Comparative Analysis of the pIgR Gene from the Antarctic Teleost Trematomus bernacchii Reveals Distinctive Features of Cold-Adapted Notothenioidei
Abstract
1. Introduction
2. Results
2.1. Analysis of T. bernacchii pIgR Gene Locus
2.2. Analysis of T. bernacchii pIgR cDNA
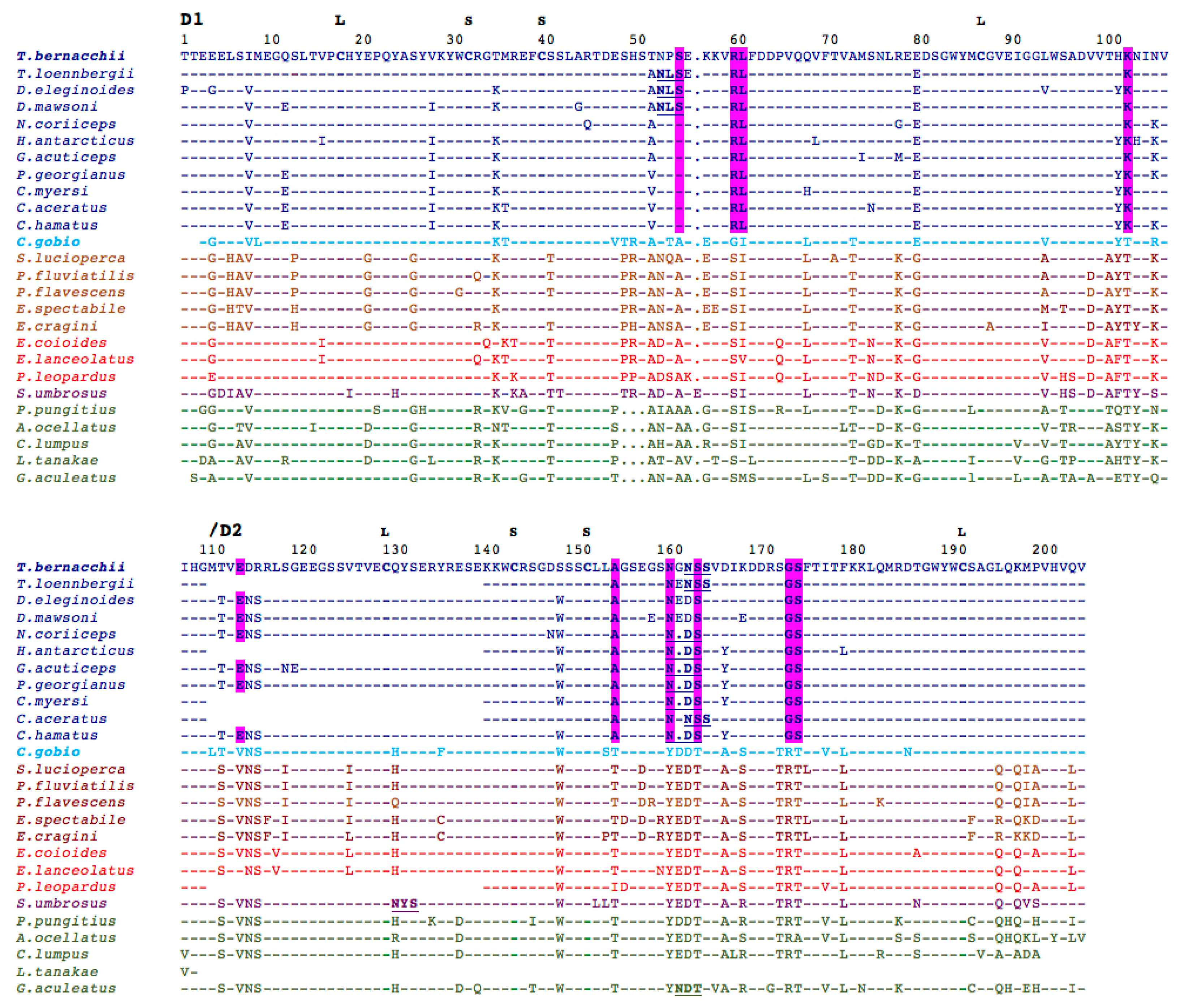
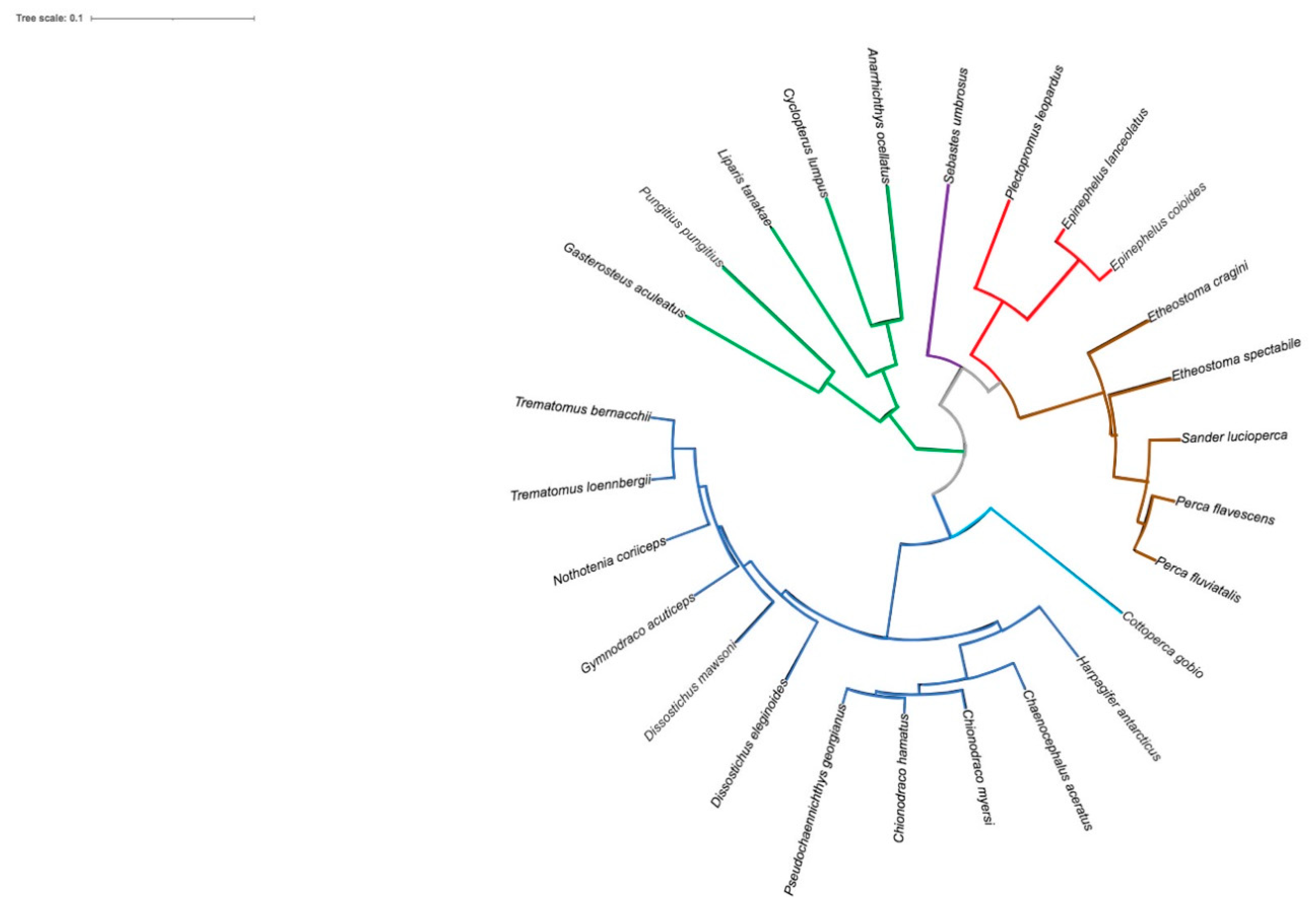
2.3. Analysis of T. bernacchii pIgR-Deduced Amino Acid Sequence
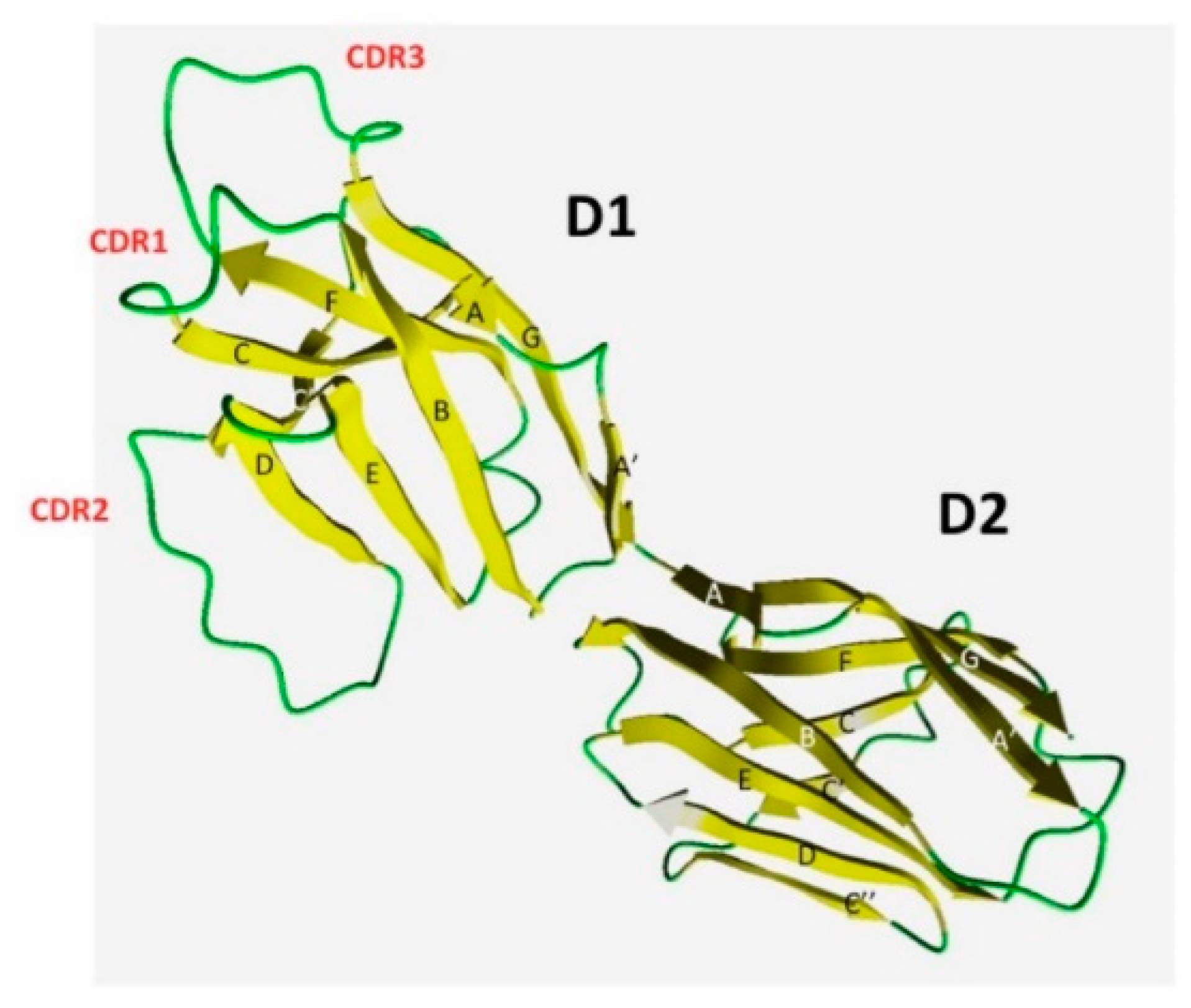
2.4. Structural Analysis of T. bernacchii pIgR
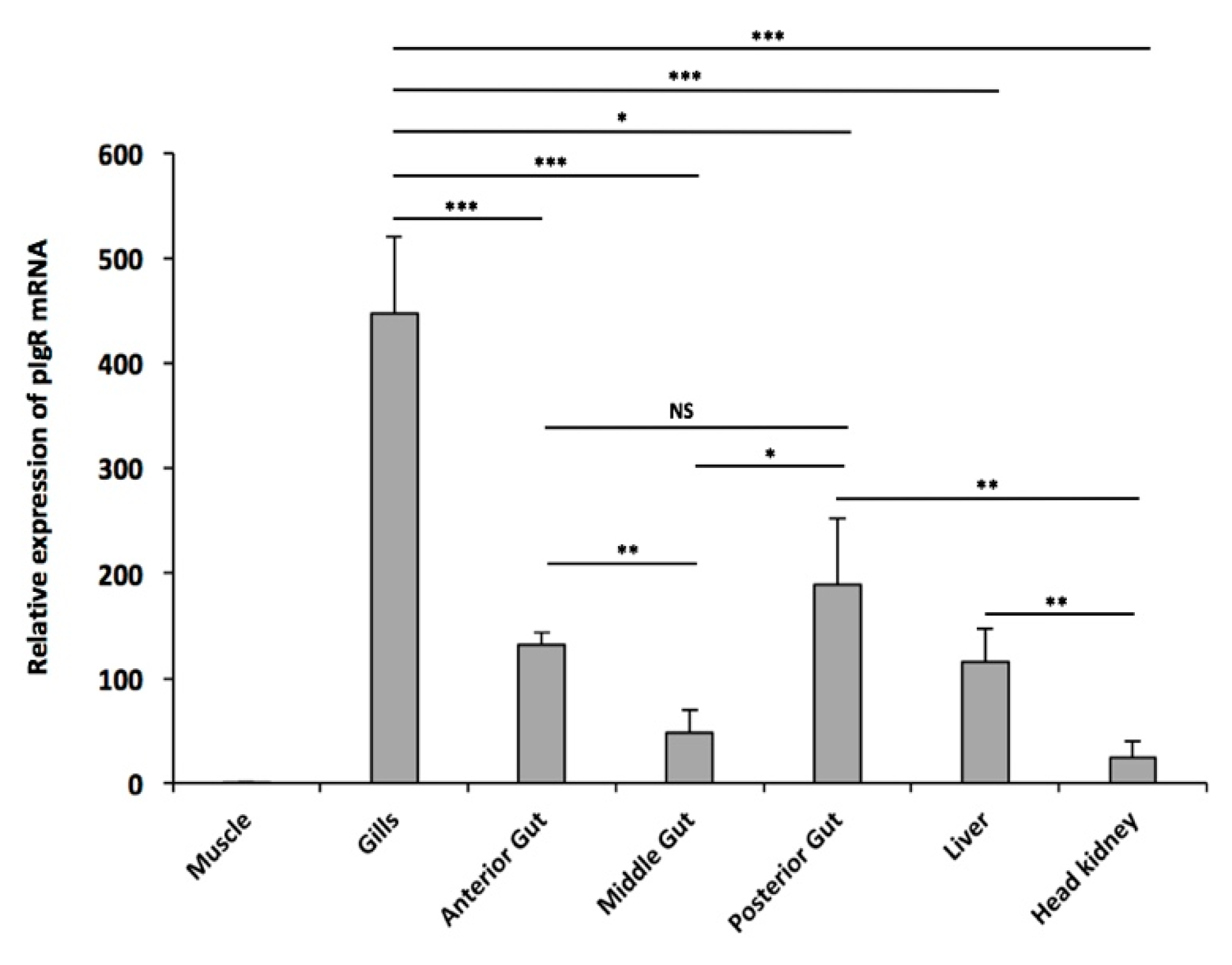
2.5. Basal Expression Analysis of pIgR Transcripts in T. bernacchii Mucosal Tissues and Lymphoid Organs
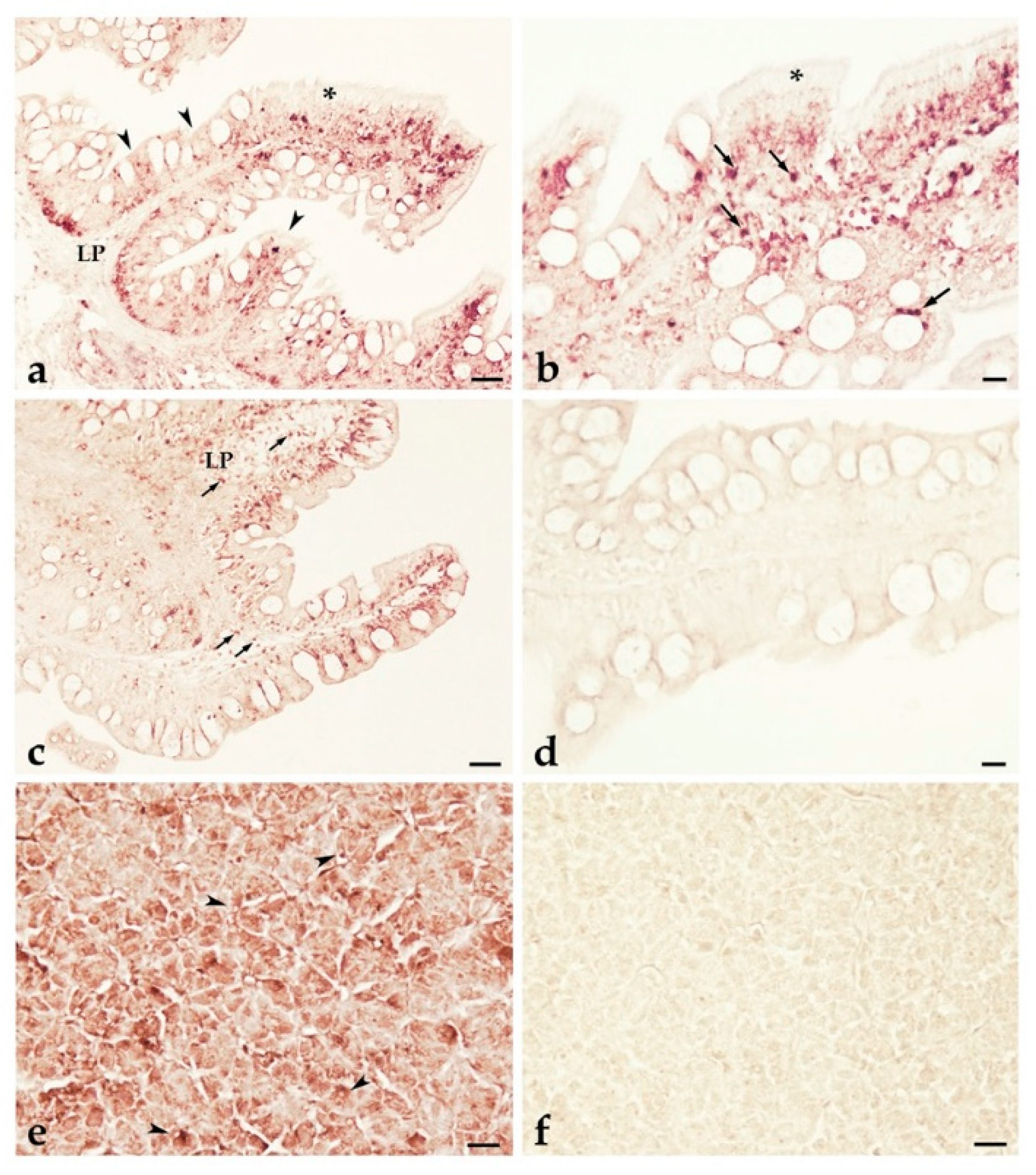
2.6. pIgR Expressing Cells in T. bernacchii Intestinal and Hepatic Tissues
3. Discussion
4. Materials and Methods
4.1. Biological Samples
4.2. Cloning of pIgR Transcript
4.3. 3′ and 5′ Rapid Amplification of cDNA Ends (RACE)
4.4. Data Availability
4.5. Gene Sequence Analyses
4.6. Deduced Amino Acid Sequence Analyses
4.7. Expression Analysis of pIgR Using Real-Time PCR
4.8. In Situ Hybridization (ISH)
4.8.1. Synthesis of RNA Probes
4.8.2. Staining Procedures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akula, S.; Mohammadamin, S.; Hellman, L. Fc receptors for immunoglobulins and their appearance during vertebrate evolution. PLoS ONE 2014, 9, e96903. [Google Scholar] [CrossRef] [PubMed]
- Kaetzel, C.S. Coevolution of mucosal immunoglobulins and the polymeric immunoglobulin receptor: Evidence that the commensal microbiota provided the driving force. ISRN Immunol. 2014, 2014, 541537. [Google Scholar] [CrossRef]
- Oreste, U.; Ametrano, A.; Coscia, M.R. On origin and evolution of the antibody molecule. Biology 2021, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Mostov, K.E.; Friedlander, M.; Blobel, G. The receptor for transepithelial transport of IgA and IgM contains multiple immunoglobulin-like domains. Nature 1984, 308, 37–43. [Google Scholar] [CrossRef]
- Kulseth, M.A.; Krajci, P.; Myklebost, O.; Rogne, S. Cloning and characterization of two forms of bovine polymeric immunoglobulin receptor cDNA. DNA Cell Biol. 1995, 14, 251–256. [Google Scholar] [CrossRef]
- Kuhn, L.C.; Kocher, H.P.; Hanly, W.C.; Cook, L.; Jaton, J.C.; Kraehenbuhl, J.P. Structural and genetic heterogeneity of the receptor mediating translocation of immunoglobulin A dimer antibodies across epithelia in the rabbit. J. Biol. Chem. 1983, 258, 6653–6659. [Google Scholar] [CrossRef]
- Wieland, W.H.; Orzaez, D.; Lammers, A.; Parmentier, H.K.; Verstegen, M.W.; Schots, A. A functional polymeric immunoglobulin receptor in chicken (Gallus gallus) indicates ancient role of secretory IgA in mucosal immunity. Biochem. J. 2004, 380, 669–676. [Google Scholar] [CrossRef]
