Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA
Abstract
1. Introduction
2. Results
2.1. Ino4p Is Required for the Stability of Ino2p
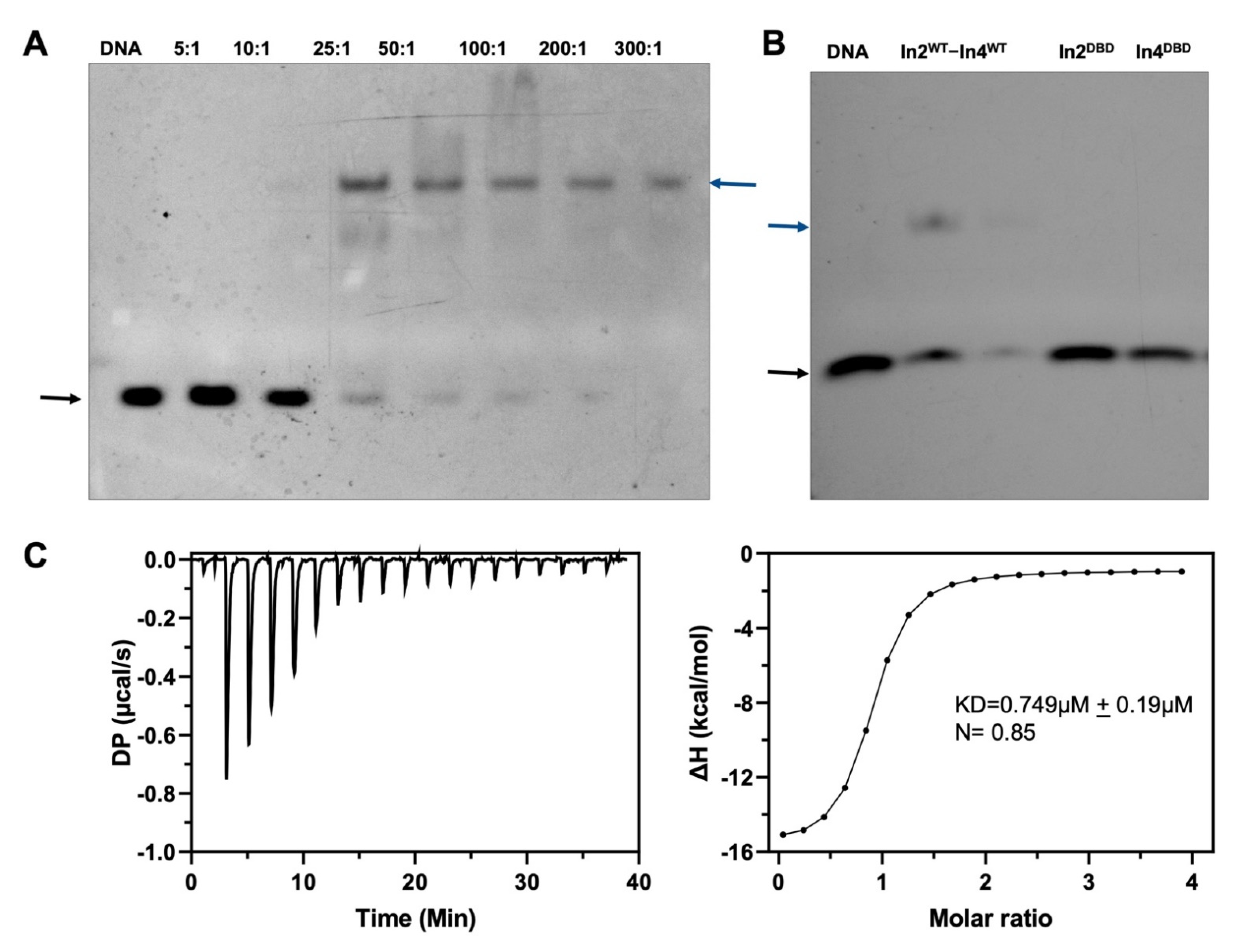
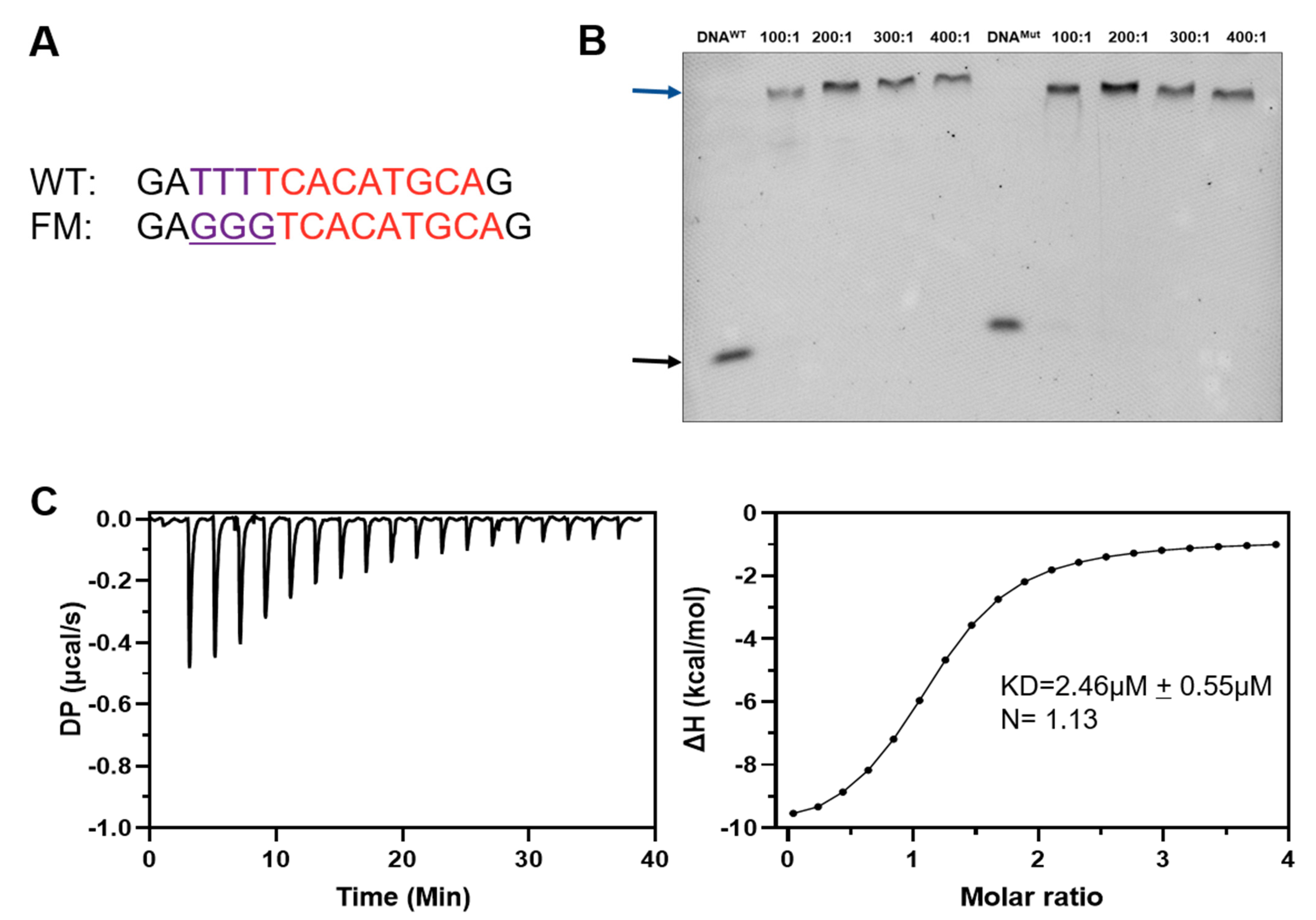
2.2. Ino2p and Ino4p Are Essential for Simultaneous Binding to Promoter DNA
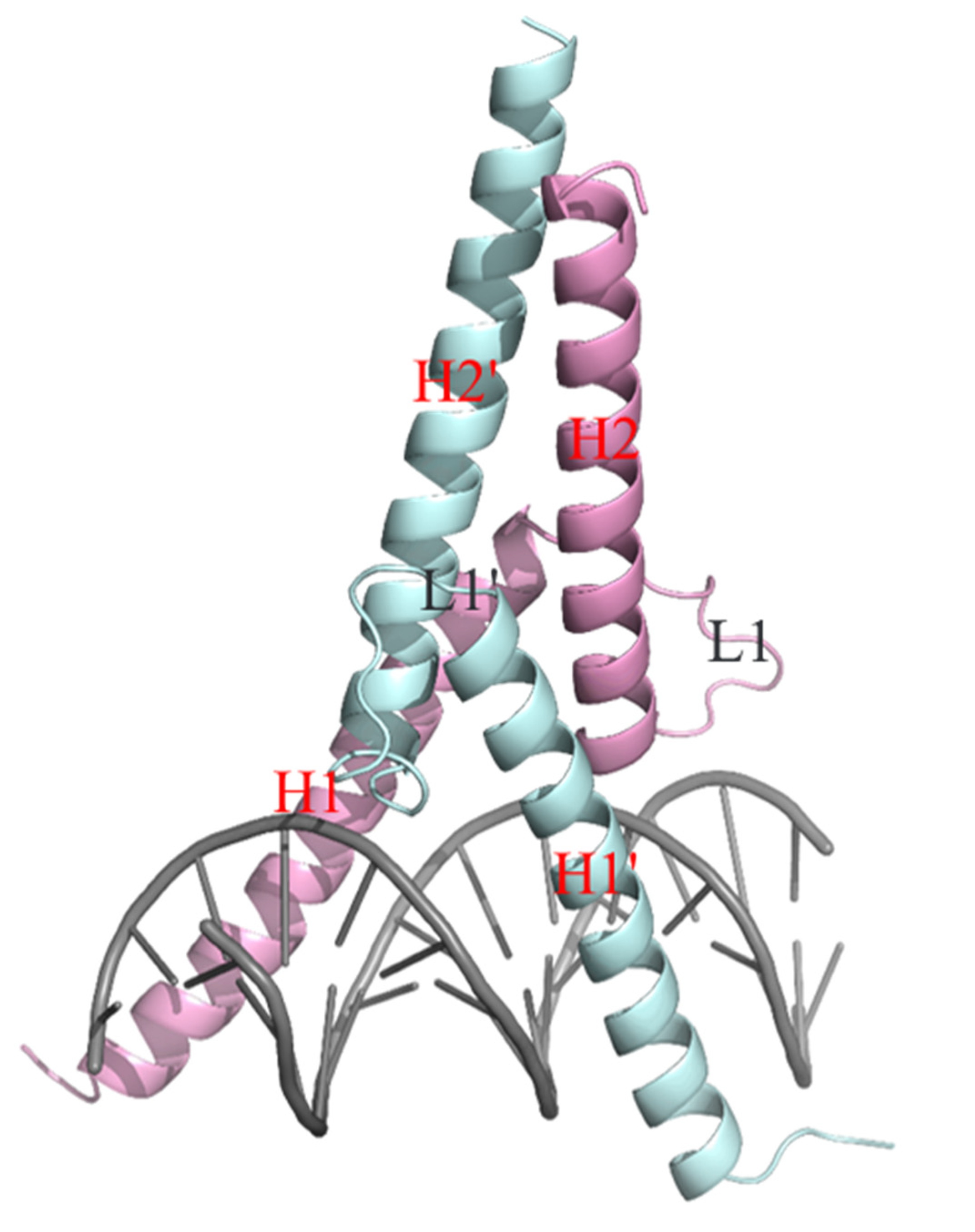
2.3. Structure of Ino2pDBD/Ino4pDBD/DNA Ternary Complex
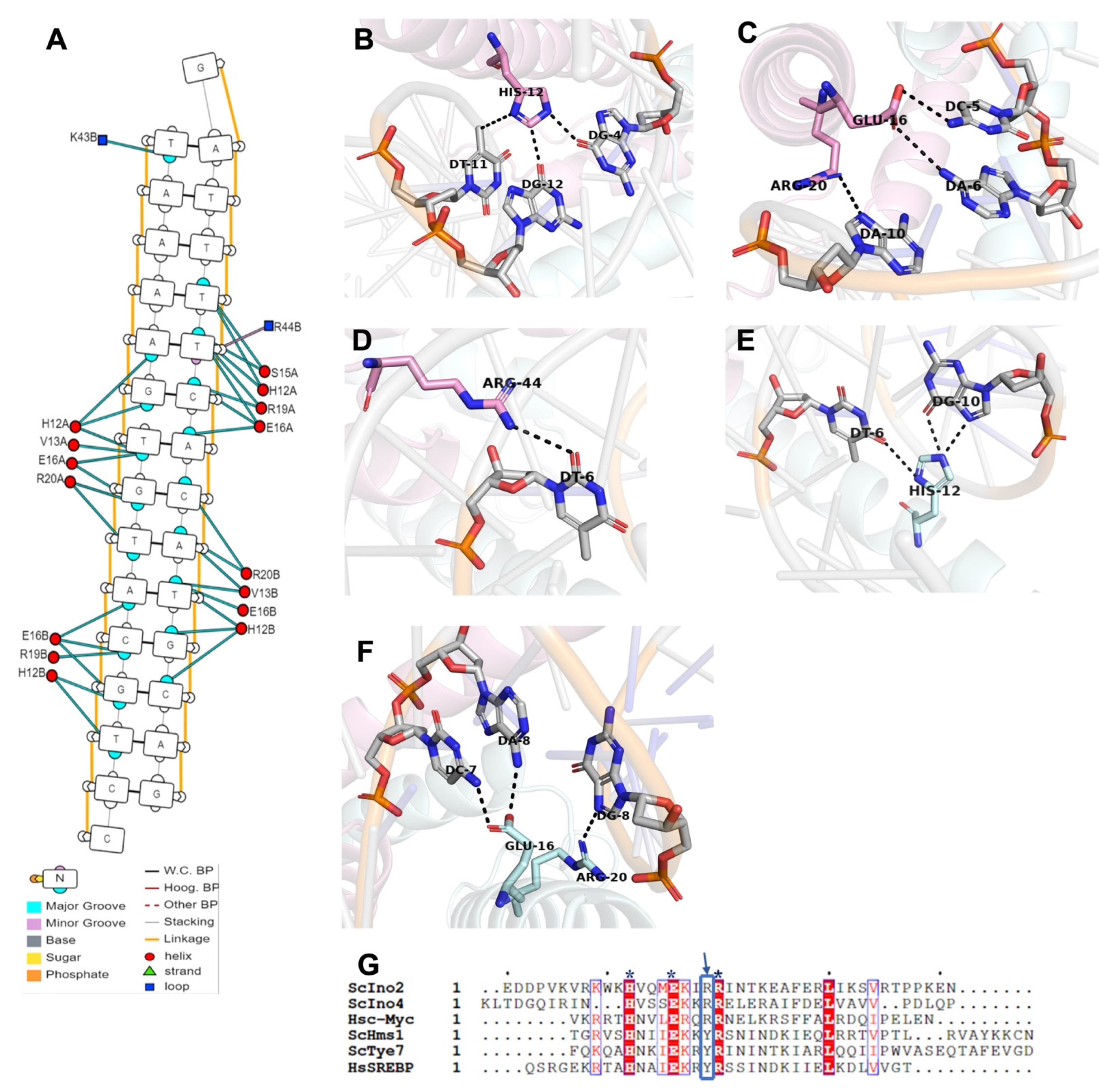
2.4. Structural Basis for the Recognition and Binding of Promoter DNA by Ino2p/Ino4p Heterodimer
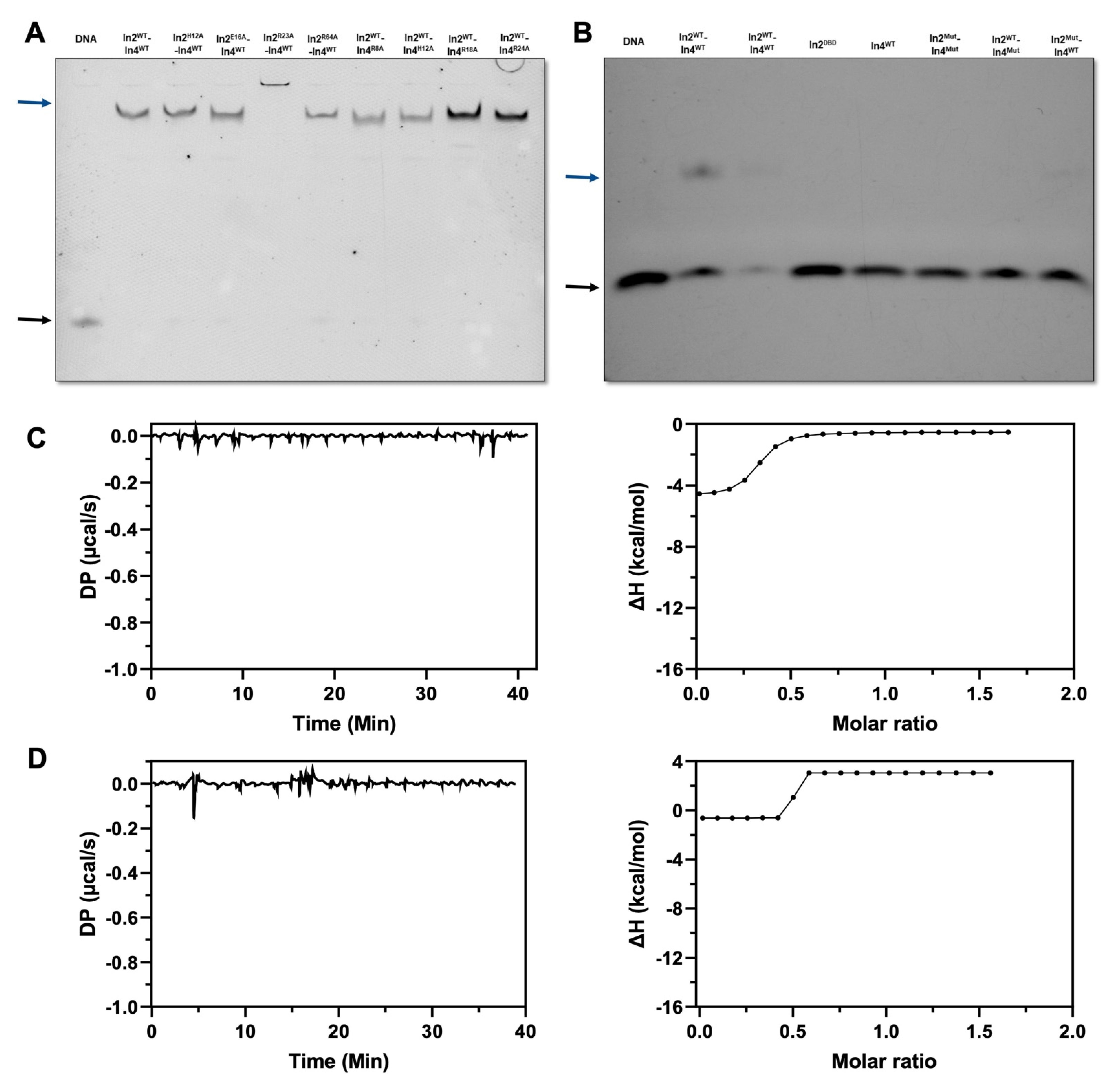
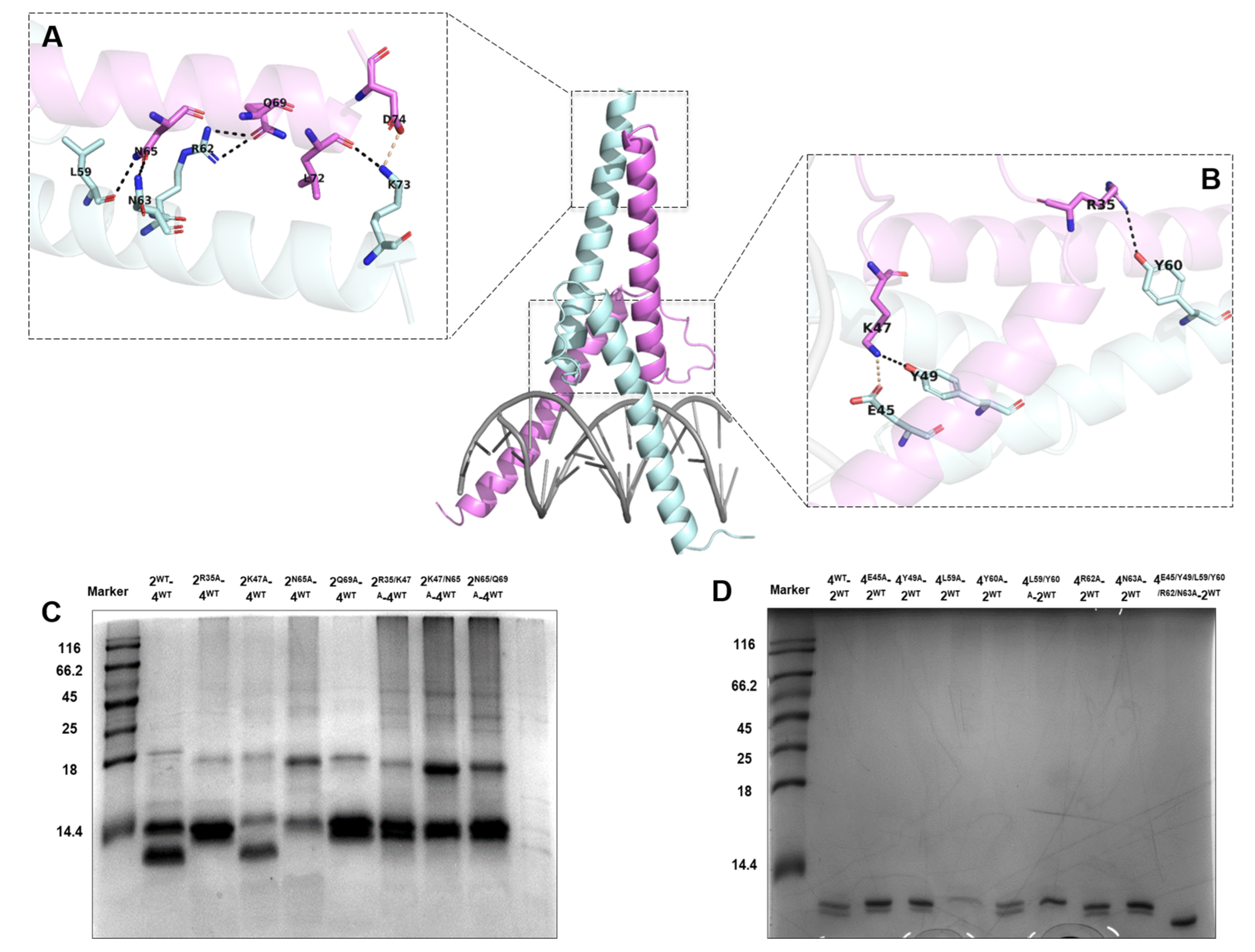
2.5. Mutual Interactions of Ino2pDBD and Ino4pDBD
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Protein Expression and Purification
4.3. Protein Crystallography
4.4. Data Collection, Structure Determination, and Refinement
4.5. Electrophoretic Shift Mobility Assay (EMSA)
4.6. Isothermal Calorimetry (ITC)
