LncRNA-Mediated Adipogenesis in Different Adipocytes
Abstract
1. Introduction
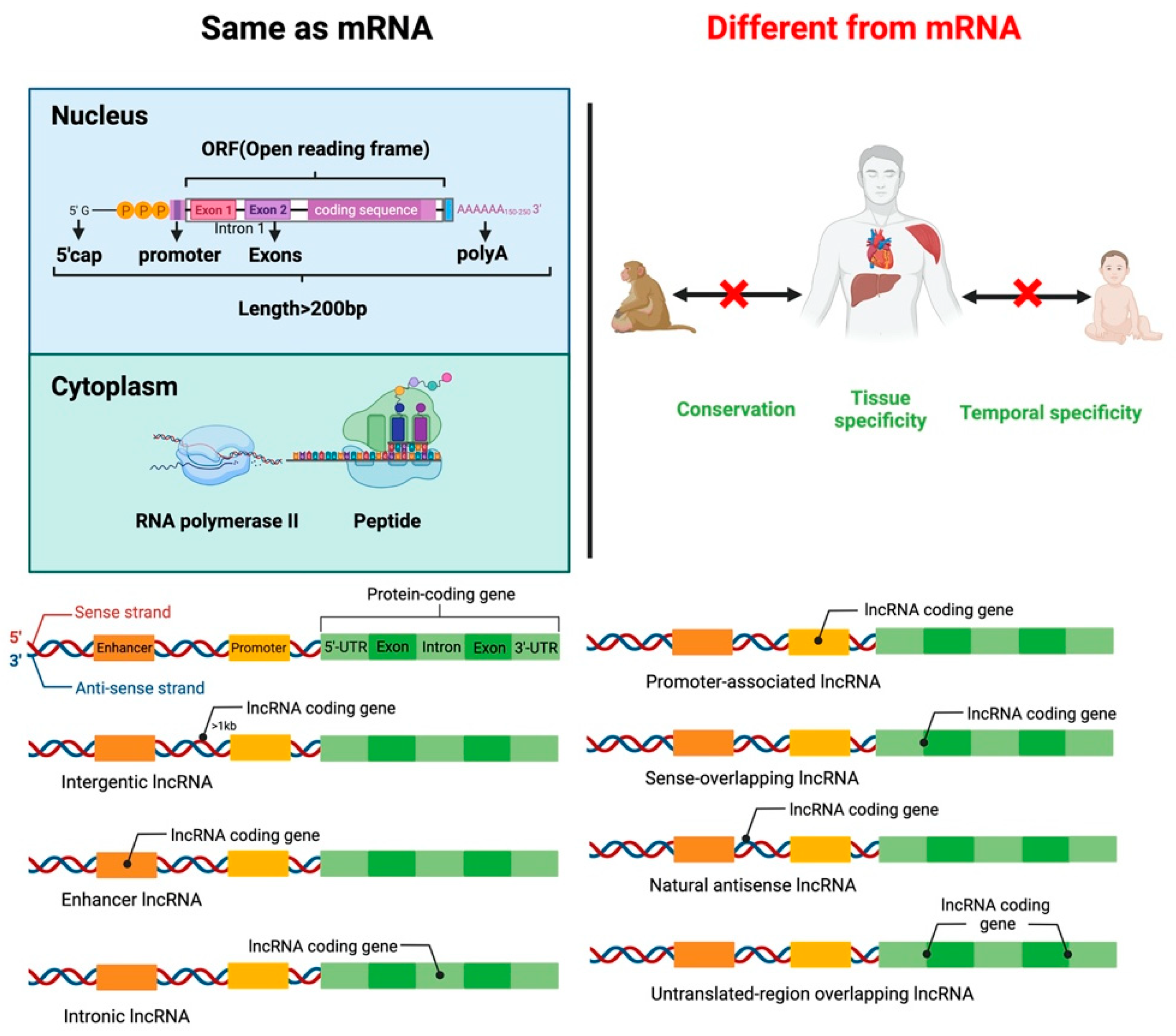
2. LncRNA
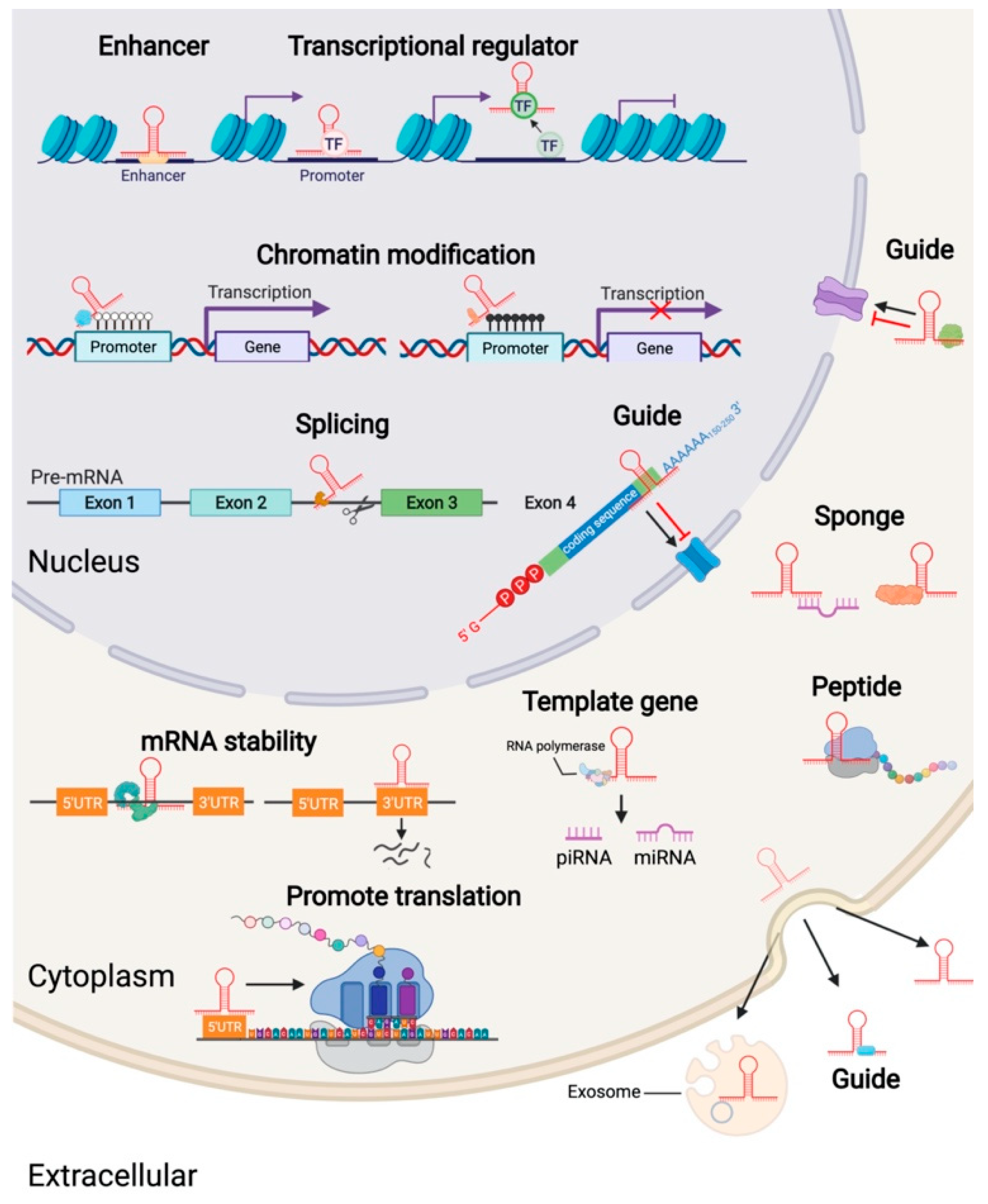
3. The Mechanism of Action of LncRNA
4. Types and Functions of Adipose Tissue
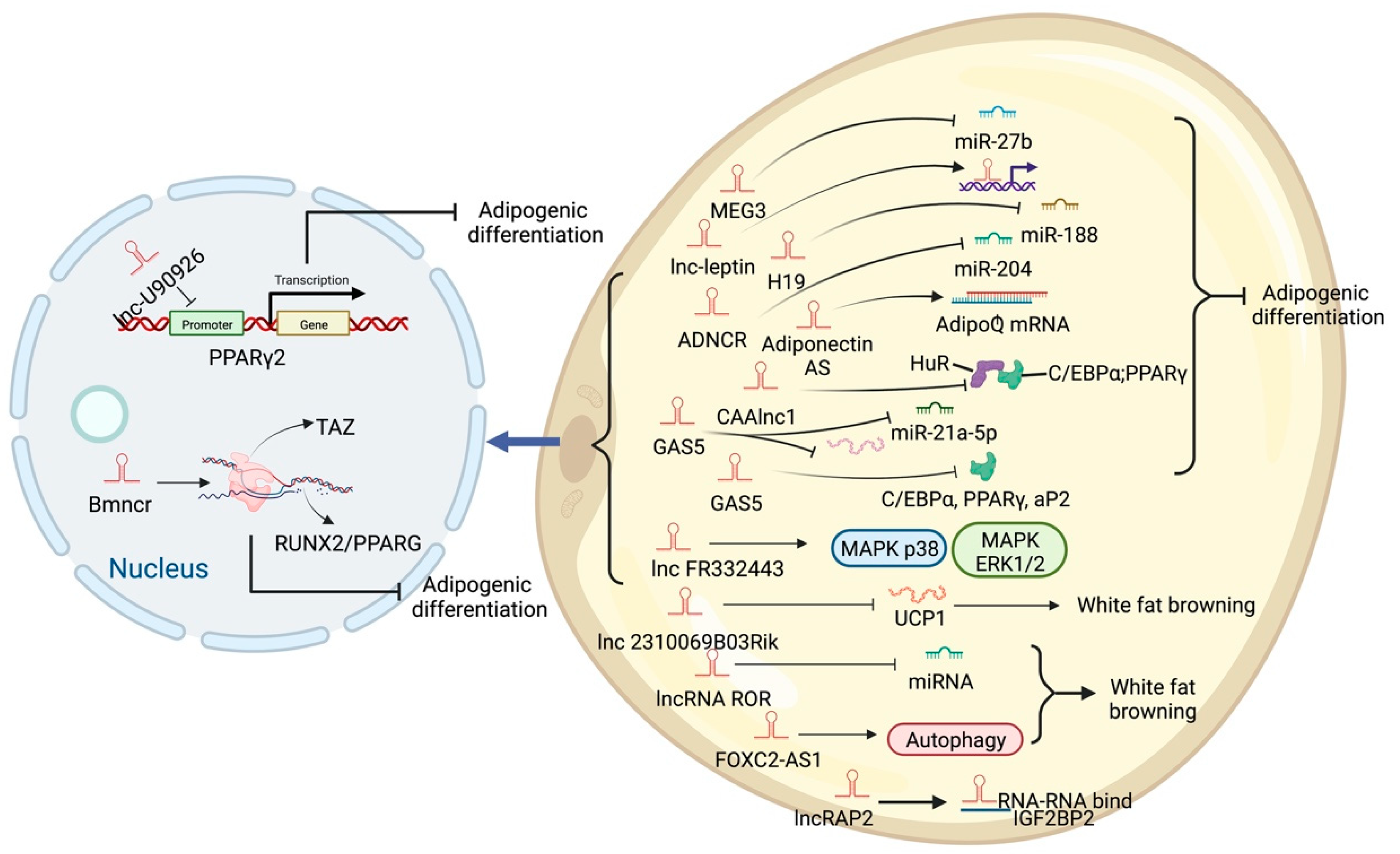
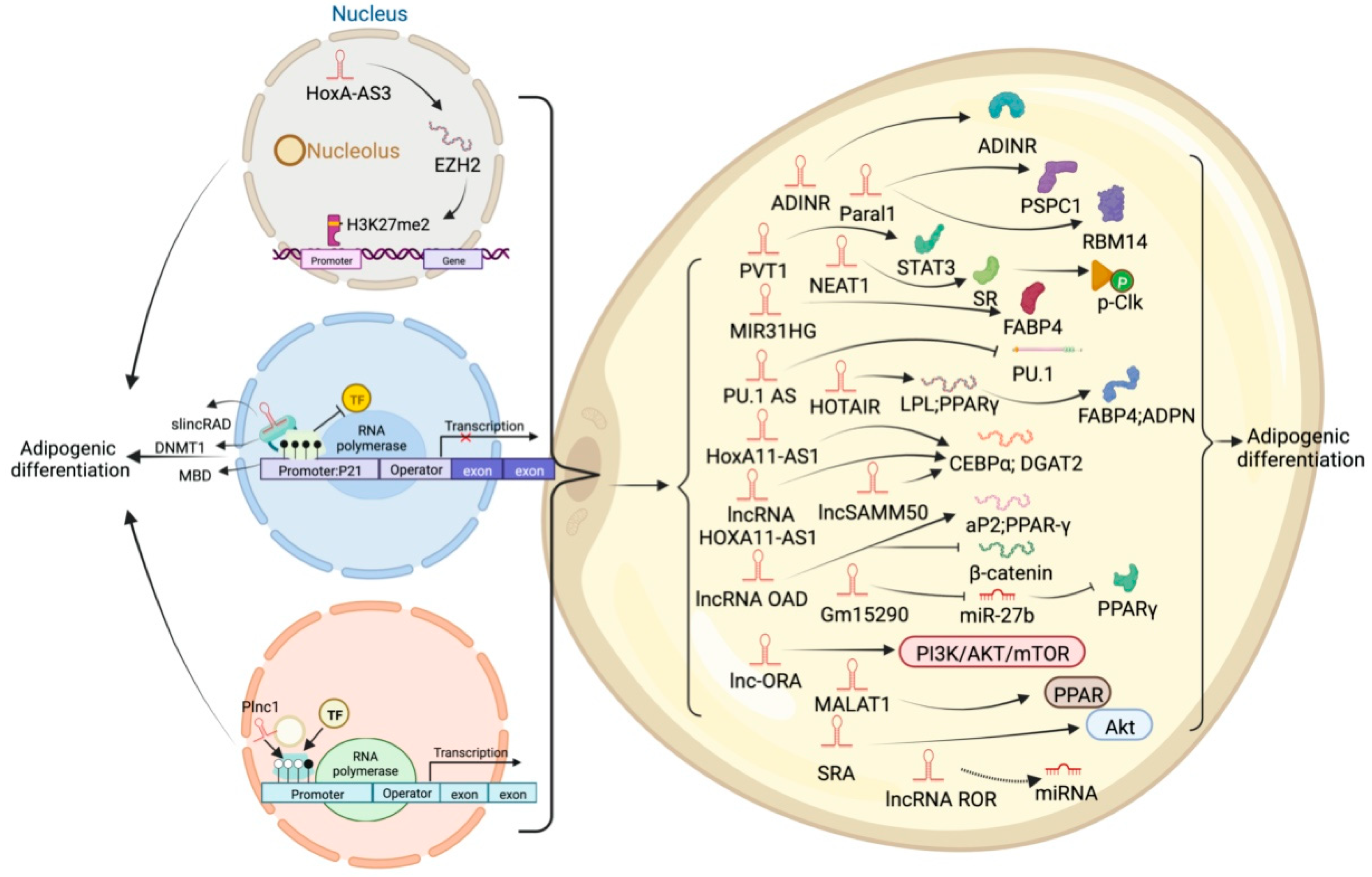
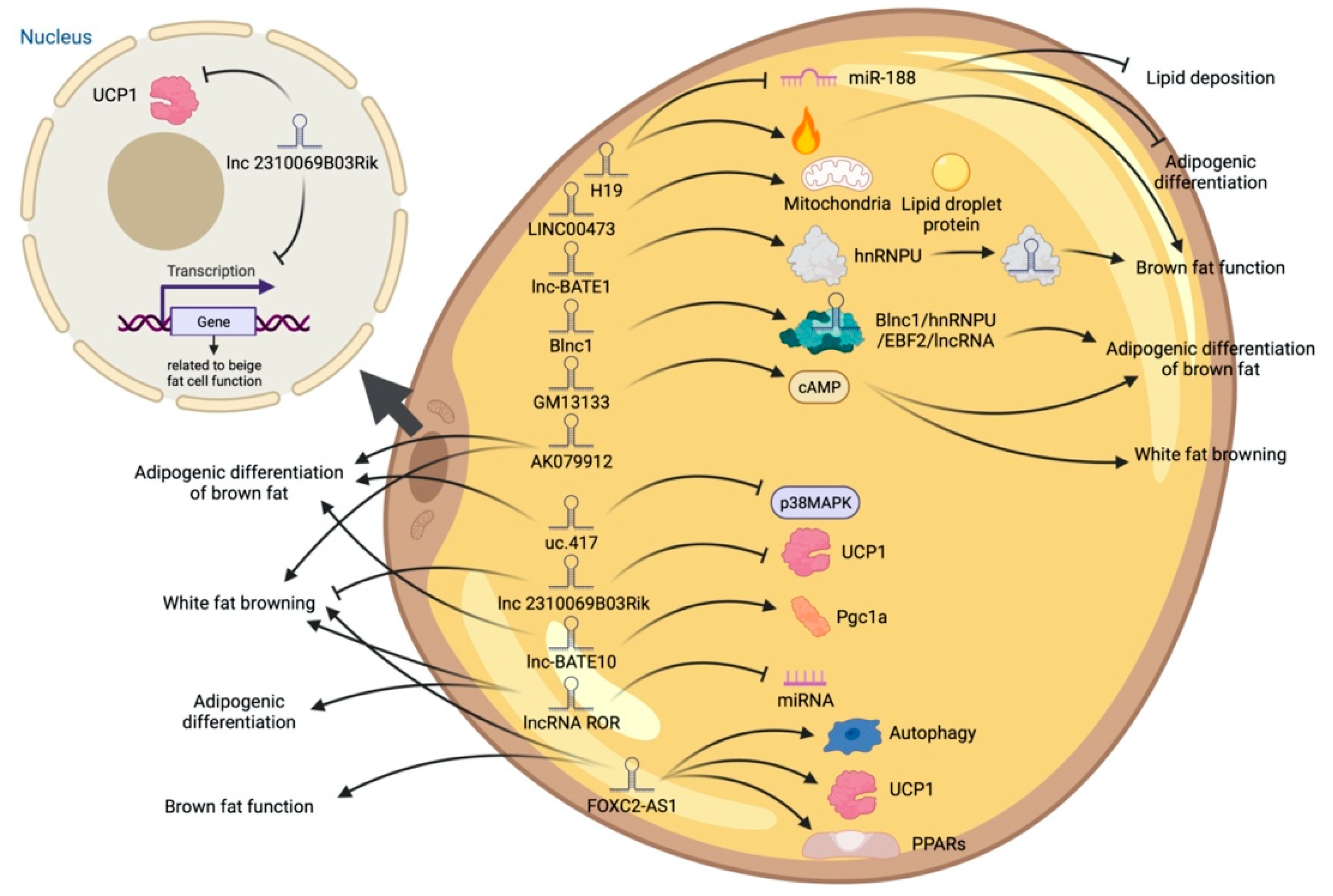
5. LncRNA and Adipose Tissue
6. Different Regulatory Modes of LncRNAs in Adipogenic Processes
6.1. Decoy Molecules
6.2. RNA-RNA Dimer
6.3. MicroRNA Sponge
6.4. Involved in Histone Modification
6.5. Scaffold
6.6. Regulate Target-Gene-Promoter Activity
6.7. Guide
6.8. Cell Signal Pathways and Regulates Downstream Genes
6.8.1. Cell Signal Pathways
6.8.2. Regulates Downstream Genes
7. The Role of LncRNA in Ectopic Fat Deposition
8. LncRNA-Mediated Adipogenesis Dysregulation and Disease
9. Summary and Outlook
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADINR | adipogenic differentiation-induced ncRNA |
| AdipoQ AS | adiponectin antisense RNA |
| ADNCR | dipocyte differentiation-associated lncRNA |
| BMI | body mass index |
| ADSCs | human adipose-derived stem cells |