- Magadán-Mompó, S.; Sánchez-Espinel, C.; Gambón-Deza, F. IgH loci of American alligator and saltwater crocodile shed light on IgA evolution. Immunogenetics 2013, 65, 531–541. [Google Scholar] [CrossRef]
- Braathen, R.; Hohman, V.S.; Brandtzaeg, P.; Johansen, F.-E. Secretory antibody formation: Conserved binding interactions between J chain and polymeric Ig receptor from humans and amphibians. J. Immunol. 2007, 178, 1589–1597. [Google Scholar] [CrossRef]
- Feng, L.-N.; Lu, D.-Q.; Bei, J.-X.; Chen, J.-L.; Liu, Y.; Zhang, Y.; Liu, X.-C.; Meng, Z.-N.; Wang, L.; Lin, H.-R. Molecular cloning and functional analysis of polymeric immunoglobulin receptor gene in orange-spotted grouper (Epinephelus coioides). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 154, 282–289. [Google Scholar] [CrossRef]
- Salinas, I.; Fernández-Montero, Á.; Ding, Y.; Sunyer, J.O. Mucosal immunoglobulins of teleost fish: A decade of advances. Dev. Comp. Immunol. 2021, 121, 104079. [Google Scholar] [CrossRef] [PubMed]
- Hohman, V.S.; Stewart, S.E.; Rumfelt, L.L.; Greenberg, A.S.; Avila, D.W.; Flajnik, M.F.; Steiner, L.A. J chain in the nurse shark: Implications for function in a lower vertebrate. J. Immunol. 2003, 170, 6016–6023. [Google Scholar] [CrossRef] [PubMed]
- Kaetzel, C.S. The polymeric immunoglobulin receptor: Bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef]
- Kong, X.; Wang, L.; Pei, C.; Zhang, J.; Zhao, X.; Li, L. Comparison of polymeric immunoglobulin receptor between fish and mammals. Vet. Immunol. Immunopath. 2018, 202, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Kortum, A.N.; Rodriguez-Nunez, I.; Yang, J.; Shim, J.; Runft, D.; O’Driscoll, M.L.; Haire, R.N.; Cannon, J.P.; Turner, P.M.; Litman, R.T.; et al. Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics 2014, 66, 267–279. [Google Scholar] [CrossRef]
- Near, T.J.; Ghezelayagh, A.; Ojeda, F.P.; Dornburg, A. Recent diversification in an ancient lineage of notothenioid fishes (Bovichtus: Notothenioidei). Polar Biol. 2019, 42, 943–952. [Google Scholar] [CrossRef]
- Eastman, J.T. Antarctic Fish Biology: Evolution in a Unique Environment; Academic Press: Cambridge, MA, USA, 1993; pp. 59–60. [Google Scholar]
- Near, T.J.; Dornburg, A.; Harrington, R.C.; Oliveira, C.; Pietsch, T.W.; Thacker, C.E.; Satoh, T.P.; Katayama, E.; Wainwright, P.C.; Eastman, J.T.; et al. Identification of the notothenioid sister lineage illuminates the biogeographic history of an Antarctic adaptive radiation. BMC Evol. Biol. 2015, 15, 109. [Google Scholar] [CrossRef]
- Coscia, M.R.; Varriale, S.; Giacomelli, S.; Oreste, U. Antarctic teleost immunoglobulins: More extreme, more interesting. Fish Shellfish Immunol. 2011, 31, 688–696. [Google Scholar] [CrossRef]
- Giacomelli, S.; Buonocore, F.; Albanese, F.; Scapigliati, G.; Gerdol, M.; Oreste, U.; Coscia, M.R. New insights into evolution of IgT genes coming from Antarctic teleosts. Mar. Genom. 2015, 24, 55–68. [Google Scholar] [CrossRef]
