Microsporidia Promote Host Mitochondrial Fragmentation by Modulating DRP1 Phosphorylation
Abstract
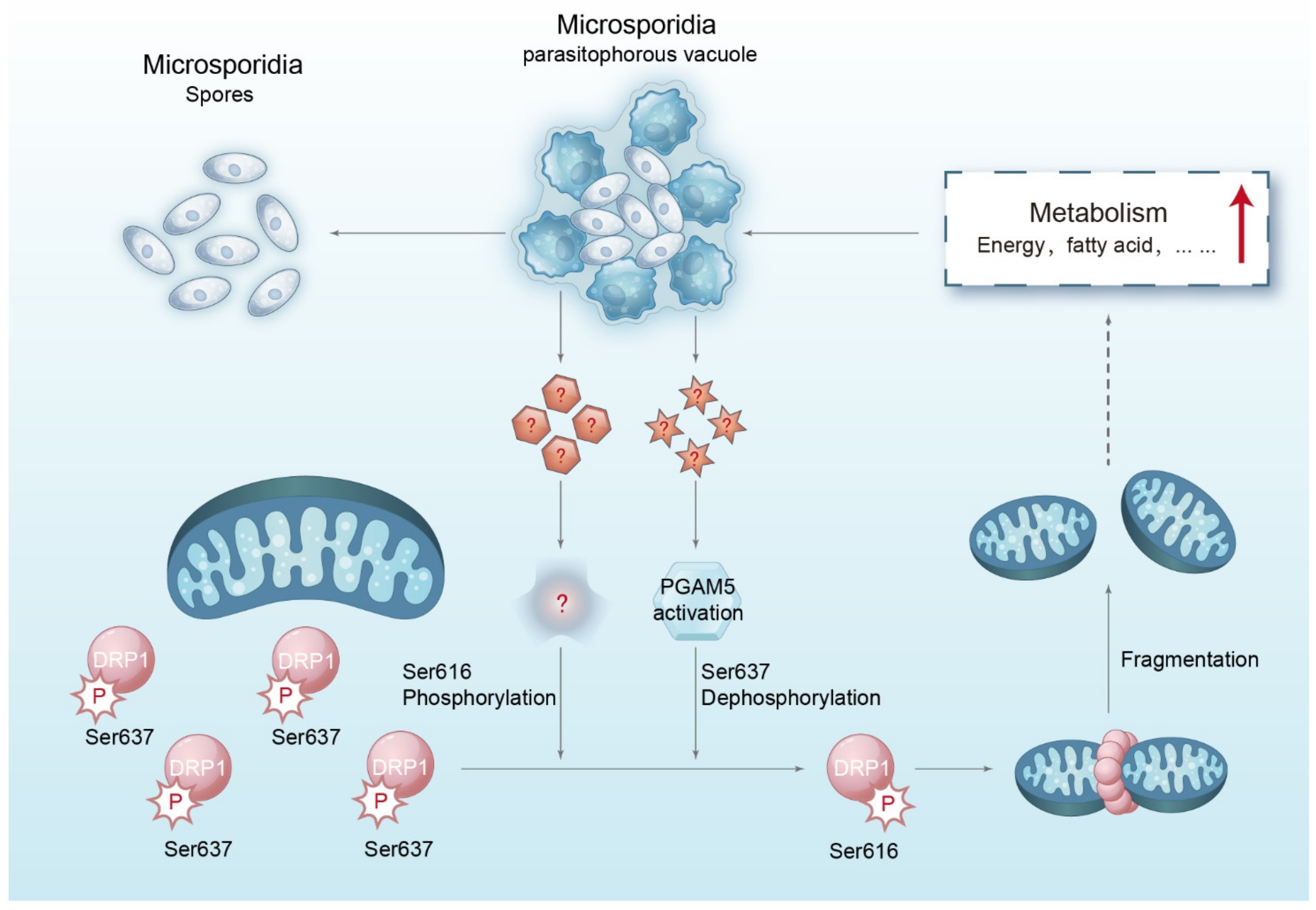
1. Introduction
2. Results
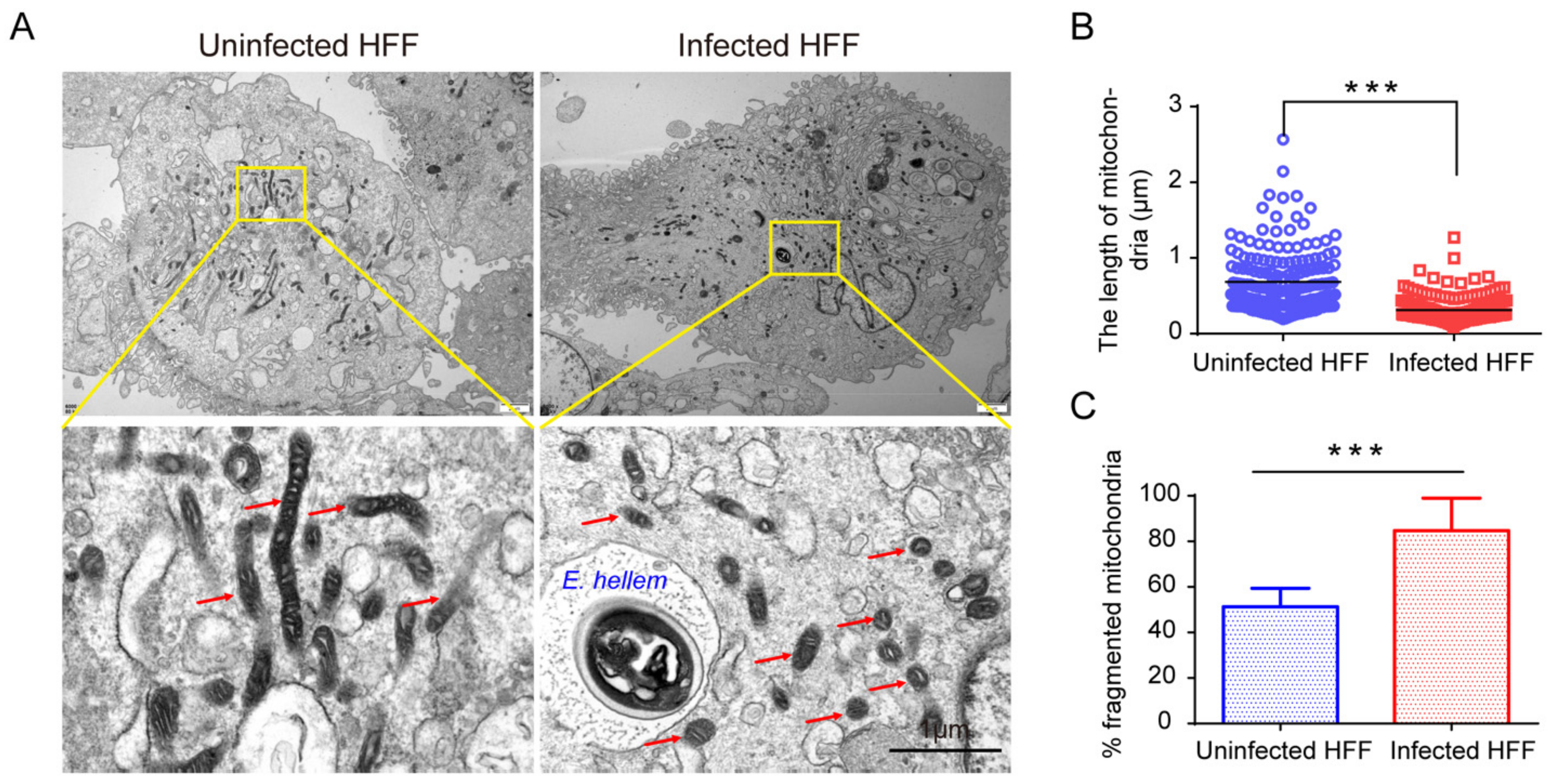
2.1. Microsporidian Infections Induced Drastic HMF in Different Cells
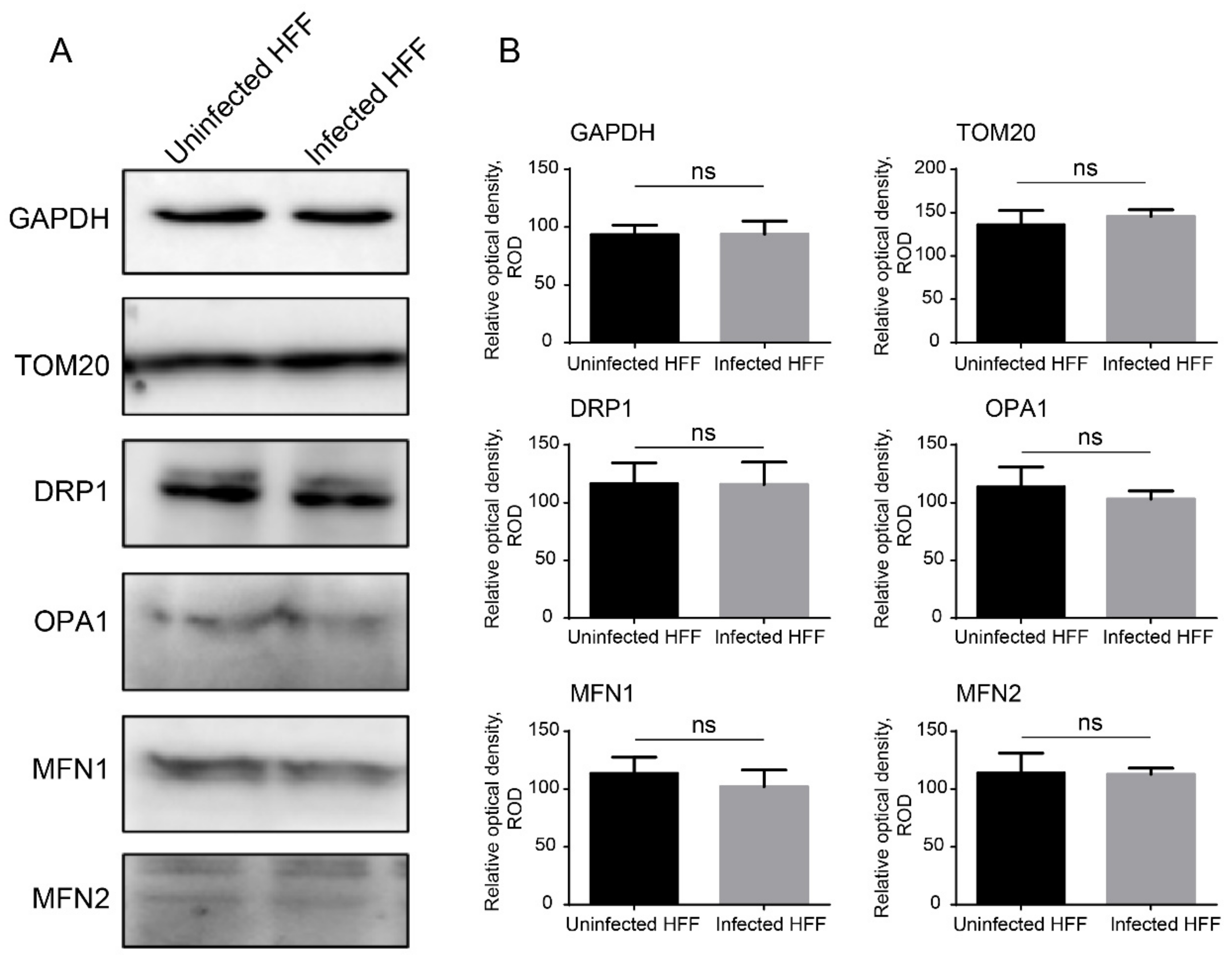
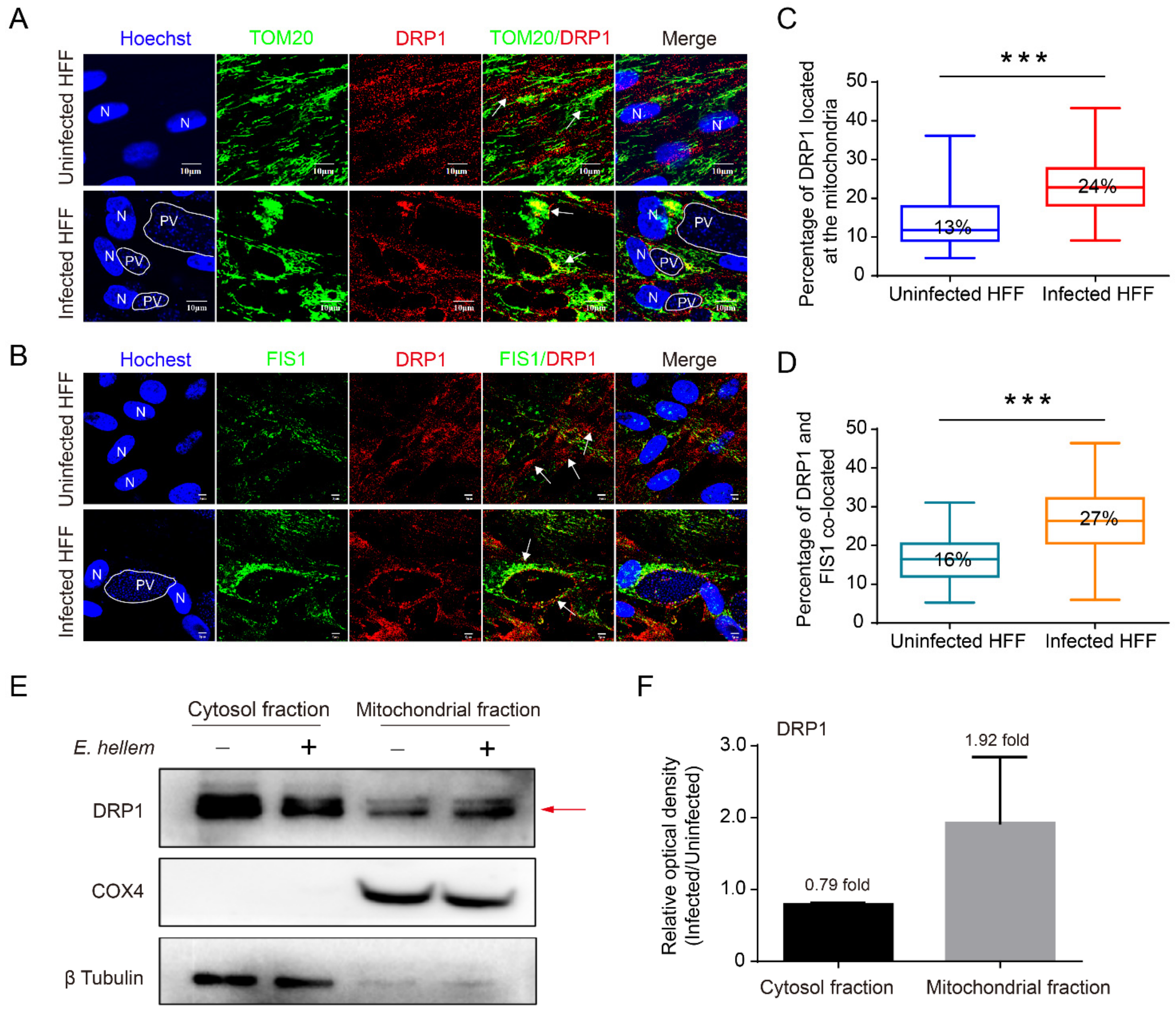
2.2. Microsporidian Infections Promoted DRP1 Translocation to Mitochondria
2.3. Microsporidia-Induced HMF Depends on DRP1 Phosphorylation
2.4. Microsporidia Infection Activated PGAM5 for Dephosphorylating the DRP1 Ser637
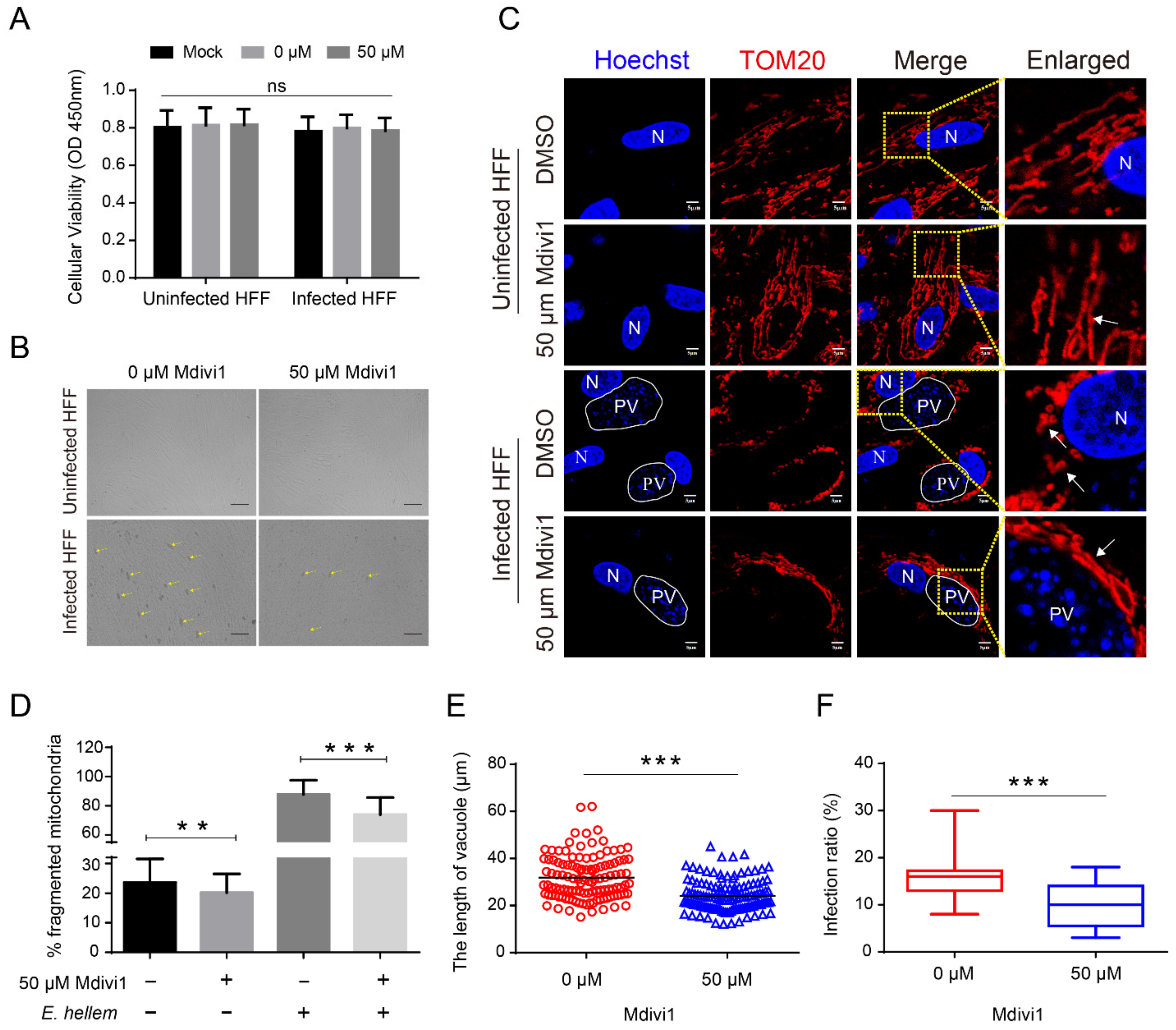
2.5. Microsporidia Proliferation Required HMF
3. Discussion
4. Materials and Methods
4.1. Cells and Reagents
4.2. E. hellem and E. cuniculi Spore Preparation and Cell Infection
4.3. Indirect Immunofluorescence Assay (IFA)
4.4. In Situ Fluorescence Hybridization (FISH)
4.5. Transmission Electron Microscopy (TEM)
4.6. Quantitative Analyses of HMF by Confocal Microscopy and TEM
4.7. Western Blot Analyses of DRP1 Expression, Phosphorylation, and PGAM5 Activation
4.8. Western Blot Analyses of DRP1 Location
4.9. Analyses of Cell Viability
4.10. RNAi of PGAM5
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desportes-Livage, I. Biology of microsporidia. Contrib. Microbiol. 2000, 6, 140–165. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L.M. Microsporidia: Emerging pathogenic protists. Acta Trop. 2001, 78, 89–102. [Google Scholar] [CrossRef]