4.7. In Vitro Binding Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chumnanpuen, P.; Nookaew, I.; Nielsen, J. Integrated analysis, transcriptome-lipidome, reveals the effects of INO-level (INO2 and INO4) on lipid metabolism in yeast. BMC Syst. Biol. 2013, 7 (Suppl. 3), S7. [Google Scholar] [CrossRef] [PubMed]
- Carman, G.M.; Henry, S.A. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 1999, 38, 361–399. [Google Scholar] [CrossRef]
- Henry, S.A.; Kohlwein, S.D.; Carman, G.M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 2012, 190, 317–349. [Google Scholar] [CrossRef] [PubMed]
- Santiago, T.C.; Mamoun, C.B. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 2003, 278, 38723–38730. [Google Scholar] [CrossRef]
- Cok, S.J.; Martin, C.G.; Gordon, J.I. Transcription of INO2 and INO4 is regulated by the state of protein N-myristoylation in Saccharomyces cerevisiae. Nucleic Acids Res. 1998, 26, 2865–2872. [Google Scholar] [CrossRef][Green Version]
- Li, Z.; Brendel, M.J.M.; Genetics, G. Co-regulation with genes of phospholipid biosynthesis of the CTR/HNM1-encoded choline/nitrogen mustard permease in Saccharomyces cerevisiae. Mol. Gen. Genet. MGG 1993, 241, 680–684. [Google Scholar] [CrossRef]