| HFD | high-fat diet |
| HOTAIR | HOX transcript antisense RNA |
| lnc-ORA | obesity-related lncRNA |
| lncRNA | long noncoding RNAs |
| MALAT1 | metastasis-associated lung adenocarcinoma transcript 1 |
| Meg3 | maternally expressed gene 3 |
| MIR31HG | miR-31 host gene |
| SRA | steroid receptor RNA activator |
| TINCR | tissue differentiation-inducing non-protein coding RNA |
| snRNA | small nuclear RNA |
| siRNA | small interfering RNA |
| miRNA | microRNA |
| WHO | World Health Organization |
| COVID-19 | Corona Virus Disease 2019 |
| lncLSTR | liver-specific triglyceride regulator RNA |
| NF-κB | nuclear factor kappa-B |
| SR | serine/arginine |
| ceRNA | competitive endogenous RNA |
| ATP | adenosine triphosphate |
| UCPl | uncoupling proteins 1 |
| NE | norepinephrine |
| β3-Ars | β3-adrenergic receptors |
| FFA | free fatty acids |
| cAMP | adenosine cyclophosphate |
| AREs. | AU-rich elements |
| TFs | transcription factors |
| EBF2 | early B cell factor 2 |
| hnRNPU | Heterogeneous nuclear ribonucleoprotein |
| IGF2BP2 | insulin-like growth factor 2 mRNA binding protein 2 |
| H3K4Me3 | trimethylated H3K4 |