- Anane, S.; Attouchi, H. Microsporidiosis: Epidemiology, clinical data and therapy. Gastroenterol. Clin. Et Biol. 2010, 34, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Weiss, L. Therapeutic targets for the treatment of microsporidiosis in humans. Expert Opin. Ther. Targets 2018, 22, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Gao, H.; Xu, J.; Luo, J.; Han, B.; Bao, J.; Pan, G.; Li, T.; Zhou, Z. Innate and Adaptive Immune Responses against Microsporidia Infection in Mammals. Front. Microbiol. 2020, 11, 1468. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Xu, X.; He, Q.; Li, L.; Guo, J.; Bao, J.; Pan, G.; Li, T.; Zhou, Z. The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasit. Vectors 2021, 14, 186. [Google Scholar] [CrossRef]
- Undeen, A.H.; Solter, L. Sugar Acquisition during the Development of Microsporidian (Microspora: Nosematidae) Spores. J. Invertebr. Pathol. 1997, 70, 106–112. [Google Scholar] [CrossRef]
- Katinka, M.D.; Duprat, S.; Cornillot, E.; Méténier, G.; Thomarat, F.; Prensier, G.; Barbe, V.; Peyretaillade, E.; Brottier, P.; Wincker, P. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414, 450–453. [Google Scholar] [CrossRef]
- Hoch, G.; Schafellner, C.; Henn, M.W.; Schopf, A. Alterations in carbohydrate and fatty acid levels of Lymantria dispar larvae caused by a microsporidian infection and potential adverse effects on a co-occurring endoparasitoid, Glyptapanteles liparidis. Arch. Insect Biochem. Physiol. 2002, 50, 109–120. [Google Scholar] [CrossRef]
- Medeiros, T.; Mehra, C.; Pernas, L. Contact and competition between mitochondria and microbes. Curr. Opin. Microbiol. 2021, 63, 189–194. [Google Scholar] [CrossRef]