- Ametrano, A.; Gerdol, M.; Vitale, M.; Greco, S.; Oreste, U.; Coscia, M.R. The evolutionary puzzle solution for the origins of the partial loss of the Cτ2 exon in notothenioid fishes. Fish Shellfish Immunol. 2021, 116, 124–139. [Google Scholar] [CrossRef]
- Abelli, L.; Coscia, M.R.; De Santis, A.; Zeni, C.; Oreste, U. Evidence for hepato-biliary transport of immunoglobulin in the Antarctic teleost fish Trematomus bernacchii. Dev. Comp. Immunol. 2005, 29, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, J.-Y. Role of polymeric immunoglobulin receptor in IgA and IgM transcytosis. Int. J. Mol. Sci. 2021, 22, 2284. [Google Scholar] [CrossRef] [PubMed]
- Hamuro, K.; Suetake, H.; Saha, N.R.; Kikuchi, K.; Suzuki, Y. A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J. Immunol. 2007, 178, 5682–5689. [Google Scholar] [CrossRef]
- Koepfli, K.-P.; Paten, B.; Genome 10K community of scientists; O’Brien, S.J. The genome 10K Project: A way forward. Annu. Rev. Anim. Biosci. 2015, 3, 57–111. [Google Scholar] [CrossRef]
- Bista, I.; Wood JM, D.; Desvignes, T.; McCarthy, S.A.; Matschiner, M.; Ning, Z.; Tracey, A.; Torrance, J.; Sims, J.; Chow, W.; et al. Genomics of cold adaptations in the Antarctic notothenioid fish radiation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bista, I.; McCarthy, S.A.; Wood JM, D.; Ning, Z.; Detrich III, H.W.; Desvignes, T.; Postlethwait, J.; Chow, W.; Howe, K.; Torrance, J.; et al. The genome sequence of the channel bull blenny, Cottoperca gobio (Gunther, 1861). Wellcome Open Res. 2020, 5, 148. [Google Scholar] [CrossRef]
- Near, T.J.; MacGuigan, D.J.; Parker, E.; Struthers, C.D.; Jones, C.D.; Dornburg, A. Phylogenetic analysis of Antarctic notothenioids illuminates the utility of RADseq for resolving Cenozoic adaptive radiations. Mol. Phylogenet. Evol. 2018, 129, 268–279. [Google Scholar] [CrossRef]
- Pei, C.; Sun, X.; Zhang, Y.; Li, L.; Gao, Y.; Wang, L.; Kong, X. Molecular cloning, expression analyses of polymeric immunoglobulin receptor gene and its variants in grass carp (Ctenopharyngodon idellus) and binding assay of the recombinant immunoglobulin-like domains. Fish Shellfish Immunol. 2019, 88, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.L.; Fu, X.-D.; Gribskov, M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA 2005, 11, 1777–1787. [Google Scholar] [CrossRef]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which transposable elements are active in the human genome? Trends Genet. 2007, 23, 183–191. [Google Scholar] [CrossRef]
- Sela, N.; Kim, E.; Ast, G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010, 11, R59. [Google Scholar] [CrossRef]