| MED1 | mediates protein complex subunit 1 |
| RBM14 | hnRNP-like binding protein 14 |
| PI3K/AKT | Phosphoinositide 3-Kinase/Protein Kinase B |
| CCND1 | Cyclin D1 |
| C/EBPα | CCAAT/enhancer-binding protein α |
| PPARγ | peroxisome proliferator-activated receptor γ |
| AdipoQ | Adiponectin |
| SIRT1 | Sirtuin type 1 |
| NEAT1 | nuclear enriched abundant transcript 1 |
| Blnc1 | brown fat lncRNA 1 |
| BAT | brown adipose tissue |
| PGC1α | peroxisome-proliferator-activated receptor gamma coactivator 1 alpha) |
| EBF2 | early B cell factor 2 |
| PRDM16 | PR-domain containing protein 16 |
| CAAlnc1 | cachexia-related anti-adipogenesis lncRNA 1 |
| HuR | Hu antigen R |
| lncXIST | long noncoding RNA X inactive specific transcript |
References
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell. Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ravasi, T. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006, 16, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Cao, Y.; Wureli, H.; Wei, N.I.; Wang, D.W.; Sheng-Wei, H.U. Cloning and Protein Encoding Ability Analysis of Sheep lncRNA-MRLN Gene. Prog. Vet. Med. 2018.
