2DTG) | ||
| CryoEM structures of IR | insulin holoreceptor (full-length receptor inclusive of transmembrane and cytoplasmic elements) | [42] |
| isolated receptor ectodomain | [41,43] | |
| an ectodomain construct (leucine-zippered receptor ectodomain) | [44] |
| Group | Compound | Pharmacological Activity | Experimental Model | References |
|---|---|---|---|---|
| 1 | Thymolphthalein (TP) weak agonist | displaces insulin from IR, binds to the IR; ↑auto- and substrate-based phosphorylation of IR | isolated primary mouse adipocytes | [183] |
| 1 | Dicholine succinate (DS) (sensitizers) | ↑IR-mediated signaling | mice | [184,185] |
| 1 | GW501516 | ↑expression of the IR (1.3-fold than insulin); ↓TNF-α (tumor necrosis factor α)-induced IR expression | differentiated 3T3-L1 adipocytes | [186] |
| 1 | DDN (activator) | ↑phosphorylation of Akt and ERK (bind to IR-TKD); ↓blood glucose | male C57BL/6J, C57BL/KsJ db/db mice; female C57BL/KsJ ob/ob mice | [187] |
| 1 | CSN (activator) | ↑IR phosphorylation (time-dependent manner) | ||
| 1 | Ceritinib (LDK378) (off-target inhibitor) | ↓IGF-1R (IC50: 8 nm) phosphorylation and downstream effector AKT; ↓IR (IC50: 7 nm) phosphorylation | human primary cell culture PhKh1 of a pediatric HGNET-BCOR patient (P1) | [188,189,190] |
| 1 | Penta-O-galloyl-D-glucose (PGG) | ↑phosphorylation of the IR and Akt (α-PGG/β-PGG isoform) | 3T3-L1 adipocytes | [191,192] |
| 1 | 6Cl-TGQ | ↑IR (without activating IGF-1R); ↑glucose uptake | 3T3-L1 adipocytes | [193] |
| 1 | Adenosine | ↑phosphorylation and activation of IR (interacted with IR-α) | HepG2 liver cells; insulin-resistant T2D Leprdb/db mice | [194] |
| 1 | Gingerenone A (Gin A) | ↑tyrosine phosphorylation of the IR; ↑translocation of GLUT4; ↑insulin-stimulated glucose uptake | murine 3T3-L1 adipocytes; rat L6 myotubes | [195] |
| 1 | Ursolic acid | ↑autophosphorylation of the β-subunit of the IR; ↑glucose uptake (dose-dependent manner) | 3T3-L1 adipocytes | [196,197] |
| 1 | Metformin | ↑autophosphorylation of the human IR (activator of AMP-activated protein kinase (AMPK)) lactic acidosis | CHO cells expressing the human IR | [198,199] |
| 1 | Rutaecarpine (activator) | ↑autophosphorylation of the human IR (bind to IR-ECD) | [200] | |
| 2 | L-783,281 (insulin mimetic) (activator) | ↑phosphorylation of the IR β subunit & IRS-1; ↑PI 3-kinase activity (13); ↑phosphorylation Akt kinase; ↑glucose uptake | Chinese hamster ovary cells (overexpress the human IR) (CHO.IR) | [201] |
| 2 | Thioctic acid (α-lipoic acid) (activator) | ↑activation of the IR (bind to IR-TKD) | mice primary hepatocytes | [202] |
| 2 | 4548-G05 (insulin mimetics) (activator) | ↑phosphorylations of IR, IRS-1, Akt | C2C12 myotubes [197] | [203] |
| 2 | TLK16998 (sensitizer) | ↑IR autophosphorylation (activates IR-TKD β-subunit); ↑IRS-1 phosphorylation;↑PI3-kinase recruitment;↑GLUT4 translocation; ↑glucose uptake | 3T3-L1 adipocytes | [204] |
| 2 | TLK19780 (activator) | ↑the amount of autophosphorylated IR; | HTC-IR cells | [205] |
| 2 | TLK19781 | ↑phosphorylation of the IR-TKD; ↑GLUT4 translocation;↑glucose transport | 3T3-L1 fibroblasts | [206,207] |
| 2 | 5-substituted isophthalamides (sensitizer) | IR sensitizer, inactive without insulin | 3T3-L1 adipocytes | [208] |
