Investigation of Biomarkers Associated with Low Platelet Counts in Normal Karyotype Acute Myeloid Leukemia
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of the Patients
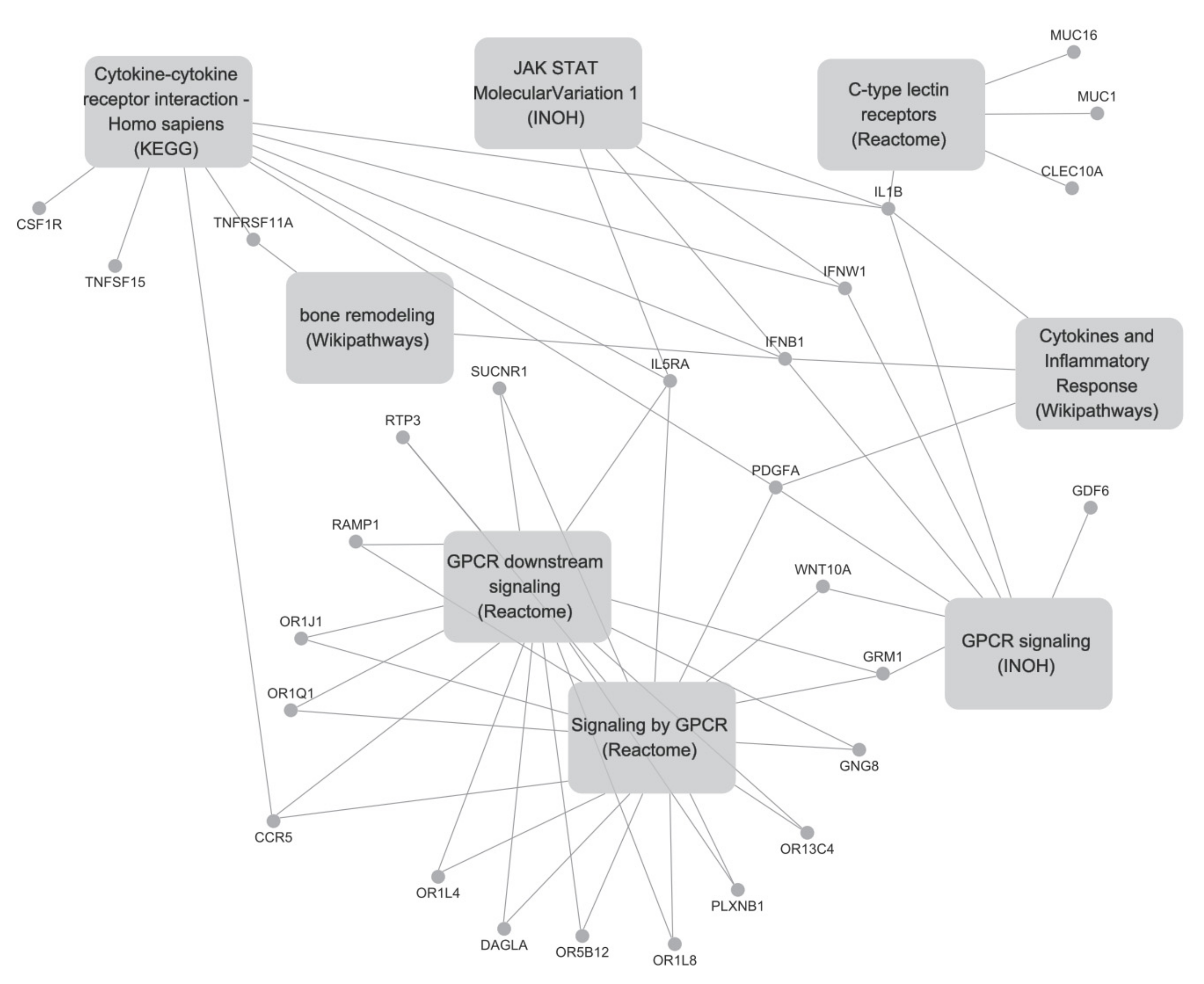
2.2. Differentially Expressed Gene Analysis, Pathway Analysis, and Network Analysis
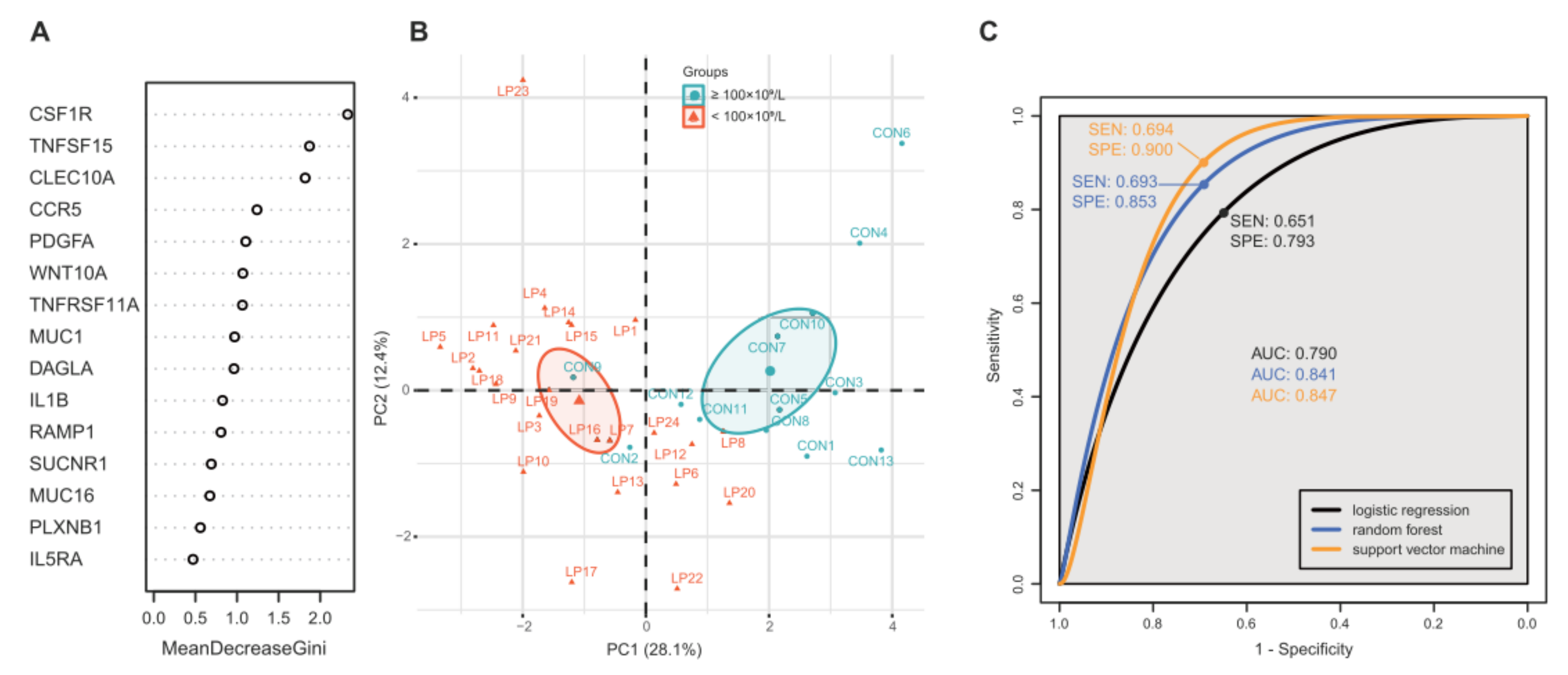
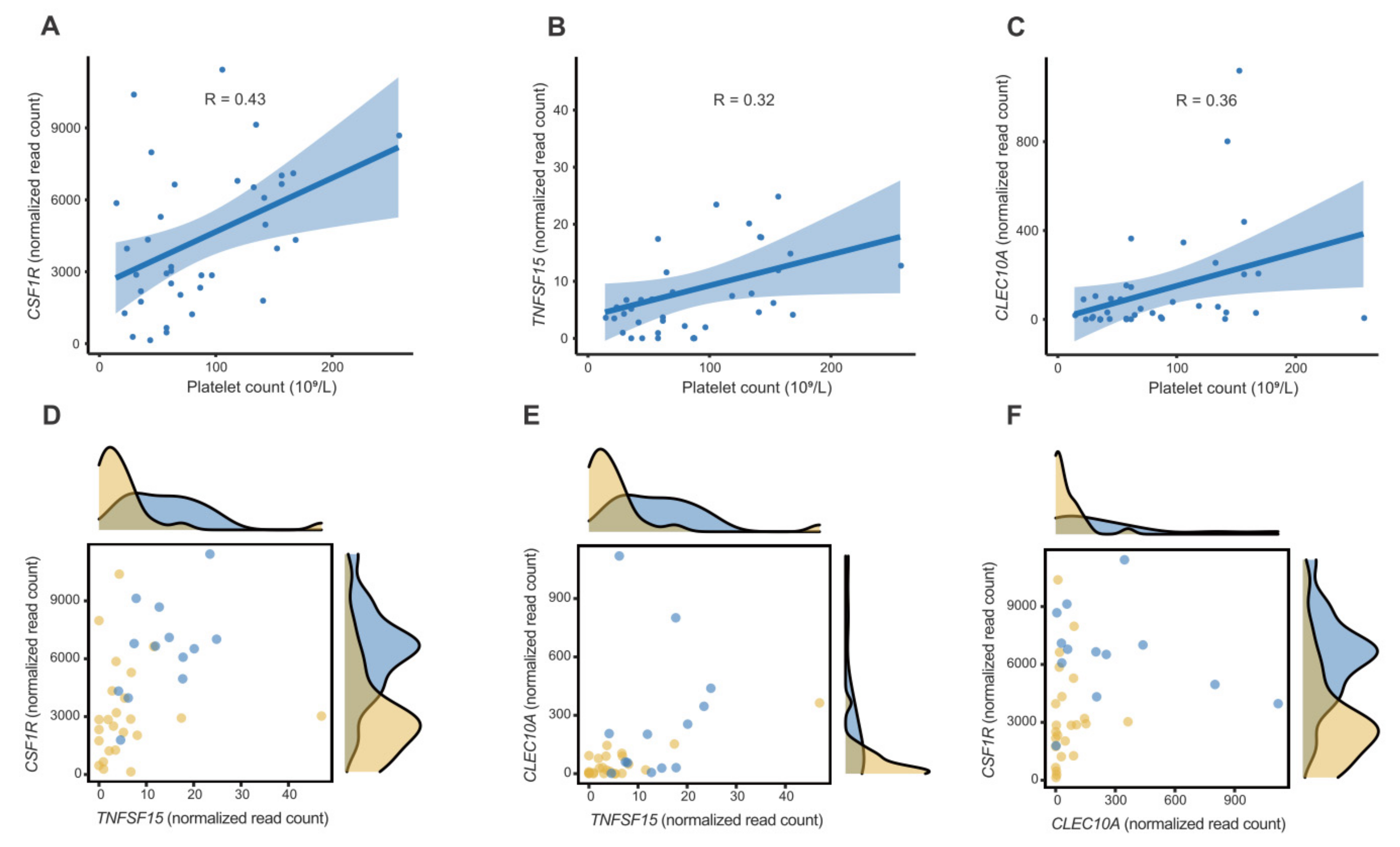
2.3. Feature Selection, Modelling, and Performance Evaluation
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Case Definition
4.2. Differentially Expressed Gene (DEG) Analysis and Pathway Analysis
4.3. Feature Selection
4.4. Computational Modelling and Validation
4.5. Statistical Analysis and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruserud, O.; Foss, B. Platelet functions and clinical effects in acute myelogenous leukemia. Thromb. Haemost. 2008, 99, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Trafalis, D.T.; Poulakidas, E.; Kapsimali, V.; Tsigris, C.; Papanicolaou, X.; Harhalakis, N.; Nikiforakis, E.; Mentzikof-Mitsouli, C. Platelet production and related pathophysiology in acute myelogenous leukemia at first diagnosis: Prognostic implications. Oncol. Rep. 2008, 19, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Amo, L.; Tamayo-Orbegozo, E.; Maruri, N.; Eguizabal, C.; Zenarruzabeitia, O.; Rinon, M.; Arrieta, A.; Santos, S.; Monge, J.; Vesga, M.A.; et al. Involvement of platelet–tumor cell interaction in immune evasion. Potential role of podocalyxin-like protein 1. Front. Oncol. 2014, 4, 245. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Che, M.; Zacharek, A.; Qiao, Y.; Li, L.; Li, X.; Lamberti, M.; Tang, K.; Cai, Y.; Guo, Y.; et al. Differential Expression of Thromboxane Synthase in Prostate Carcinoma: Role in Tumor Cell Motility. Am. J. Pathol. 2004, 164, 429–439. [Google Scholar] [CrossRef]
- Zarà, M.; Canobbio, I.; Visconte, C.; Canino, J.; Torti, M.; Guidetti, G.F. Molecular mechanisms of platelet activation and aggregation induced by breast cancer cells. Cell. Signal. 2018, 48, 45–53. [Google Scholar] [CrossRef]