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, J.; Terai, G.; Hamada, M. Computational prediction of lncRNA-mRNA interactionsby integrating tissue specificity in human transcriptome. Biol. Direct 2017, 12, 1–8. [Google Scholar] [CrossRef]
- Kosinska-Selbi, B.; Mielczarek, M.; Szyda, J. Review: Long non-coding RNA in livestock. Animal 2020, 14, 2003–2013. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220. [Google Scholar] [CrossRef]
- Rey, F.; Urrata, V.; Gilardini, L.; Bertoli, S.; Calcaterra, V.; Zuccotti, G.V.; Cancello, R.; Carelli, S. Role of long non-coding RNAs in adipogenesis: State of the art and implications in obesity and obesity-associated diseases. Obes. Rev. 2021, 22, e13203. [Google Scholar] [CrossRef]
- Zhu, J.J.; Fu, H.J.; Wu, Y.G.; Zheng, X.F. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef]
- Chen, X.; Yan, G.-Y. Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics 2013, 29, 2617–2624. [Google Scholar] [CrossRef]
- Verma, M.; Rajput, M.; Kishore, K.; Kathirvel, S. Asian BMI criteria are better than WHO criteria in predicting Hypertension: A cross-sectional study from rural India. J. Fam. Med. Prim. Care 2019, 8, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Després, J.; Lemieux, I.; Alméras, N. Abdominal Obesity and the Metabolic Syndrome; Springer: New York, NY, USA, 2006. [Google Scholar]
- Fernandez, X.; Monin, G.; Talmant, A.; Mourot, J.; Lebret, B. Influence of intramuscular fat content on the quality of pig meat—1. Composition of the lipid fraction and sensory characteristics of m. longissimus lumborum. Meat Sci. 1999, 53, 59–65. [Google Scholar] [CrossRef]
- Karampela, Ι.; Vallianou, N.; Magkos, F.; Apovian, C.M.; Dalamaga, Μ. Obesity, Hypovitaminosis D, and COVID-19: The Bermuda Triangle in Public Health. Curr. Obes. Rep. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dalamaga, M.; Christodoulatos, G.S.; Karampela, I.; Vallianou, N.; Apovian, C.M. Understanding the Co-Epidemic of Obesity and COVID-19: Current Evidence, Comparison with Previous Epidemics, Mechanisms, and Preventive and Therapeutic Perspectives. Curr. Obes. Rep. 2021, 10, 214–243. [Google Scholar] [CrossRef] [PubMed]
- Pardo, M.; Roca-Rivada, A.; Seoane, L.M.; Casanueva, F. Obesidomics: Contribution of adipose tissue secretome analysis to obesity research. Endocrine 2012, 41, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, V.; Labbé, S.; Blondin, D.P.; Phoenix, S.; Guérin, B.; Haman, F.; Turcotte, E.E.; Richard, D.; Carpentier, A.C. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Investig. 2012, 122, 545–552. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Zhang, C.L.; Zhu, K.P.; Ma, X.L. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017, 396, 66–75. [Google Scholar] [CrossRef]
- Hamazaki, N.; Nakashima, K.; Hayashi, K.; Imamura, T. Detection of Bidirectional Promoter-Derived lncRNAs from Small-Scale Samples Using Pre-Amplification-Free Directional RNA-seq Method. Methods Mol. Biol. 2017, 1605, 83. [Google Scholar]
- Alvarez-Dominguez, J.R.; Knoll, M.; Gromatzky, A.A.; Lodish, H.F. The Super-Enhancer-Derived alncRNA-EC7/Bloodlinc Potentiates Red Blood Cell Development in trans. Cell Rep. 2017, 19, 2503. [Google Scholar] [CrossRef]
- Blythe, A.J.; Fox, A.; Bond, C.S. The ins and outs of lncRNA structure: How, why and what comes next? Biochim. Biophys. Acta 2016, 1859, 46–58. [Google Scholar] [CrossRef]
- Chengtao, D.U.; Wang, H.; Yang, S.; Xiangchen, L.I.; Zhou, X.; Zhao, A. Study on the Role of lncRNA in the Process of Japanese Encephalitis Virus Infecting PK15 Cells. China Anim. Husb. Vet. Med. 2019, 46, 2045–2052. [Google Scholar]
- Zhang, K.; Shi, Z.M.; Chang, Y.N.; Hu, Z.M.; Qi, H.X.; Hong, W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014, 547, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Sun, S.G.; Yue, Z.Q.; Bai, F. Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother. 2021, 134, 111158. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Yang, Y.; Yan, L.; Wang, J. LncRNA XIST facilitates cell growth, migration and invasion via modulating H3 histone methylation of DKK1 in neuroblastoma. Cell Cycle 2019, 18, 1882–1892. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; Mcdonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S. The Xist lncRNA Exploits Three-Dimensional Genome Architecture to Spread Across the X Chromosome. Science 2013, 341, 767. [Google Scholar] [CrossRef]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915.e2904. [Google Scholar] [CrossRef]
- Stewart, S. Transcriptional interference at tandem lncRNA and protein-coding genes: An emerging theme in regulation of cellular nutrient homeostasis. Nucleic Acids Res. 2020, 48, 8243–8254. [Google Scholar]
- Dahariya, S.; Pa Ddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [PubMed]
- Maria, T.; Kalliopi, S.; Georgia, C. The Nucleolus: In Genome Maintenance and Repair. Int. J. Mol. Sci. 2017, 18, 1411. [Google Scholar]
- Fortini, E.; Li, R.; Fox, A.H. Long Non-Coding RNAs and Nuclear Body Formation and Function; Springer: New York, NY, USA, 2013. [Google Scholar]
- Pirogov, S.A.; Gvozdev, V.A.; Klenov, M.S. Long Noncoding RNAs and Stress Response in the Nucleolus. Cells 2019, 8, 668. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Wang, X.; Hu, Y.; Du, H.; Dong, B.; Ao, S.; Zhang, L.; Sun, Z.; Zhang, L.; Lv, G.; et al. Solamargine inhibits gastric cancer progression by regulating the expressionof lncNEAT1_2 via the MAPK signaling pathway. Int. J. Oncol. 2019, 54, 1545–1554. [Google Scholar]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Hussen, B.M.; Azimi, T.; Hidayat, H.J.; Taheri, M.; Ghafouri-Fard, S. NF-KappaB interacting LncRNA: Review of its roles in neoplastic and non-neoplastic conditions. Biomed. Pharmacother. 2021, 139, 111604. [Google Scholar] [CrossRef]
- Hu, M.; Wang, R.; Li, X.; Fan, M.; Lin, J.; Zhen, J.; Chen, L.; Lv, Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J. Cell. Mol. Med. 2017, 21, 2732–2747. [Google Scholar] [CrossRef]