- Daane, J.M.; Detrich, H.W., III. Adaptation and diversity of Antarctic fishes: A genomic perspective. Annu. Rev. Anim. Biosci. 2022, 10, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Cocca, E.; Iorio, S.D.; Capriglione, T. Identification of a novel helitron transposon in the genome of Antarctic fish. Mol. Phylogenet. Evol. 2011, 58, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Han, M.; Peng, Z. Evolution and diversity of transposable elements in fish genomes. Sci. Rep. 2019, 9, 15399. [Google Scholar] [CrossRef] [PubMed]
- Auvinet, J.; Graça, P.; Belkadi, L.; Petit, L.; Bonnivard, E.; Dettaï, A.; Detrich, W.H.; Ozouf-Costaz, C.; Higuet, D. Mobilization of retrotransposons as a cause of chromosomal diversification and rapid speciation: The case for the Antarctic teleost genus Trematomus. BMC Genom. 2018, 19, 339. [Google Scholar] [CrossRef]
- Pisano, E.; Coscia, M.R.; Mazzei, F.; Ghigliotti, L.; Coutanceau, J.-P.; Ozouf-Costaz, C.; Oreste, U. Cytogenetic mapping of immunoglobulin heavy chain genes in Antarctic fish. Genetica 2007, 130, 9–17. [Google Scholar] [CrossRef]
- Coscia, M.R.; Varriale, S.; De Santi, C.; Giacomelli, S.; Oreste, U. Evolution of the Antarctic teleost immunoglobulin heavy chain gene. Mol. Phylogenet. Evol. 2010, 55, 226–233. [Google Scholar] [CrossRef]
- Dwyer, K.; Agarwal, N.; Pile, L.; Ansari, A. Gene architecture facilitates intron-mediated enhancement of transcription. Front. Mol. Biosci. 2021, 8, 276. [Google Scholar] [CrossRef]
- Kushiro, A.; Sato, T. Polymeric immunoglobulin receptor gene of mouse: Sequence, structure and chromosomal location. Gene 1997, 204, 277–282. [Google Scholar] [CrossRef]
- Lau, D.T.; Saeed-Kothe, A.; Parker, S.K.; Detrich, H.W., III. Adaptive evolution of gene expression in Antarctic fishes: Divergent transcription of the 5′-to-5′ linked adult α1- and β-globin genes of the Antarctic teleost Notothenia coriiceps is controlled by dual promoters and intergenic enhancers1. Am. Zool. 2001, 41, 113–132. [Google Scholar] [CrossRef][Green Version]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Varriale, A.; Bernardi, G. DNA methylation and body temperature in fishes. Gene 2006, 385, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Coscia, M.R.; Morea, V.; Tramontano, A.; Oreste, U. Analysis of a cDNA sequence encoding the immunoglobulin heavy chain of the Antarctic teleost Trematomus bernacchii. Fish Shellfish Immunol. 2000, 10, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Rombout, J.; Vandertuin, S.; Yang, G.; Schopman, N.; Mroczek, A.; Hermsen, T.; Tavernethiele, J. Expression of the polymeric immunoglobulin receptor (PIgR) in mucosal tissues of common carp (Cyprinus carpio L.). Fish Shellfish Immunol. 2008, 24, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Tadiso, T.M.; Sharma, A.; Hordvik, I. Analysis of polymeric immunoglobulin receptor- and CD300-like molecules from Atlantic salmon. Mol. Immunol. 2011, 49, 462–473. [Google Scholar] [CrossRef]