- Schmied, L.; Höglund, P.; Meinke, S. Platelet-Mediated Protection of Cancer Cells from Immune Surveillance—Possible Implications for Cancer Immunotherapy. Front. Immunol. 2021, 12, 640578. [Google Scholar] [CrossRef]
- Haemmerle, M.; Stone, R.L.; Menter, D.G.; Afshar-Kharghan, V.; Sood, A.K. The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell 2018, 33, 965–983. [Google Scholar] [CrossRef]
- Li, N.; Hu, H.; Lindqvist, M.; Wikström-Jonsson, E.; Goodall, A.H.; Hjemdahl, P. Platelet-Leukocyte Cross Talk in Whole Blood. Arter. Thromb. Vasc. Biol. 2000, 20, 2702–2708. [Google Scholar] [CrossRef]
- Cluxton, C.D.; Spillane, C.; O’Toole, S.; Sheils, O.; Gardiner, C.M.; O’Leary, J.J. Suppression of Natural Killer cell NKG2D and CD226 anti-tumour cascades by platelet cloaked cancer cells: Implications for the metastatic cascade. PLoS ONE 2019, 14, e0211538. [Google Scholar] [CrossRef]
- Foss, B.; Ulvestad, E.; Hervig, T. Effects of cytarabine and various anthracyclins on platelet activation: Characterization ofin vitro effects and their possible clinical relevance in acute myelogenous leukemia. Int. J. Cancer 2001, 97, 106–114. [Google Scholar] [CrossRef]
- Zhang, Q.; Dai, K.; Bi, L.; Jiang, S.; Han, Y.; Yu, K.; Zhang, S. Pretreatment platelet count predicts survival outcome of patients with de novo non-M3 acute myeloid leukemia. PeerJ 2017, 5, e4139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bruserud, B.F.O. Effects of normal platelets on proliferation and constitutive cytokine secretion by human acute myelogenous leukaemia blasts. Platelets 1997, 8, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Foss, B.; Mentzoni, L.; Bruserud, Ø. Effects of Vascular Endothelial Growth Factor on Acute Myelogenous Leukemia Blasts. J. Hematotherapy 2001, 10, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Shen, Y.; Li, Y.; Wang, B.; Wang, B.; Wu, D.; Ruan, C.; Wang, Y. Ectopic ATP synthase β subunit proteins on human leukemia cell surface interact with platelets by binding glycoprotein IIb. Haematologica 2019, 104, e364–e368. [Google Scholar] [CrossRef]
- Hoyle, C.; Hayhoe, F.G.J. Abnormal Megakaryopoiesis in Acute Leukaemia. Br. J. Haematol. 1988, 68, 393–394. [Google Scholar] [CrossRef] [PubMed]