- Hu, G.; Lou, Z.; Mamta, G.; Ki, K.Y. The Long Non-Coding RNA GAS5 Cooperates with the Eukaryotic Translation Initiation Factor 4E to Regulate c-Myc Translation. PLoS ONE 2014, 9, e107016. [Google Scholar] [CrossRef]
- BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 2019, 98, 87–96. [CrossRef]
- Wu, D.M.; Wang, S.; Wen, X.; Han, X.R.; Wang, Y.J.; Shen, M.; Fan, S.H.; Zhang, Z.F.; Shan, Q.; Li, M.Q. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018, 9, 947. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Q.; Wang, Y.; Xiong, Y.; Chen, X.C.; Ma, M.L.; Cai, R.; Gao, Y.; Sun, Y.M.; Yang, G.S.; Pang, W.J. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 2016, 6, 21865. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Chen, S.; Li, W.; Chen, W.; Gu, W. LncRNA MACC1-AS1 sponges multiple miRNAs and RNA-binding protein PTBP1. Oncogenesis 2019, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Ferrer, L.O.; Steinbach, B.; Pantel, K.; Schwarzenbach, H. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol. Oncol. 2019, 13, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Daas, S.I.; Rizeq, B.R.; Nasrallah, G.K. Adipose tissue dysfunction in cancer cachexia. J. Cell. Physiol. 2018, 234, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Chen, Y. Latest Advancements on Combating Obesity by Targeting Human Brown/Beige Adipose Tissues. Front. Endocrinol. 2022, 13, 884944. [Google Scholar] [CrossRef]
- Barra, N.G.; Henriksbo, B.D.; Anhê, F.F.; Schertzer, J.D. The NLRP3 inflammasome regulates adipose tissue metabolism. Biochem. J. 2020, 477, 1089–1107. [Google Scholar] [CrossRef]
- White, U.; Ravussin, E. Dynamics of adipose tissue turnover in human metabolic health and disease. Diabetologia 2019, 62, 17–23. [Google Scholar] [CrossRef]
- Symonds, M.E.; Pope, M.; Budge, H. The Ontogeny of Brown Adipose Tissue. Annu. Rev. Nutr. 2015, 35, 295–320. [Google Scholar] [CrossRef]
- Kaisanlahti, A.; Glumoff, T. Browning of white fat: Agents and implications for beige adipose tissue to type 2 diabetes. J. Physiol. Biochem. 2019, 75, 1–10. [Google Scholar] [CrossRef]
- Roden, M.; Shulman, G.I. The integrative biology of type 2 diabetes. Nature 2019, 576, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Kita, S.; Maeda, N.; Shimomura, I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J. Clin. Investig. 2019, 129, 4041–4049. [Google Scholar] [CrossRef] [PubMed]
- Ioanna, M.; Christos, Y.; Flora, B.; George, C. The Potential Role of Exosomes in Child and Adolescent Obesity. Children 2021, 8, 196. [Google Scholar]
- Chen, Y.; Buyel, J.J.; Hanssen, M.; Siegel, F.; Pan, R.; Naumann, J.; Schell, M.; Lans, A.; Schlein, C.; Froehlich, H. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat. Commun. 2016, 7, 11420. [Google Scholar] [CrossRef]
- Rui, L. Brown and Beige Adipose Tissues in Health and Disease. Compr. Physiol. 2017, 7, 1281. [Google Scholar]
- Lizcano, F. The Beige Adipocyte as a Therapy for Metabolic Diseases. Int. J. Mol. Sci. 2019, 20, 5058. [Google Scholar] [CrossRef] [PubMed]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus White Adipose Tissue: A Mini-Review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef]
- Hao, R.; Yuan, L.; Zhang, N.; Li, C.; Yang, J. Brown adipose tissue: Distribution and influencing factors on FDG PET/CT scan. J. Pediatric Endocrinol. Metab. 2012, 25, 233–237. [Google Scholar] [CrossRef]
- Wang, G.X.; Zhao, X.Y.; Lin, J.D. The brown fat secretome: Metabolic functions beyond thermogenesis. Trends Endocrinol. Metab. 2015, 26, 231–237. [Google Scholar] [CrossRef]
- Pan, R.; Zhu, X.; Maretich, P.; Chen, Y. Metabolic Improvement via Enhancing Thermogenic Fat-Mediated Non-shivering Thermogenesis: From Rodents to Humans. Front. Endocrinol. 2020, 11, 633. [Google Scholar] [CrossRef]
- Nicolas, S.-Z.; Salvatore, F.; Claire, C.; Ozren, S.; Colin, D.J.; Ana, S.; Christelle, V.-D.; Valentina, T.; Dorothée, R.; Stéphane, G.; et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015, 21, 1497–1501. [Google Scholar]
- Xu, Z.; You, W.; Zhou, Y.; Chen, W.; Shan, T. Cold-induced lipid dynamics and transcriptional programs in white adipose tissue. BMC Biol. 2019, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, J.; Dai, H.; Duan, Y.; Zhao, B. Brown and beige adipose tissue: A novel therapeutic strategy for obesity and type 2 diabetes mellitus. Adipocyte 2021, 10, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Ruiz, H.H.; Jhun, K.; Finan, B.; Oberlin, D.J.; Verena, V.; Kalinovich, A.V.; Petrovic, N.; Wolf, Y.; Clemmensen, C. Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat. Med. 2017, 23, 623–630. [Google Scholar] [CrossRef]
- Matesanz, N.; Bernardo, E.; Acín-Pérez, R.; Manieri, E.; Pérez-Sieira, S.; Hernández-Cosido, L.; Montalvo-Romeral, V.; Mora, A.; Rodríguez, E.; Leiva-Vega, L. MKK6 controls T3-mediated browning of white adipose tissue. Nat. Commun. 2017, 8, 856. [Google Scholar] [CrossRef]
- Porter, C.; Herndon, D.N.; Bhattarai, N.; Ogunbileje, J.O.; Szczesny, B.; Szabo, C.; Toliver-Kinsky, T.; Sidossis, L.S. Severe Burn Injury Induces Thermogenically Functional Mitochondria in Murine White Adipose Tissue. Shock 2015, 44, 258–264. [Google Scholar] [CrossRef]
- Tumova, J.; Andel, M.; Trnka, J. Excess of free fatty acids as a cause of metabolic dysfunction in skeletal muscle. Physiol. Res. 2015, 65, 193. [Google Scholar] [CrossRef]
- Wu, L.; Mo, W.; Feng, J.; Li, J.; Yu, Q.; Li, S.; Zhang, J.; Chen, K.; Ji, J.; Dai, W.; et al. Astaxanthin Attenuates Hepatic Damages and Mitochondrial Dysfunction in Nonalcoholic Fatty Liver Disease by Regulating the FGF21/PGC-1α Pathway. Br. J. Pharmacol. 2021, 23. [Google Scholar] [CrossRef]
- Nagayama, D.; Shirai, K. Hypertriglyceridemia-induced pancreatitis. Nihon Rinsho Jpn. J. Clin. Med. 2013, 71, 1602. [Google Scholar]
- Zhang, Q.; Kong, X.; Yuan, H.; Guan, H.; Li, Y.; Niu, Y. Mangiferin Improved Palmitate-Induced-Insulin Resistance by Promoting Free Fatty Acid Metabolism in HepG2 and C2C12 Cells via PPAR α: Mangiferin Improved Insulin Resistance. J. Diabetes Res. 2019, 2019, 1–13. [Google Scholar]