- Nikawa, J.I.; Hosaka, K.J.M.M. Isolation and characterization of genes that promote the expression of inositol transporter gene ITR1 in Saccharomyces cerevisiae. Mol. Microbiol. 2010, 16, 301–308. [Google Scholar] [CrossRef]
- Dietz, M.; Heyken, W.T.; Hoppen, J.; Geburtig, S.; Schüller, H.J. TFIIB and subunits of the SAGA complex are involved in transcriptional activation of phospholipid biosynthetic genes by the regulatory protein Ino2 in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 2003, 48, 1119–1130. [Google Scholar] [CrossRef]
- Ye, C.; Bandara, W.M.; Greenberg, M.L. Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 24898–24908. [Google Scholar] [CrossRef]
- Carman, G.M.; Han, G.S. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 2011, 80, 859–883. [Google Scholar] [CrossRef]
- Dong, H.; Yu, D.; Wang, B.; Pan, L. Identification and Characterization of a Novel Basic Helix-Loop-Helix Transcription Factor of Phospholipid Synthesis Regulation in Aspergillus niger. Front. Microbiol. 2020, 10, 2985. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.A.; Lopes, J.M. Survey and Summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 2000, 28, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.; Henry, S.A. Regulation of the yeast INO1 gene: The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics 2000, 154, 1485–1495. [Google Scholar] [CrossRef] [PubMed]
- Heyken, W.T.; Repenning, A.; Kumme, J.; Schüller, H.J. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol. Microbiol. 2010, 56, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Ambroziak, J.; Henry, S.A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 1994, 269, 15344–15349. [Google Scholar] [CrossRef]
- Kumme, J.; Dietz, M.; Wagner, C.; Schüller, H.J. Dimerization of yeast transcription factors INO2 and INO4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor. Curr. Genet. 2008, 54, 35–45. [Google Scholar] [CrossRef]
- Dettmann, A.; Jäschke, Y.; Triebel, I.; Bogs, J.; Schröder, I.; Schüller, H.J. Mediator subunits and histone methyltransferase Set2 contribute to Ino2-dependent transcriptional activation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol. Genet. Genom. 2010, 283, 211–221. [Google Scholar] [CrossRef]