| 2 | Tyrphostin (AG-1024) | ↓autophosphorylation IGF-1R (IC50:0.4 μM)/IR (IC50:0.1 μM) | NIH-3T3 fibroblasts | [20] |
| 2 | Picropodophyllin (PPP) | ↓IGF-1R autophosphorylation at the substrate level | mice | [209] |
| 2 | Nordihydroguaiaretic acid (INSM-18) | ↓activation of the IGF-1R (inhibitor); ↓phosphorylation of the Akt/PKB serine kinase | MCF-7 human breast cancer cells | [210] |
| 3 | 5-benzylidenethiazolidine-2,4-dione | ↓IGF-IR and IR kinase activity | MCF-7 human breast cancer cell line | [211] |
| 3 | 5-(furan-2-ylmethylene) thiazolidine-2,4-dione | ↓IGF-IR and IR kinase activity | ||
| 3 | PQIP | ↓autophosphorylation of the IGF-1R (IC50: 19 nmol/L) | 3T3/huIGF1R fibrosarcoma cells | [212] |
| 3 | linsitinib (OSI-906) | ↓IR/IGF-1R kinase; ↓autophosphorylation of IR and IGF-1R (human) | 3T3/huIGF-1R fibrosarcoma cells; GEO human colorectal cancer cells | [213] |
| 3 | Compound 9b | ↓phosphorylation of IR and IGF-1R; ↓pAkt | GEO human colorectal tumor cell line/tumor xenograft models | [214] |
| 3 | BMS-536924 | ↓phosphorylation of the IR and IGF-1R tyrosine kinase; ↓Akt and MAPK phosphorylation | Sal tumor model | [215] |
| 3 | BMS-554417 | ↓IGF-IR and IR kinase activity and proliferation | carcinoma cell lines (Colo205 and OV202) | [216] |
| 3 | BMS-754807 | ↓phosphorylation of IGF-1R and IR | postmenopausal, estrogen-dependent breast cancer | [217] |
| 3 | KW-2450 | ↓tyrosine kinase of IR (IC50: 5.64 nM)/IGF-1R (IC50: 7.39 nM) | HT-29/GFP colon cancer xenograft model | [218,219] |
| 3 | XL228 | ↓tyrosine kinase of IGF-1R and other protein kinases | patients with advanced malignancies | [220] |
| 3 | pyrrole-5-carboxaldehyde analogues | ↓tyrosine kinase of IR and IGF-1R; ↓autophosphorylation of IR and IGF-1R | human embryonic kidney cells (HEK-293) | [221] |
| 3 | Phenobarbital (PB) (antagonist) | ↓dephosphorylate-activated IR; ↓dephosphorylation of phosphorylated Akt/FOXO1 | primary hepatocytes and HepG2 cells; mouse | [222] |
| 3 | Uric acid (UA) | induced ENPP1 binding to IR α-subunit; ↓autophosphorylation of IR and IRS; ↓glucose transport | human umbilical vein endothelial cell | [223,224,225] |
| 3 | 2′-O-methylperlatolic acid (sensitizer) | ↑insulin signaling pathway (binds to IR-ECD); ↑cellular glucose uptake | Hepa and C2C12 myotubes | [226] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhu, X.; Bi, X.; Huang, J.; Zhou, L. The Insulin Receptor: An Important Target for the Development of Novel Medicines and Pesticides. Int. J. Mol. Sci. 2022, 23, 7793. https://doi.org/10.3390/ijms23147793
Zhang X, Zhu X, Bi X, Huang J, Zhou L. The Insulin Receptor: An Important Target for the Development of Novel Medicines and Pesticides. International Journal of Molecular Sciences. 2022; 23(14):7793. https://doi.org/10.3390/ijms23147793
Chicago/Turabian StyleZhang, Xiaohong, Xuezhen Zhu, Xiaoyang Bi, Jiguang Huang, and Lijuan Zhou. 2022. "The Insulin Receptor: An Important Target for the Development of Novel Medicines and Pesticides" International Journal of Molecular Sciences 23, no. 14: 7793. https://doi.org/10.3390/ijms23147793
APA StyleZhang, X., Zhu, X., Bi, X., Huang, J., & Zhou, L. (2022). The Insulin Receptor: An Important Target for the Development of Novel Medicines and Pesticides. International Journal of Molecular Sciences, 23(14), 7793. https://doi.org/10.3390/ijms23147793