- Terry, R.S.; AM, D.; JE, S. Segregation of a microsporidian parasite during host cell mitosis. Parasitology 1999, 118, 43–48. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hacker, C.; Howell, M.; Bhella, D.; Lucocq, J. Strategies for maximizing ATP supply in the microsporidian Encephalitozoon cuniculi: Direct binding of mitochondria to the parasitophorous vacuole and clustering of the mitochondrial porin VDAC. Cell. Microbiol. 2014, 16, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Kyei-Poku, G.; Sokolova, Y. The microsporidium Nosema disstriae (Thomson 1959): Fine structure and phylogenetic position within the N. bombycis clade. J. Invertebr. Pathol. 2017, 143, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, Y.; Tu, V.; Tomita, T.; Mayoral, J.; Williams, T.; Horta, A.; Huang, H.; Weiss, L.M. Microsporidia Interact with Host Cell Mitochondria via Voltage-Dependent Anion Channels Using Sporoplasm Surface Protein 1. mBio 2019, 10, e01944-19. [Google Scholar] [CrossRef]
- Ding, Z.; Pan, J.; Huang, H.; Jiang, G.; Chen, J.; Zhu, X.; Wang, R.; Xu, G. An integrated metabolic consequence of Hepatospora eriocheir infection in the Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2018, 72, 443–451. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Wang, L.; Zhou, Z. Molecular and biochemical responses in the midgut of the silkworm, Bombyx mori, infected with Nosema bombycis. Parasites Vectors 2018, 11, 147. [Google Scholar] [CrossRef]
- He, X.; He, X.; Liu, H.; Li, M.; Cai, S.; Fu, Z.; Lu, X. Proteomic analysis of BmN cells (Bombyx mori) in response to infection with Nosema bombycis. Acta Biochim. Et Biophys. Sin. 2014, 46, 982–990. [Google Scholar] [CrossRef]