- Keating, M.J.; Smith, T.L.; Gehan, E.A.; McCredie, K.B.; Bodey, G.P.; Freireich, E.J. A prognostic factor analysis for use in development of predictive models for response in adult acute leukemia. Cancer 1982, 50, 457–465. [Google Scholar] [CrossRef]
- Lee, E.-J.; Lee, A.I. Thrombocytopenia. Prim. Care 2016, 43, 543–557. [Google Scholar] [CrossRef]
- Semple, J.W.; Italiano, J.E., Jr.; Freedman, J. Platelets and the immune continuum. Nat. Rev. Immunol. 2011, 11, 264–274. [Google Scholar] [CrossRef]
- Morrell, C.N.; Aggrey, A.A.; Chapman, L.M.; Modjeski, K.L. Emerging roles for platelets as immune and inflammatory cells. Blood 2014, 123, 2759–2767. [Google Scholar] [CrossRef]
- Kaushansky, K. The molecular mechanisms that control thrombopoiesis. J. Clin. Investig. 2005, 115, 3339–3347. [Google Scholar] [CrossRef]
- Chen, Y.; Zhong, H.; Zhao, Y.; Luo, X.; Gao, W. Role of platelet biomarkers in inflammatory response. Biomark. Res. 2020, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Offermanns, S. Activation of Platelet Function Through G Protein–Coupled Receptors. Circ. Res. 2006, 99, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Estevez, B.; Du, X. New Concepts and Mechanisms of Platelet Activation Signaling. Physiology 2017, 32, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Astarita, J.L.; Acton, S.E.; Turley, S.J. Podoplanin: Emerging functions in development, the immune system, and cancer. Front. Immunol. 2012, 3, 283. [Google Scholar] [CrossRef]
- Lavallée, V.-P.; Chagraoui, J.; Macrae, T.; Marquis, M.; Bonnefoy, A.; Krosl, J.; Lemieux, S.; Marinier, A.; Pabst, C.; Rivard, G.-É.; et al. Transcriptomic landscape of acute promyelocytic leukemia reveals aberrant surface expression of the platelet aggregation agonist Podoplanin. Leukemia 2018, 32, 1349–1357. [Google Scholar] [CrossRef]
- Ruhnke, L.; Stölzel, F.; Wagenführ, L.; Altmann, H.; Platzbecker, U.; Herold, S.; Rump, A.; Schröck, E.; Bornhäuser, M.; Schetelig, J.; et al. Case Report: ANXA2 Associated Life-Threatening Coagulopathy with Hyperfibrinolysis in a Patient With Non-APL Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 666014. [Google Scholar] [CrossRef]