- Xu, Z.; Parra, D.; Gómez, D.; Salinas, I.; Zhang, Y.-A.; von Gersdorff Jørgensen, L.; Heinecke, R.D.; Buchmann, K.; LaPatra, S.; Sunyer, J.O. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA 2013, 110, 13097–13102. [Google Scholar] [CrossRef]
- Palm, H.W. Ecology of Pseudoterranova decipiens (Krabbe, 1878) (Nematoda: Anisakidae) from Antarctic Waters. Parasitol. Res. 1999, 85, 638–646. [Google Scholar] [CrossRef][Green Version]
- Orecchia, P.; Mattiucci, S.; D’Amelio, S.; Paggi, L.; Plötz, J.; Cianchi, R.; Nascetti, G.; Arduino, P.; Bullini, L. Two new members in the Contracaecum osculatum Complex (Nematoda, Ascaridoidea) from the Antarctic. Int. J. Parasitol. 1994, 24, 367–377. [Google Scholar] [CrossRef]
- Rombout JH, W.M.; Taverne-Thiele, A.J.; Villena, M.I. The gut-associated lymphoid tissue (galt) of carp (Cyprinus carpio l.): An immunocytochemical analysis. Dev. Comp. Immunol. 1993, 17, 55–66. [Google Scholar] [CrossRef]
- Sheng, X.; Qian, X.; Tang, X.; Xing, J.; Zhan, W. Polymeric immunoglobulin receptor mediates immune excretion of mucosal igm–antigen complexes across intestinal epithelium in flounder (Paralichthys olivaceus). Front. Immunol. 2018, 9, 1562. [Google Scholar] [CrossRef]
- Salinas, I.; Parra, D. 6-Fish mucosal immunity: Intestine. In Mucosal Health in Aquaculture; Beck, B.H., Peatman, E., Eds.; Academic Press: San Diego, CA, USA, 2015. [Google Scholar] [CrossRef]
- Brandl, K.; Kumar, V.; Eckmann, L. Gut-liver axis at the frontier of host-microbial interactions. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G413–G419. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhan, W.; Ding, B.; Sheng, X. Molecular cloning and expression analysis of polymeric immunoglobulin receptor in flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2013, 35, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Y.; Li, H.; Dong, S.; Wang, Q.; Huang, Z.; Kong, W.; Zhang, X.; Xu, Y.; Chen, X.; et al. Polymeric immunoglobulin receptor in dojo loach (Misgurnus anguillicaudatus): Molecular characterization and expression analysis in response to bacterial and parasitic challenge. Fish Shellfish Immunol. 2018, 73, 175–184. [Google Scholar] [CrossRef]
- Nguinkal, J.A.; Brunner, R.M.; Verleih, M.; Rebl, A.; de Los Ríos-Pérez, L.; Schäfer, N.; Hadlich, F.; Stüeken, M.; Wittenburg, D.; Goldammer, T. The first highly contiguous genome assembly of pikeperch (Sander lucioperca), an emerging aquaculture species in Europe. Genes 2019, 10, 708. [Google Scholar] [CrossRef]
- Roques, C.; Zahm, M.; Cabau, C.; Klopp, C.; Bouchez, O.; Donnadieu, C.; Kuhl, H.; Gislard, M.; Guendouz, S.; Journot, L.; et al. A chromosome-scale genome assembly of the European perch, Perca fluviatilis. EMBL/GenBank/DDBJ Databases 2019. submitted. [Google Scholar]