- Labbé, K.; Murley, A.; Nunnari, J. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 2014, 30, 357–391. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Kleele, T.; Rey, T.; Winter, J.; Zaganelli, S.; Mahecic, D.; Perreten Lambert, H.; Ruberto, F.; Nemir, M.; Wai, T.; Pedrazzini, T.; et al. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 2021, 593, 435–439. [Google Scholar] [CrossRef]
- Chang, C.; Blackstone, C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 2010, 1201, 34–39. [Google Scholar] [CrossRef]
- Sharp, W.W.; Fang, Y.H.; Han, M.; Zhang, H.J.; Hong, Z.; Banathy, A.; Morrow, E.; Ryan, J.J.; Archer, S.L. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: Therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J. 2014, 28, 316. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, H.; Chen, S.; Du, F.; Wang, X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012, 148, 228–243. [Google Scholar] [CrossRef]
- Yu, B.; Ma, J.; Li, J.; Wang, D.; Wang, Z.; Wang, S. Mitochondrial phosphatase PGAM5 modulates cellular senescence by regulating mitochondrial dynamics. Nat. Commun. 2020, 11, 2549. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Tan, Y.; Du, W.; Li, Y.; Toan, S.; Mui, D.; Tian, F.; Zhou, H. Phosphoglycerate mutase 5 exacerbates cardiac ischemia-reperfusion injury through disrupting mitochondrial quality control. Redox Biol. 2021, 38, 101777. [Google Scholar] [CrossRef] [PubMed]