- Gao, B.; Wang, H.; Lafdil, F.; Feng, D. STAT proteins—Key regulators of anti-viral responses, inflammation, and tumorigenesis in the liver. J. Hepatol. 2012, 57, 430–441. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Plenge, R. JAK and STAT Signaling Molecules in Immunoregulation and Immune-Mediated Disease. Immunity 2012, 36, 542–550. [Google Scholar] [CrossRef]
- Lu, W.-J.; Lin, K.-C.; Huang, S.-Y.; Thomas, P.A.; Wu, Y.-H.; Wu, H.-C.; Lin, K.-H.; Sheu, J.-R. Role of a Janus kinase 2-dependent signaling pathway in platelet activation. Thromb. Res. 2014, 133, 1088–1096. [Google Scholar] [CrossRef]
- Sharif, P.S.; Abdollahi, M. The role of platelets in bone remodeling. Inflamm. Allergy-Drug Targets 2010, 9, 393–399. [Google Scholar] [CrossRef]
- Khan, F.A.; Parayaruthottam, P.; Roshan, G.; Menon, V.; Fidha, M.; Fernandes, A.K. Platelets and Their Pathways in Dentistry: Systematic Review. J. Int. Soc. Prev. Community Dent. 2017, 7, S55–S60. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D. M1 and M2 Macrophages: Oracles of Health and Disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, R.; Kratochvill, F.; Murray, P.J.; Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 2015, 36, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.B.; Danielsen, A.V.; Hamilton-Dutoit, S.J.; Bendix, K.; Nørgaard, P.; Møller, M.B.; Steiniche, T.; D’Amore, F. High intratumoral macrophage content is an adverse prognostic feature in anaplastic large cell lymphoma. Histopathology 2014, 65, 490–500. [Google Scholar] [CrossRef]
- Stanley, E.R.; Chitu, V. CSF-1 Receptor Signaling in Myeloid Cells. Cold Spring Harb. Perspect. Biol. 2014, 6, a021857. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Liu, L.; Gong, C.-Y.; Shi, H.-S.; Zeng, Y.-H.; Wang, X.-Z.; Zhao, Y.-W.; Wei, Y.-Q. Prognostic Significance of Tumor-Associated Macrophages in Solid Tumor: A Meta-Analysis of the Literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef]
- Sapi, E. The Role of CSF-1 in Normal Physiology of Mammary Gland and Breast Cancer: An Update. Exp. Biol. Med. 2004, 229, 1–11. [Google Scholar] [CrossRef]