- Feron, R.; Morvezen, R.; Bestin, A.; Haffray, P.; Klopp, C.; Zahm, M.; Cabau, C.; Roques, C.; Donnadieu, C.; Bouchez, O.; et al. A chromosome-scale genome assembly of the yellow perch, Perca flavescens. EMBL/GenBank/DDBJ Databases 2019. submitted. [Google Scholar]
- Moran, R.L.; Catchen, J.M.; Fuller, R.C. A chromosome-level genome assembly, high-density linkage maps, and genome scans reveal the genomic architecture of hybrid incompatibilities underlying speciation via character displacement in darters (Percidae: Etheostominae). EMBL/GenBank/DDBJ Databases 2019. submitted. [Google Scholar]
- Reid, B.N.; Moran, R.L.; Kopack, C.J.; Fitzpatrick, S.W. Rapture-ready darters: Choice of reference genome and genotyping method (whole-genome or sequence capture) influence population genomic inference in Etheostoma. Mol. Ecol. Resour. 2021, 21, 404–420. [Google Scholar] [CrossRef]
- Zhou, Q.; Gao, H.; Zhang, Y.; Fan, G.; Xu, H.; Zhai, J.; Xu, W.; Chen, Z.; Zhang, H.; Liu, S.; et al. A chromosome-level genome of the giant grouper (Epinephelus lanceolatus) provides insights into its innate immunity and rapid growth. Mol. Ecol. Resour. 2019, 19, 1322–1332. [Google Scholar] [CrossRef]
- Zhou, Q.; Guo, X.; Huang, Y.; Gao, H.; Xu, H.; Liu, S.; Zheng, W.; Zhang, T.; Tian, C.; Zhu, C.; et al. De novo sequencing and chromosomal-scale genome assembly of leopard coral grouper, Plectropomus leopardus. Mol. Ecol. Resour. 2020, 20, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Varadharajan, S. Pungitius pungitius genome assembly, contig: LG2, whole genome shotgun sequence. EMBL/GenBank/DDBJ Databases 2019. submitted. [Google Scholar]
- Culibrk, L.; Leelakumari, S.; Taylor, G.A.; Tse, K.; Cheng, D.; Chuah, E.; Kirk, H.; Pandoh, P.; Troussard, A.; Zhao, Y.; et al. The genome of the wolf eel (Anarrhichthys ocellatus). EMBL/GenBank/DDBJ Databases 2019. submitted. [Google Scholar]
- Nath, S.; Shaw, D.E.; White, M.A. Improved contiguity of the threespine stickleback genome using long-read sequencing. G3 2021, 11, jkab007. [Google Scholar] [CrossRef] [PubMed]
- Touma, J.; García, K.K.; Bravo, S.; Leiva, F.; Moya, J.; Vargas-Chacoff, L.; Reyes, A.; Vidal, R. De novo assembly and characterization of patagonian toothfish transcriptome and develop of EST-SSR markers for population genetics. Front. Mar. Sci. 2019, 6, 720. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.J.; Lee, J.K.; Ahn, D.H.; Kim, M.G.; Lee, H.; Lee, J.; Kim, B.-K.; Park, H. Transcriptomics and comparative analysis of three Antarctic notothenioid fishes. PLoS ONE 2012, 7, e43762. [Google Scholar] [CrossRef]