- Losón, O.; Song, Z.; Chen, H.; Chan, D. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 2013, 24, 659–667. [Google Scholar] [CrossRef]
- Escoll, P.; Song, O.; Viana, F.; Steiner, B.; Lagache, T.; Olivomarin, J.; Impens, F.; Brodin, P.; Hilbi, H.; Buchrieser, C. Legionella pneumophila Modulates Mitochondrial Dynamics to Trigger Metabolic Repurposing of Infected Macrophages. Cell Host Microbe 2017, 22, 302. [Google Scholar] [CrossRef]
- Lobet, E.; Willemart, K.; Ninane, N.L.; Demazy, C.; Sedzicki, J.; Lelubre, C.; De Bolle, X.; Renard, P.; Raes, M.; Dehio, C. Mitochondrial fragmentation affects neither the sensitivity to TNFα-induced apoptosis of Brucella-infected cells nor the intracellular replication of the bacteria. Sci. Rep. 2018, 8, 5173. [Google Scholar] [CrossRef]
- Aguilar-López, B.; Correa, F.; Moreno-Altamirano, M.; Espitia, C.; Hernández-Longoria, R.; Oliva-Ramírez, J.; Padierna-Olivos, J.; Sánchez-García, F. LprG and PE_PGRS33 Mycobacterium tuberculosis virulence factors induce differential mitochondrial dynamics in macrophages. Scand. J. Immunol. 2019, 89, e12728. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Itoh, R.; Shimizu, A.; Walenna, N.; Chou, B.; Ishii, K.; Soejima, T.; Fujikane, A.; Hiromatsu, K. Chlamydia trachomatis targets mitochondrial dynamics to promote intracellular survival and proliferation. Cell. Microbiol. 2019, 21, e12962. [Google Scholar] [CrossRef]
- Jain, P.; Luo, Z.Q.; BIanke, S.R. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc. Natl. Acad. Sci. USA 2011, 108, 16032–16037. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.; Pypaert, M.; Flavell, R.; Galán, J. A Salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 2003, 163, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Chan, D. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012, 46, 265–287. [Google Scholar] [CrossRef]