- Renaville, B.; Bacciu, N.; Lanzoni, M.; Corazzin, M.; Piasentier, E. Polymorphism of fat metabolism genes as candidate markers for meat quality and production traits in heavy pigs. Meat Sci. 2015, 110, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; He, X.; Sheng, Z.; Kai, W.; Wu, H.; Lin, C. LncRNA TINCR/miR-31-5p/C/EBP-α feedback loop modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. Stem Cell Res. 2018, 32, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Xu, H.; Li, D.; Li, H.; Lu, D. A novel long noncoding RNA, AC092834.1, regulates the adipogenic differentiation of human adipose-derived mesenchymal stem cells via the DKK1/Wnt/β-catenin signaling pathway. Biophys. Res. Commun. 2020, 525, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, Y.; Fan, L.; Deng, L.; Fan, J.; Li, D.; Li, H.; Zhao, R.C. Lnc13728 facilitates human mesenchymal stem cell adipogenic differentiation via positive regulation of ZBED3 and downregulation of the WNT/β-catenin pathway. Stem Cell Res. Ther. 2021, 12, 176. [Google Scholar] [CrossRef]
- Ba, I.Z.; Chai, X.R.; Yoon, M.J.; Kim, H.J.; Sun, L. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. 2017, 15, e2002176. [Google Scholar] [CrossRef]
- Jules, J.; Chen, W.; Li, Y.P. C/EBPα and PU.1 exhibit different responses to RANK signaling for osteoclastogenesis. Bone 2017, 107, 104. [Google Scholar] [CrossRef]
- Shen, L.; Han, J.; Wang, H.; Meng, Q.; Wu, G. Cachexia-related long noncoding RNA, CAAlnc1, suppresses adipogenesis by blocking the binding of HuR to adipogenic transcription factor mRNAs. Int. J. Cancer 2019, 145, 1809–1821. [Google Scholar] [CrossRef]
- Wu, C.; Fang, S.; Zhang, H.; Li, X.; Du, Y.; Zhang, Y.; Lin, X.; Wang, L.; Ma, X.; Xue, Y.; et al. Long noncoding RNA XIST regulates brown preadipocytes differentiation and combats high-fat diet induced obesity by targeting C/EBPα. Mol. Med. 2022, 28, 6. [Google Scholar] [CrossRef]
- Tang, H.Q.; Meng, Y.L.; Lu, Q.L.; Dou, Y.Y.; Liang, L.L.; Luo, Y. Decreased long noncoding RNA ADIPOQ promoted cell proliferation and metastasis via miR-219c-3p/TP53 pathway in colorectal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7645–7654. [Google Scholar]
- Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Et Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 420–432.
- Xu-Yun, Z.; Siming, L.; Guo-Xiao, W.; Qi, Y.; Lin, J.D. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell 2014, 55, 372–382. [Google Scholar]
- Alvarez-Dominguez, J.R.; Winther, S.; Hansen, J.B.; Lodish, H.F.; Knoll, M. An adipose lncRAP2-Igf2bp2 complex enhances adipogenesis and energy expenditure by stabilizing target mRNAs. iScience 2022, 25, 103680. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Duan, Y.; Sang, Y.; Li, Y.; Zhang, H.; Liang, Y.; Liu, Y.; Zhang, N.; Yang, Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J. Cell. Physiol. 2018, 234, 9105–9117. [Google Scholar]
- Li, M.; Sun, X.; Cai, H.; Sun, Y.; Plath, M.; Li, C.; Lan, X.; Lei, C.; Lin, F.; Chen, H. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim. Et Biophys. Acta 2016, 1859, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ma, C.; Yang, B.; Yin, C.; Zhang, B.; Xiao, Y. LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem. Biophys. Res. Commun. 2017, 493, 1168–1175. [Google Scholar] [CrossRef]
- Elena, S.; Ines, D.; Isabella, G.; Matteo, O.; Paul, K.; Motoharu, A.; Gerfried, M.; Eduardo, F.R.; Marta, P.J.; Wolfgang, W. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 2018, 9, 3622. [Google Scholar]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef]
- Huang, Y.; Jin, C.; Zheng, Y.; Li, X.; Zhang, S.; Zhang, Y.; Jia, L.; Li, W. Knockdown of lncRNA MIR31HG inhibits adipocyte differentiation of human adipose-derived stem cells via histone modification of FABP4. Sci. Rep. 2017, 7, 8080. [Google Scholar] [CrossRef]
- Xiao, T.; Liu, L.; Li, H.; Sun, Y.; Luo, H. Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPα. Stem Cell Rep. 2015. [Google Scholar]
- Zhang, H.; Liu, Y.; Yan, L.; Min, Z.; Yu, X.; Wei, D.; Wang, S.; Li, Q.; Chen, H.; Zhang, Y. Increased levels of the long noncoding RNA, HOXA-AS3, promote proliferation of A549 cells. Cell Death Dis. 2018, 9, 707. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.; Bai, Z.; Xu, D.; Yuan, B.; Lo, K.; Yoon, M.; Lim, Y.; Knoll, M.; Slavov, N.; Chen, S. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab. 2015, 21, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiao, Y.; Yang, M.; Su, T.; Sun, X.; Guo, Q.; Huang, Y.; Luo, X.H. Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. J. Clin. Investig. 2018, 128, 5219–5221. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.V.; Brown, E.L.; Desouza, T.; Jespersen, N.Z.; Nielsen, S. Human thermogenic adipocyte regulation by the long noncoding RNA LINC00473. Nat. Metab. 2020, 2, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lin, S.; Li, J.L.; Wei, N.; Wu, L. CRTC1-MAML2 fusion-induced lncRNA LINC00473 expression maintains the growth and survival of human mucoepidermoid carcinoma cells. Oncogene 2018, 37, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xue, C.; Lin, J.; Ferguson, J.F.; Weiner, A.; Liu, W.; Han, Y.; Hinkle, C.; Li, W.; Jiang, H. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci. Transl. Med. 2018, 10, eaar5987. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mi, L.; Yu, L.; Yu, Q.; Liu, T.; Wang, G.-X.; Zhao, X.-Y.; Wu, J.; Lin, J.D. Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc. Natl. Acad. Sci. USA 2017, 114, E7111. [Google Scholar]
- Chen, J.; Liu, Y.; Lu, S.; Yin, L.; Zong, C.; Cui, S.; Qin, D.; Yang, Y.; Guan, Q.; Li, X. The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int. J. Obes. 2017, 41, 299–308. [Google Scholar] [CrossRef]
- Zhu, E.; Zhang, J.; Li, Y.; Yuan, H.; Wang, B. Long noncoding RNA Plnc1 controls adipocyte differentiation by regulating peroxisome proliferator–activated receptor γ. FASEB J. 2018, 33, 2396–2408. [Google Scholar] [CrossRef]
- Chen, Y.T.; Yang, Q.Y.; Hu, Y.; Liu, X.D.; de Avila, J.M.; Zhu, M.J.; Nathanielsz, P.W.; Du, M. Imprinted lncRNA Dio3os preprograms intergenerational brown fat development and obesity resistance. Nat. Commun. 2021, 12, 6845. [Google Scholar] [CrossRef]