- Song, W.; Li, L.; Huang, H.; Jiang, K.; Zhang, F.; Wang, L.; Zhao, M.; Ma, L. Tissue-based transcriptomics of Chionodraco hamatus: Sequencing, de novo assembly, annotation and marker discovery. J. Fish Biol. 2019, 94, 251–260. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, J.-H.; Jo, E.; Choi, E.; Kim, J.; Choi, S.-G.; Chung, S.; Kim, H.-W.; Park, H. Chromosomal assembly of the Antarctic toothfish (Dissostichus mawsoni) genome using third-generation DNA sequencing and Hi-C technology. Zool. Res. 2021, 42, 124–129. [Google Scholar] [CrossRef]
- Bargelloni, L.; Babbucci, M.; Ferraresso, S.; Papetti, C.; Vitulo, N.; Carraro, R.; Pauletto, M.; Santovito, G.; Lucassen, M.; Mark, F.C.; et al. Draft genome assembly and transcriptome data of the icefish Chionodraco myersi reveal the key role of mitochondria for a life without hemoglobin at subzero temperatures. Commun. Biol. 2019, 2, 443. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, S.K.; Tørresen, K.O.; Boessenkool, S.; Malmstrøm, M.; Star, B.; Jakobsen, S.K.; Riiser, S.E. The Chaenocephalus aceratus whole genome shotgun project. EMBL/GenBank/DDBJ Database 2018. submitted. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-3.0; 1996, Unpublished Data. Available online: https://www.repeatmasker.org/ (accessed on 8 July 2021).
- Hubley, R.; Finn, R.D.; Clements, J.; Eddy, S.R.; Jones, T.A.; Bao, W.; Smit, A.F.A.; Wheeler, T.J. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016, 44, D81–D89. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D. Object-Oriented Transcription Factors Database (OoTFD). Nucleic Acids Res. 2000, 28, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Marín, A. Characterization and prediction of alternative splice sites. Gene 2006, 366, 219–227. [Google Scholar] [CrossRef]
- Liu, H.; Han, H.; Li, J.; Wong, L. DNAFSMiner: A web-based software toolbox to recognize two types of functional sites in DNA sequences. Bioinformatics 2005, 21, 671–673. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Cuthbertson, J.M.; Doyle, D.A.; Sansom, M.S.P. Transmembrane helix prediction: A comparative evaluation and analysis. Protein Eng. Des. Sel. 2005, 18, 295–308. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 6.0 achieves signal peptide prediction across all types using protein language models. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. In Multiple Sequence Alignment Methods; Russell, D.J., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 105–116. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 2001, 310–322. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. Models@Home: Distributed computing in bioinformatics using a screensaver based approach. Bioinformatics 2002, 18, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Picchietti, S.; Abelli, L.; Guerra, L.; Randelli, E.; Proietti Serafin, F.; Belardinelli, M.C.; Buonocore, F.; Bernini, C.; Fausto, A.M.; Scapigliati, G. MHC II-β chain gene expression studies define the regional organization of the thymus in the developing bony fish Dicentrarchus labrax (L.). Fish Shellfish Immunol. 2015, 42, 483–493. [Google Scholar] [CrossRef] [PubMed]