- Cereghetti, G.; Stangherlin, A.; Martins de Brito, O.; Chang, C.; Blackstone, C.; Bernardi, P.; Scorrano, L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. USA 2008, 105, 15803–15808. [Google Scholar] [CrossRef] [PubMed]
- Wikstrom, J.; Mahdaviani, K.; Liesa, M.; Sereda, S.; Si, Y.; Las, G.; Twig, G.; Petrovic, N.; Zingaretti, C.; Graham, A.; et al. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO J. 2014, 33, 418–436. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; He, Q.; Xu, J.; Xu, C.; Han, Y.; Gao, H.; Meng, X.; Pan, G.; Li, T.; Zhou, Z. Microsporidia infection upregulates host energy metabolism but maintains ATP homeostasis. J. Invertebr. Pathol. 2021, 186, 107596. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.; Gaume, B.; Bergmann-Leitner, E.; Leitner, W.; Robert, E.; Catez, F.; Smith, C.; Youle, R. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 2001, 1, 515–525. [Google Scholar] [CrossRef]
- del Aguila, C.; Izquierdo, F.; Granja, A.; Hurtado, C.; Fenoy, S.; Fresno, M.; Revilla, Y. Encephalitozoon microsporidia modulates p53-mediated apoptosis in infected cells. Int. J. Parasitol. 2006, 36, 869–876. [Google Scholar] [CrossRef]
- Nevárez-Garza, A.M.; Castillo-Velázquez, U.; Soto-Domínguez, A.; Montes-de-Oca-Luna, R.; Zamora-Ávila, D.E.; Wong-González, A.; Rodríguez-Tovar, L.E. Quantitative analysis of TNF-α, IL-4, and IL-10 expression, nitric oxide response, and apoptosis in Encephalitozoon cuniculi-infected rabbits. Dev. Comp. Immunol. 2018, 81, 235–243. [Google Scholar] [CrossRef]