- Alice, L.K.; Shiqi, H.; Esther, W.A.C.; Zhi-Chun, Z.; Khee-Shing, L.M.; Meihui, L.; Lei, S. Adipocyte Long-Noncoding RNA Transcriptome Analysis of Obese Mice Identified Lnc-Leptin, Which Regulates Leptin. Diabetes 2018, 67, 1045–1056. [Google Scholar]
- Yi, F.; Zhang, P.; Wang, Y.; Xu, Y.; Du, Q. Long non-coding RNA slincRAD functions in methylation regulation during the early stage of mouse adipogenesis. RNA Biol. 2019, 16, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
- Irmin, F.F.; Oger, F.; Gheeraert, C.; Dubois-Chevalier, J.; Lefebvre, P. The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci. Rep. 2017, 7, 14087. [Google Scholar] [CrossRef] [PubMed]
- Magun, R.; Burgering, B.M.; Coffer, P.J.; Pardasani, D.; Lin, Y.; Chabot, J.; Sorisky, A. Expression of a constitutively activated form of protein kinase B (c-Akt) in 3T3-L1 preadipose cells causes spontaneous differentiation. Endocrinology 1996, 137, 3590–3593. [Google Scholar] [CrossRef]
- Collado, M.; Medema, R.H.; Garcia-Cao, I.; Dubuisson, M.L.; Barradas, M.; Glassford, J.; Rivas, C.; Burgering, B.M.; Serrano, M.; Lam, E.W. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J. Biol. Chem. 2000, 275, 21960–21968. [Google Scholar] [CrossRef]
- Belmonte, N.; Phillips, B.W.; Massiera, F.; Villageois, P.; Wdziekonski, B.; Saint-Marc, P.; Nichols, J.; Aubert, J.; Saeki, K.; Yuo, A.; et al. Activation of extracellular signal-regulated kinases and CREB/ATF-1 mediate the expression of CCAAT/enhancer binding proteins beta and -delta in preadipocytes. Mol. Endocrinol. 2001, 15, 2037–2049. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prusty, D.; Park, B.H.; Davis, K.E.; Farmer, S.R. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J. Biol. Chem. 2002, 277, 46226–46232. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Mushiake, S.; Bessho, K.; Murakami, M.; Namba, N.; Kokubu, C.; Michigami, T.; Ozono, K. Wnt/Lrp/beta-catenin signaling suppresses adipogenesis by inhibiting mutual activation of PPARgamma and C/EBPalpha. Biochem. Biophys Res. Commun. 2007, 363, 276–282. [Google Scholar] [CrossRef]
- Ikeda, S.; Kishida, S.; Yamamoto, H.; Murai, H.; Koyama, S.; Kikuchi, A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. Embo J. 1998, 17, 1371–1384. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef]
- Cai, R.; Tang, G.; Zhang, Q.; Yong, W.; Pang, W. A Novel lnc-RNA, Named lnc-ORA, Is Identified by RNA-Seq Analysis, and Its Knockdown Inhibits Adipogenesis by Regulating the PI3K/AKT/mTOR Signaling Pathway. Cells 2019, 8, 477. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, Z.; Dai, Z.; Zhong, Y.; Liu, X.; Zuo, C. Long non-coding RNA lnc-OAD is required for adipocyte differentiation in 3T3-L1 preadipocytes. Biochem. Biophys. Res. Commun. 2019, 511, 753–758. [Google Scholar] [CrossRef]
- You, L.H.; Zhou, Y.H.; Cui, X.W.; Wang, X.Y.; Sun, Y.Z.; Gao, Y.; Wang, X.; Wen, J.; Xie, K.; Tang, R.R. GM13133 is a negative regulator in mouse white adipocytes differentiation and drives the characteristics of brown adipocytes. J. Cell. Physiol. 2017, 233, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; You, L.; Li, Y.; Zhu, L.; Zhang, F.; Xie, K.; Cao, Y.; Ji, C.; Guo, X. A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J. 2016, 4301–4312. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Tang, C.Y.; Tang, H.N.; Wu, H.X.; Zhou, H.D. Long Non-coding RNA 332443 Inhibits Preadipocyte Differentiation by Targeting Runx1 and p38-MAPK and ERK1/2-MAPK Signaling Pathways. Front. Cell Dev. Biol. 2021, 9, 663959. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.B.; Mckenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17. [Google Scholar] [CrossRef]
- Tello-Flores, V.A.; Beltrán-Anaya, F.; Ramírez-Vargas, M.; Esteban-Casales, B.E.; Flores-Alfaro, E. Role of Long Non-Coding RNAs and the Molecular Mechanisms Involved in Insulin Resistance. Int. J. Mol. Sci. 2021, 22, 7256. [Google Scholar] [CrossRef]
- Liu, S.; Xu, R.; Isabelle, G.; Cawthorn, W.P.; Macdougald, O.A.; Chen, X.W.; Saltiel, A.R.; Koenig, R.J.; Xu, B.; Agoulnik, I.U. SRA Regulates Adipogenesis by Modulating p38/JNK Phosphorylation and Stimulating Insulin Receptor Gene Expression and Downstream Signaling. PLoS ONE 2014, 9, e95416. [Google Scholar] [CrossRef]
- Denise, C.; Gay, C.; Li, P.; Rehka, P.; James, W.; Niketa, P. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARγ2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 2014, 5, 1050–1063. [Google Scholar]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Non-Coding RNA 2020, 6, 22. [Google Scholar] [CrossRef]
- Han, J.; Shen, L.; Zhan, Z.; Liu, Y.; Wu, G. The long noncoding RNA MALAT1 modulates adipose loss in cancer-associated cachexia by suppressing adipogenesis through PPAR-γ. Nutr. Metab. 2021, 18, 27. [Google Scholar] [CrossRef]
- Gong, D.; Zhao, Z.W.; Qiang, Z.M.; Yu, X.H.; Tang, C.K. The Long Noncoding RNA Metastasis-Associated Lung Adenocarcinoma Transcript-1 Regulates CCDC80 Expression by Targeting miR-141-3p/miR-200a-3p in Vascular Smooth Muscle Cells. J. Cardiovasc. Pharmacol. 2020, 75, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Bast-Habersbrunner, A.; Kiefer, C.; Weber, P.; Fromme, T.; Schießl, A.; Schwalie, P.C.; Deplancke, B.; Li, Y.; Klingenspor, M. LncRNA Ctcflos orchestrates transcription and alternative splicing in thermogenic adipogenesis. EMBO Rep. 2021, 22, e51289. [Google Scholar] [CrossRef] [PubMed]
- Thunen, A.; La Placa, D.; Zhang, Z.; Shively, J.E. Role of lncRNA LIPE-AS1 in adipogenesis. Adipocyte 2022, 11, 11–27. [Google Scholar] [CrossRef]
- Zhu, R.; Feng, X.; Wei, Y.; Guo, D.; Huang, J. lncSAMM50 Enhances Adipogenic Differentiation of Buffalo Adipocytes With No Effect on Its Host Gene. Front. Genet. 2021, 12, 626158. [Google Scholar] [CrossRef] [PubMed]