- Valero, J.G.; Matas-Céspedes, A.; Arenas, F.; Rodriguez, V.; Carreras, J.; Serrat, N.; Guerrero-Hernández, M.; Yahiaoui, A.; Balagué, O.; Martin, S.; et al. The receptor of the colony-stimulating factor-1 (CSF-1R) is a novel prognostic factor and therapeutic target in follicular lymphoma. Leukemia 2021, 35, 2635–2649. [Google Scholar] [CrossRef]
- Ho, W.J.; Jaffee, E.M. Macrophage-Targeting by CSF1/1R Blockade in Pancreatic Cancers. Cancer Res. 2021, 81, 6071–6073. [Google Scholar] [CrossRef]
- Zhao, C.-C.; Han, Q.-J.; Ying, H.-Y.; Gu, X.-X.; Yang, N.; Li, L.-Y.; Zhang, Q.-Z. TNFSF15 facilitates differentiation and polarization of macrophages toward M1 phenotype to inhibit tumor growth. OncoImmunology 2022, 11, 2032918. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; NI, J.; Jiang, G.; Lu, J.; Xing, L.; Lincoln, C.; Carter, K.C.; Janat, F.; Kozak, D.; Xu, S.; et al. VEGI, a novel cytokine of the tumor necrosis factor family, is an angiogenesis inhibitor that suppresses the growth of colon carcinomas in vivo. FASEB J. 1999, 13, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, C.; Lovato, O.; Bertolaso, A.; Zoratti, E.; Malpeli, G.; Mimiola, E.; Tinelli, M.; Aprili, F.; Tecchio, C.; Perbellini, O.; et al. Expression and function of the TL1A/DR3 axis in chronic lymphocytic leukemia. Oncotarget 2015, 6, 32061–32074. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, L.; Zhang, L.; Li, J.; Liao, L.; Huang, L.; Li, W.; Yang, J. Immunological role and prognostic potential of CLEC10A in pan-cancer. Am. J. Transl. Res. 2022, 14, 2844–2860. [Google Scholar] [CrossRef]
- He, M.; Han, Y.; Cai, C.; Liu, P.; Chen, Y.; Shen, H.; Xu, X.; Zeng, S. CLEC10A is a prognostic biomarker and correlated with clinical pathologic features and immune infiltrates in lung adenocarcinoma. J. Cell. Mol. Med. 2021, 25, 3391–3399. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, H.; Chen, Q.; Zhang, Y.; Cheng, H.; Yang, J.; Hu, X. Low Platelet Counts at Diagnosis Predict Better Survival for Patients with Intermediate-Risk Acute Myeloid Leukemia. Acta Haematol. 2019, 143, 9–18. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- May, F.; Hagedorn, I.; Pleines, I.; Bender, M.; Vögtle, T.; Eble, J.; Elvers, M.; Nieswandt, B. CLEC-2 is an essential platelet-activating receptor in hemostasis and thrombosis. Blood 2009, 114, 3464–3472. [Google Scholar] [CrossRef]