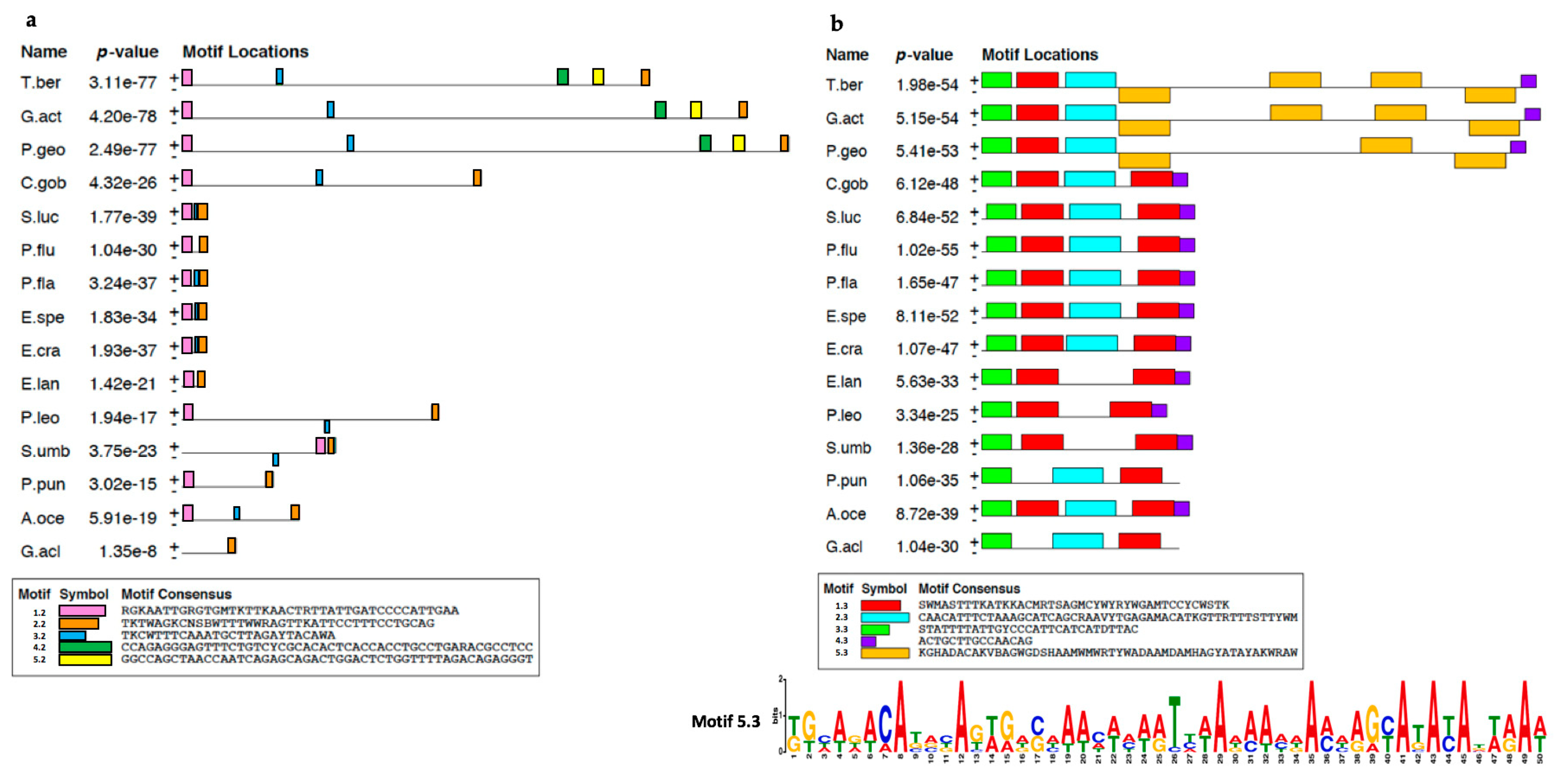
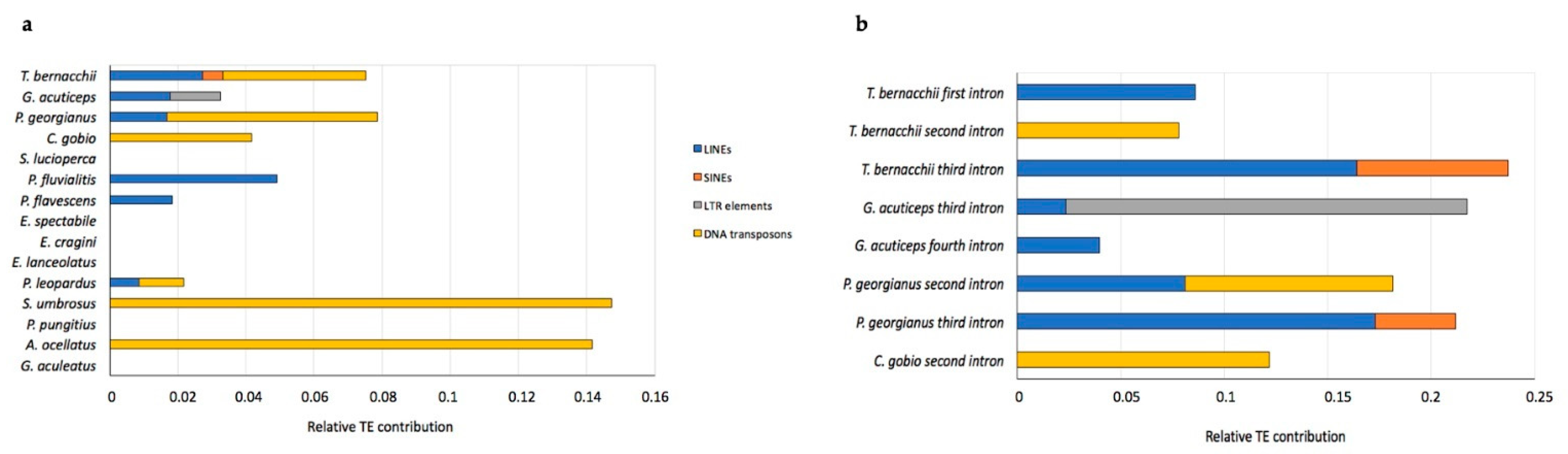
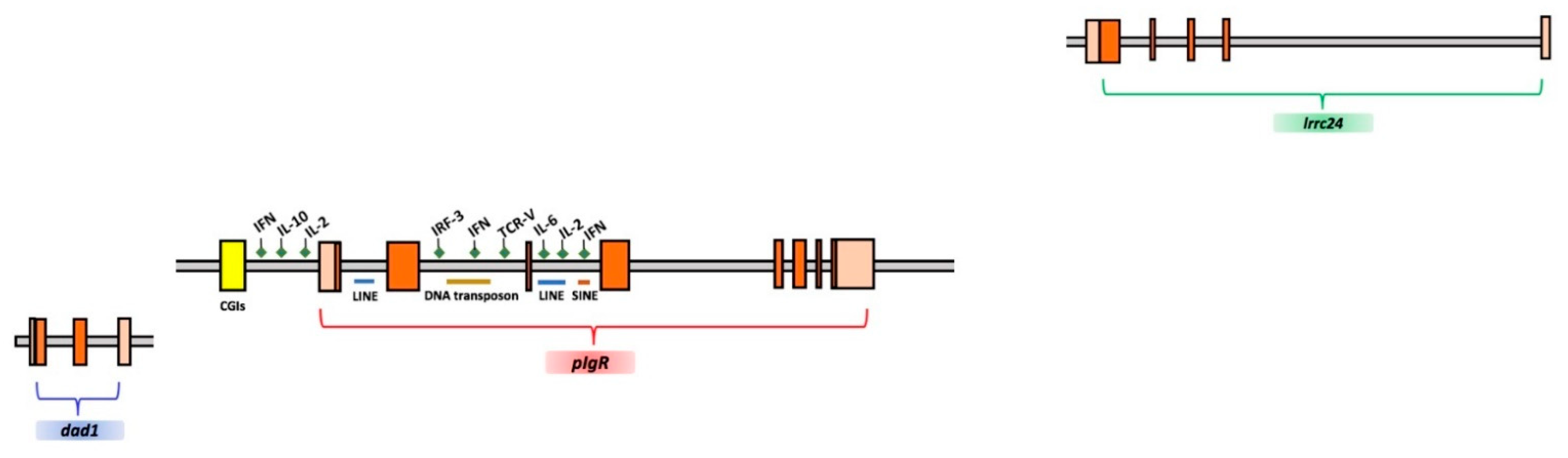
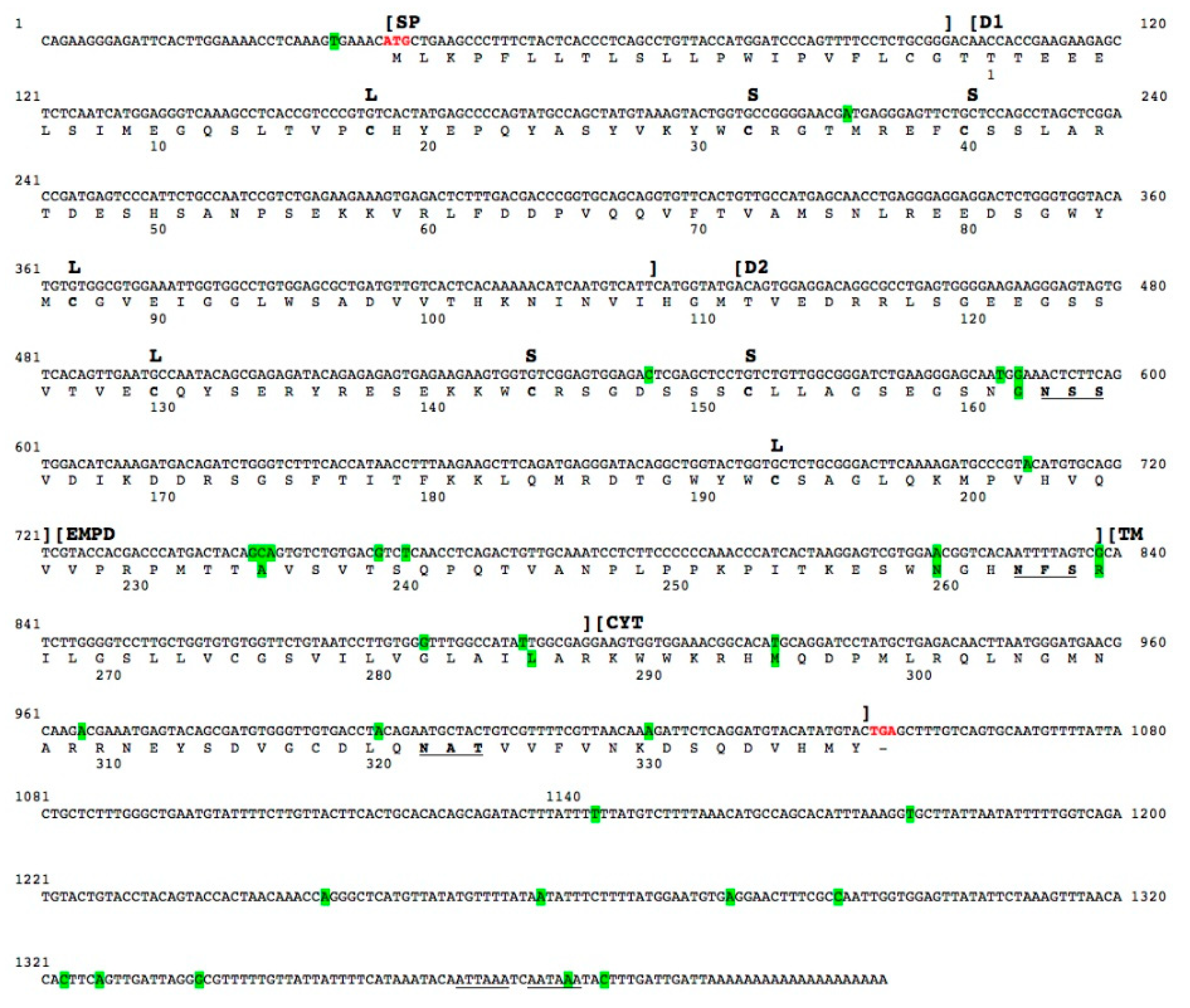




| Suborder | Species | pIgR Gene Size | 1st Intron | 2nd Intron | 3rd Intron | 4th Intron | 5th Intron | 6th Intron | 7th Intron |
|---|---|---|---|---|---|---|---|---|---|
| Notothenioidei | Trematomus bernacchii Gymnodraco acuticeps Pseudochaennichthys georgianus Cottoperca gobio | 8310 8235 9013 5930 | 628 583 582 596 | 2372 2866 3080 1519 | 548 552 538 204 | 2825 2906 2939 1713 | 124 124 133 156 | 146 85 85 129 | 111 111 111 115 |
| Percoidei | Sander lucioperca Perca fluviatilis Perca flavescens Etheostoma spectabile Etheostoma cragini | 4818 3976 6669 5939 3700 | 649 590 554 579 595 | 124 126 125 124 124 | 211 211 211 153 207 | 1080 1237 1229 1014 985 | 161 171 171 143 143 | 155 137 156 137 145 | 115 122 115 115 117 |
| Serranoidei | Epinephelus lanceolatus Plectropomus leopardus | 5831 7090 | 588 545 | 117 1304 | 206 183 | 2969 2135 | 163 161 | 93 150 | 107 118 |
| Scorpaenoidei | Sebastes umbrosus | 6887 | 601 | 779 | 208 | 2631 | 634 | 133 | 118 |
| Cottoidei | Pungitius pungitius Anarrhichthys ocellatus Gasterosteus aculeatus | 5126 5432 4095 | 579 595 577 | 461 593 274 | 196 205 195 | 872 1369 1210 | 136 176 89 | 167 153 159 | 96 106 99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ametrano, A.; Picchietti, S.; Guerra, L.; Giacomelli, S.; Oreste, U.; Coscia, M.R. Comparative Analysis of the pIgR Gene from the Antarctic Teleost Trematomus bernacchii Reveals Distinctive Features of Cold-Adapted Notothenioidei. Int. J. Mol. Sci. 2022, 23, 7783. https://doi.org/10.3390/ijms23147783
Ametrano A, Picchietti S, Guerra L, Giacomelli S, Oreste U, Coscia MR. Comparative Analysis of the pIgR Gene from the Antarctic Teleost Trematomus bernacchii Reveals Distinctive Features of Cold-Adapted Notothenioidei. International Journal of Molecular Sciences. 2022; 23(14):7783. https://doi.org/10.3390/ijms23147783
Chicago/Turabian StyleAmetrano, Alessia, Simona Picchietti, Laura Guerra, Stefano Giacomelli, Umberto Oreste, and Maria Rosaria Coscia. 2022. "Comparative Analysis of the pIgR Gene from the Antarctic Teleost Trematomus bernacchii Reveals Distinctive Features of Cold-Adapted Notothenioidei" International Journal of Molecular Sciences 23, no. 14: 7783. https://doi.org/10.3390/ijms23147783
APA StyleAmetrano, A., Picchietti, S., Guerra, L., Giacomelli, S., Oreste, U., & Coscia, M. R. (2022). Comparative Analysis of the pIgR Gene from the Antarctic Teleost Trematomus bernacchii Reveals Distinctive Features of Cold-Adapted Notothenioidei. International Journal of Molecular Sciences, 23(14), 7783. https://doi.org/10.3390/ijms23147783