- Sokolova, Y.Y.; Bowers, L.; Alvarez, X.; Didier, E. Encephalitozoon cuniculi and Vittaforma corneae (Phylum Microsporidia) inhibit staurosporine-induced apoptosis in human THP-1 macrophages in vitro. Parasitology 2019, 146, 569–579. [Google Scholar] [CrossRef]
- Çetinkaya, Ü.; Caner, A.; Charyyeva, A.; Şentürk, M.; Eren, M. Encephalitozoon intestinalis Infection Impacts the Expression of Apoptosis-Related Genes in U937 Macrophage Cells. Acta Parasitol. 2021, 66, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Gao, H.; Xu, J.; Luo, J.; Chen, J.; Han, B.; Li, T.; Zhou, Z. Microsporidian Encephalitozoon hellem secretes EhPTP4 to regulate host endoplasmic reticulum-associated degradation. Acta Microbiol. Sin. 2022, 61, 357–373. [Google Scholar] [CrossRef]
- Hester, J.; Lindquist, H.; Bobst, A.; Schaefer, F. Fluorescent in situ detection of Encephalitozoon hellem spores with a 6-carboxyfluorescein-labeled ribosomal RNA-targeted oligonucleotide probe. J. Eukaryot. Microbiol. 2000, 47, 299–308. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Johansson, M.A.; Leena, T.; Visvesvara, G.S.; Moura, L.S.; Dasilva, A.J.; Girouard, A.S.; Olga, M. Retrospective species identification of microsporidian spores in diarrheic fecal samples from human immunodeficiency virus/AIDS patients by multiplexed fluorescence in situ hybridization. J. Clin. Microbiol. 2007, 45, 1255–1260. [Google Scholar] [CrossRef]