- Herold, F.; Kleps, J.; Wolska, I.; Nowak, G. Synthesis of new hexahydro- and octahydropyrido[1,2-c]pyrimidine derivatives with an arylpiperazine moiety as ligands for 5-HT1A and 5-HT2A receptors. Il Farmaco 2002, 57, 959–971. [Google Scholar] [CrossRef]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Groups | PD-AML | PND-AML | p Value |
|---|---|---|---|
| Number | 24 | 13 | |
| Age (yrs) | 61.1 ± 14.2 | 63.7 ± 14.8 | 0.608 |
| Male ratio | 54.2% (13/24) | 46.2% (6/13) | 0.904 |
| Ethnicity | 0.550 | ||
| Asian | 4.3% (1/23) | 0% (0/13) | |
| Black or African American | 4.3% (1/23) | 0% (0/13) | |
| White | 91.3% (21/23) | 100% (13/13) | |
| Laboratory findings | |||
| Hb (g/L) | 97 ± 18 | 107 ± 13 | 0.078 |
| Platelet (×109/L) | 51.3 ± 22.3 | 151.3 ± 36.5 | 0.000 |
| BM blast (%) | 61.0 ± 24.7 | 46.8 ± 32.6 | 0.145 |
| BM cellularity (%) | 76.1 ± 21.5 | 67.7 ± 28.3 | 0.323 |
| CD34 (+) † | 94.4% (17/18) | 90.0% (9/10) | 1.000 |
| CD117 (+) † | 100% (21/21) | 81.8% (9/11) | 0.212 |
| FAB classification | 0.854 | ||
| M0 | 12.5% (3/24) | 15.4% (2/13) | |
| M1 | 8.3% (2/24) | 23.1% (3/13) | |
| M2 | 25.0% (6/24) | 23.1% (3/13) | |
| M4 | 29.2% (7/24) | 23.1% (3/13) | |
| M5 | 16.7% (4/24) | 15.4% (2/13) | |
| M7 | 4.2% (1/24) | 0% (0/13) | |
| Not classified | 4.2% (1/24) | 0% (0/13) | |
| Mutation profile | |||
| FLT3 | 25.0% (6/24) | 30.8% (4/13) | 1.000 |
| IDH1 | 4.3% (1/23) | 7.7% (1/13) | 1.000 |
| IDH2 | 21.7% (5/23) | 23.1% (3/13) | 1.000 |
| WT1 | 8.7% (2/23) | 7.7% (1/13) | 1.000 |
| TP53 | 4.3% (1/23) | 0% (0/13) | 1.000 |
| RUNX1 | 21.7% (5/23) | 46.2% (6/13) | 0.250 |
| ASXL1 | 0% (0/23) | 7.7% (1/13) | 0.769 |
| Pathway Name | p | q | Genes (Fold Change) | Pathway Source |
|---|---|---|---|---|
| Cytokine-cytokine receptor interaction-Homo sapiens (human) | 0.0001 | 0.0219 | IFNW1(5.37), IL5RA(6.69), TNFSF15(0.44), CSF1R(0.49), GDF6(300), IFNB1(2.73), TNFRSF11A(0.46), IL1B(2.06), CCR5(0.36) | KEGG |