- Ashburner, B.P.; Lopes, J.M. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol. Cell. Biol. 1995, 15, 1709–1715. [Google Scholar] [CrossRef][Green Version]
- Ji, X.; Wang, L.; Zang, D.; Wang, Y. Transcription Factor-Centered Yeast One-Hybrid Assay. Methods Mol. Biol. 2018, 1794, 183–194. [Google Scholar]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Xue, L.; Yue, J.; Ke, J.; Khan, M.H.; Wen, W.; Sun, B.; Zhu, Z.; Niu, L. Distinct oligomeric structures of the YoeB-YefM complex provide insights into the conditional cooperativity of type II toxin-antitoxin system. Nucleic Acids Res. 2020, 48, 10527–10541. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Khan, M.H.; Yue, J.; Zhu, Z.; Niu, L. The two paralogous copies of the YoeB-YefM toxin-antitoxin module in Staphylococcus aureus differ in DNA binding and recognition patterns. J. Biol. Chem. 2022, 298, 101457. [Google Scholar] [CrossRef]
- Bouhlel, M.A.; Lambert, M.; David-Cordonnier, M.H. Targeting Transcription Factor Binding to DNA by Competing with DNA Binders as an Approach for Controlling Gene Expression. Curr. Top. Med. Chem. 2015, 15, 1323–1358. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66 Pt 1, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.F.; Xu, Y.P.; Li, L.F.; Su, X.D. Crystal Structure of Tetrameric Arabidopsis MYC2 Reveals the Mechanism of Enhanced Interaction with DNA. Cell Rep. 2017, 19, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Zwart, P.H.; Afonine, P.V.; Grosse-Kunstleve, R.W.; Hung, L.W.; Ioerger, T.R.; McCoy, A.J.; McKee, E.; Moriarty, N.W.; Read, R.J.; Sacchettini, J.C.; et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008, 426, 419–435. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Laskowski, R.A.; Macarthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Delano, W.L. PyMOL: An Open-Source Molecular Graphics Tool. Protein Cryst. 2002, 40, 82–92. [Google Scholar]
| Data Set | Ino2pDBD/Ino4pDBD/DNA |
|---|---|