- Troemel, E.R.; Felix, M.A.; Whiteman, N.K.; Barriere, A.; Ausubel, F.M. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008, 6, e309. [Google Scholar] [CrossRef]
- Otera, H.; Miyata, N.; Kuge, O.; Mihara, K. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 2016, 212, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Choi, S.E.; Koh, H.C. PGAM5 regulates PINK1/Parkin-mediated mitophagy via DRP1 in CCCP-induced mitochondrial dysfunction. Toxicol. Lett. 2018, 284, 120–128. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Xu, J.; Xie, C.; Zhao, Z.; Guo, J.; Wen, Y.; Li, T.; Zhou, Z. Microsporidia Promote Host Mitochondrial Fragmentation by Modulating DRP1 Phosphorylation. Int. J. Mol. Sci. 2022, 23, 7746. https://doi.org/10.3390/ijms23147746
Luo J, Xu J, Xie C, Zhao Z, Guo J, Wen Y, Li T, Zhou Z. Microsporidia Promote Host Mitochondrial Fragmentation by Modulating DRP1 Phosphorylation. International Journal of Molecular Sciences. 2022; 23(14):7746. https://doi.org/10.3390/ijms23147746
Chicago/Turabian StyleLuo, Jian, Jinzhi Xu, Chaolu Xie, Zuoming Zhao, Junrui Guo, Yuan Wen, Tian Li, and Zeyang Zhou. 2022. "Microsporidia Promote Host Mitochondrial Fragmentation by Modulating DRP1 Phosphorylation" International Journal of Molecular Sciences 23, no. 14: 7746. https://doi.org/10.3390/ijms23147746
APA StyleLuo, J., Xu, J., Xie, C., Zhao, Z., Guo, J., Wen, Y., Li, T., & Zhou, Z. (2022). Microsporidia Promote Host Mitochondrial Fragmentation by Modulating DRP1 Phosphorylation. International Journal of Molecular Sciences, 23(14), 7746. https://doi.org/10.3390/ijms23147746