| GPCR downstream signaling | 0.0020 | 0.0393 | DAGLA(0.42), OR13C4(7.33), IL5RA(6.69), PLXNB1(2.47), CCR5(0.36), OR1L8(3.82), GNG8(300), OR5B12(0.09), OR1Q1(3.10), RAMP1(5.35), OR1J1(5.49), OR1L4(2.98), GRM1(3.57), SUCNR1(2.36), RTP3(300) | Reactome |
| Signalling by GPCR | 0.0049 | 0.0499 | DAGLA(0.42), PDGFA(0.39), IL5RA(6.69), OR13C4(7.33), OR1L8(3.82), GNG8(300), RTP3(300), OR5B12(0.09), OR1Q1(3.10), PLXNB1(2.47), OR1J1(5.49), WNT10A(0.41), RAMP1(5.35), OR1L4(2.98), GRM1(3.57), SUCNR1(2.36), CCR5(0.36) | Reactome |
| Cytokines and Inflammatory Response | 0.0007 | 0.0393 | PDGFA(0.39), IL1B(2.06), IFNB1(2.73) | Wikipathways |
| GPCR signaling | 0.0022 | 0.0393 | IFNW1(5.37), PDGFA(0.39), WNT10A(0.41), GDF6(300), IFNB1(2.73), IL1B(2.06), GRM1(3.57) | INOH |
| C-type lectin receptors | 0.0020 | 0.0393 | CLEC10A(0.20), MUC16(3.07), IL1B(2.06), MUC1(2.40) | Reactome |
| JAK-STAT Molecular Variation 1 | 0.0034 | 0.0440 | IFNW1(5.37), IL1B(2.06), IL5RA(6.69), IFNB1(2.73) | INOH |
| Bone remodeling | 0.0045 | 0.0499 | TNFRSF11A(0.46), IFNB1(2.73) | Wikipathways/ BioCarta |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, C.-H.; Yun, J.W. Investigation of Biomarkers Associated with Low Platelet Counts in Normal Karyotype Acute Myeloid Leukemia. Int. J. Mol. Sci. 2022, 23, 7772. https://doi.org/10.3390/ijms23147772
Park C-H, Yun JW. Investigation of Biomarkers Associated with Low Platelet Counts in Normal Karyotype Acute Myeloid Leukemia. International Journal of Molecular Sciences. 2022; 23(14):7772. https://doi.org/10.3390/ijms23147772
Chicago/Turabian StylePark, Chang-Hun, and Jae Won Yun. 2022. "Investigation of Biomarkers Associated with Low Platelet Counts in Normal Karyotype Acute Myeloid Leukemia" International Journal of Molecular Sciences 23, no. 14: 7772. https://doi.org/10.3390/ijms23147772
APA StylePark, C.-H., & Yun, J. W. (2022). Investigation of Biomarkers Associated with Low Platelet Counts in Normal Karyotype Acute Myeloid Leukemia. International Journal of Molecular Sciences, 23(14), 7772. https://doi.org/10.3390/ijms23147772