| Data collection | |
| Beamline | BL18U |
| Wavelength (Å) | 0.97847 |
| Space group | P3121 |
| Unit cell parameters | |
| a, b, c (Å) | 97.89, 97.89, 89.78 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution range (Å) | 48.95–2.25 (2.35–2.25) |
| Subunit in an asymmetric unit | 1 |
| Unique reflections | 23,995 (2177) |
| Average redundancy | 19.4 (19.0) |
| Completeness (%) | 100 (99.9) |
| Rmerge a | 0.073 (1.592) |
| I/σ(I) | 30.3 (2.4) |
| Refinement Statistics | |
| Resolution range (Å) | 48.95–2.25 |
| Rfactor (%) b | 21.45 |
| Rfree (%) c | 23.66 |
| RMSD bond lengths (Å) | 0.01 |
| RMSD bond angles (°) | 1.11 |
| Average B factors (Å2) | |
| Ino2p | 52.82 |
| Ino4p | 60.16 |
| DNA d | 51.18 |
| Water | 60.2 |
| Ramachandran plot e | |
| Favored (%) | 98.6 |
| Allowed (%) | 1.4 |
| Outliers (%) | 0 |
| PDB entry | 7XQ5 |
| Protein | Promoter DNA | N (Site) | Kd (µM) | ΔH (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| Ino2pWT/Ino4pWT | Native | 0.939 | 0.749 | −14.9 | −7.91 |
| Ino2pWT/Ino4pWT | Flanking mutant | 1.13 | 2.46 | −9.67 | −7.27 |
| Ino2pMut/Ino4pWT | Native | NB | |||
| Ino2pWT/Ino4pMut | Native | NB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.H.; Xue, L.; Yue, J.; Schüller, H.-J.; Zhu, Z.; Niu, L. Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA. Int. J. Mol. Sci. 2022, 23, 7600. https://doi.org/10.3390/ijms23147600
Khan MH, Xue L, Yue J, Schüller H-J, Zhu Z, Niu L. Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA. International Journal of Molecular Sciences. 2022; 23(14):7600. https://doi.org/10.3390/ijms23147600
Chicago/Turabian StyleKhan, Muhammad Hidayatullah, Lu Xue, Jian Yue, Hans-Joachim Schüller, Zhongliang Zhu, and Liwen Niu. 2022. "Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA" International Journal of Molecular Sciences 23, no. 14: 7600. https://doi.org/10.3390/ijms23147600
APA StyleKhan, M. H., Xue, L., Yue, J., Schüller, H.-J., Zhu, Z., & Niu, L. (2022). Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA. International Journal of Molecular Sciences, 23(14), 7600. https://doi.org/10.3390/ijms23147600