- Iwase, M.; Sakai, S.; Seno, S.; Yeh, Y.S.; Kuo, T.; Takahashi, H.; Nomura, W.; Jheng, H.F.; Horton, P.; Osato, N.; et al. Long non-coding RNA 2310069B03Rik functions as a suppressor of Ucp1 expression under prolonged cold exposure in murine beige adipocytes. Biosci. Biotechnol. Biochem. 2020, 84, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Kimura, A.P.; Ohmura, K.; Naito, S.; Ieko, M. Knockdown of long noncoding RNA dreh facilitates cell surface GLUT4 expression and glucose uptake through the involvement of vimentin in 3T3-L1 adipocytes. Gene 2020, 735, 144404. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jia, R.; Wei, X.; Luo, X. Time-sequential expression of lnc AK079912 during adipose tissue development and browning in mice. Nan Fang Yi Ke Da Xue Xue Bao J. South. Med. Univ. 2019, 39, 1494–1499. [Google Scholar]
- Nuermaimaiti, N.; Jie, L.; Liang, X.; Yi, J.; Guan, Y. Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem. Biophys. Res. Commun. 2018, 495, 1878–1884. [Google Scholar] [CrossRef]
- Wang, M.; Hao, C.; Xin, H.; Bao, H.; Wei, Y. Aberrant Expression of lncRNA (HOXA11-AS1) and Homeobox A ( HOXA9, HOXA10, HOXA11, and HOXA13) Genes in Infertile Women With Endometriosis. Reprod. Sci. 2017, 25, 654–661. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.; Qin, Z.Y.; Li, J.; Shen, Z.Y. The role and possible mechanism of long noncoding RNA PVT1 in modulating 3T3-L1 preadipocyte proliferation and differentiation. IUBMB Life 2020, 72, 1460–1467. [Google Scholar] [CrossRef]
- Divoux, A.; Karastergiou, K.; Xie, H.; Guo, W.; Perera, R.J.; Fried, S.K.; Smith, S.R. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity 2014, 22, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Ting, L.I.; Ding-Guo, R.; Gao, J.; Wang, H.; Xiao-Yang, X.U. Role of skeletal muscle fat ectopic deposition in insulin resistance induced by high-fat diet. Sheng Li Xue Bao Acta Physiol. Sin. 2018, 70, 433–444. [Google Scholar]
- Cobbina, E.; Akhlaghi, F. Non-alcoholic fatty liver disease (NAFLD)—Pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Ys, A.; Yu, S.A.; Cl, B.; Jg, A. LncRNA NEAT1-MicroRNA-140 axis exacerbates nonalcoholic fatty liver through interrupting AMPK/SREBP-1 signaling. Biochem. Biophys. Res. Commun. 2019, 514, 584–590. [Google Scholar]
- Ye, J.; Lin, Y.; Yu, Y.; Sun, D. LncRNA NEAT1/microRNA-129-5p/SOCS2 axis regulates liver fibrosis in alcoholic steatohepatitis. J. Transl. Med. 2020, 18, 445. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Yang, W.; Ruan, X.; Kiesewetter, K.; Zhu, J.; Cao, H. Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell Metab. 2016, 627–639. [Google Scholar] [CrossRef]
- Li, D.; Guo, L.; Deng, B.; Min, L.; Yang, T.; Fan, Y.; Yang, Z. Long noncoding RNA HR1 participates in the expression of SREBP1c through phosphorylation of the PDK1/AKT/FoxO1 pathway. Mol. Med. Rep. 2018, 18, 2850–2856. [Google Scholar]
- Lan, X.; Wu, L.; Wu, N.; Chen, Q.; Li, Y.; Du, X.; Wei, C.; Feng, L.; Li, Y.; Osoro, E.K. Long Noncoding RNA lnc-HC Regulates PPARγ-Mediated Hepatic Lipid Metabolism through miR-130b-3p. Mol. Therapy. Nucleic Acids 2019, 18, 954–965. [Google Scholar] [CrossRef]
- Gui, W.; Zhu, W.F.; Zhu, Y.; Tang, S.; Li, H. LncRNAH19 improves insulin resistance in skeletal muscle by regulating heterogeneous nuclear ribonucleoprotein A1. Cell Commun. Signal. 2020, 18, 173. [Google Scholar] [CrossRef]
- Lawrence, V.J.; Kopelman, P.G. Medical consequences of obesity. Clin. Dermatol. 2004, 22, 296–302. [Google Scholar] [CrossRef]
- Liu, S.; Sheng, L.; Miao, H.; Saunders, T.L.; MacDougald, O.A.; Koenig, R.J.; Xu, B. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J. Biol. Chem. 2014, 289, 13000–13009. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cui, Q.; Zhang, X.; Luo, X.; Liu, Y.; Zuo, J.; Peng, Y. Long non-coding RNAs regulation in adipogenesis and lipid metabolism: Emerging insights in obesity. Cell Signal. 2018, 51, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, Y.; Hou, Y.; Yang, L.; Wan, X.; Qin, Y.; Liu, Y.; Wang, R.; Zhu, P.; Teng, Y.; et al. A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J. Hematol. Oncol. 2021, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Peng, J.; Song, J.; He, J.; Jiang, M.; Wang, J.; Ma, L.; Wang, Y.; Lin, M.; Wu, H.; et al. Loss of Hilnc prevents diet-induced hepatic steatosis through binding of IGF2BP2. Nat. Metab. 2021, 3, 1569–1584. [Google Scholar] [CrossRef]
- Yu, X.H.; Deng, W.Y.; Chen, J.J.; Xu, X.D.; Liu, X.X.; Chen, L.; Shi, M.W.; Liu, Q.X.; Tao, M.; Ren, K. LncRNA kcnq1ot1 promotes lipid accumulation and accelerates atherosclerosis via functioning as a ceRNA through the miR-452-3p/HDAC3/ABCA1 axis. Cell Death Dis. 2020, 11, 1043. [Google Scholar] [CrossRef]
- Ruan, Y.; Lin, N.; Ma, Q.; Chen, R.; Zhang, Z.; Wen, W.; Chen, H.; Sun, J. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function. Cell Physiol. Biochem. 2018, 46, 335–350. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Abdelghany, A.A.; Toraih, E.A.; Mohamed, A.M. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosn. J. Basic Med. Sci. 2020, 20, 365–371. [Google Scholar] [CrossRef]
- Goyal, B.; Yadav, S.; Awasthee, N.; Gupta, S.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Et Biophys. Acta Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef]
- Bao, M.H.; Szeto, V.; Yang, B.B.; Zhu, S.Z.; Sun, H.S.; Feng, Z.P. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018, 9, 281. [Google Scholar] [CrossRef]
- Bai, H.L.; Lu, Z.F.; Zhao, J.J.; Ma, X.; Li, X.H.; Xu, H.; Wu, S.G.; Kang, C.M.; Lu, J.B.; Xu, Y.J.; et al. Microarray profiling analysis and validation of novel long noncoding RNAs and mRNAs as potential biomarkers and their functions in atherosclerosis. Physiol. Genom. 2019, 51, 644–656. [Google Scholar] [CrossRef]
- Bierhoff, H. Analysis of lncRNA-Protein Interactions by RNA-Protein Pull-Down Assays and RNA Immunoprecipitation (RIP); Humana Press: New York, NY, USA, 2018. [Google Scholar]
- Wei, G.H.; Wang, X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur. Rev. Med Pharmacol. Sci. 2017, 21, 3850–3856. [Google Scholar] [PubMed]
- Liao, K.; Xu, J.; Yang, W.; You, X.; Zhong, Q.; Wang, X. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol. Immunol. 2018, 101, 182. [Google Scholar] [CrossRef] [PubMed]
| LncRNA | Target miRNA | Species | Year |
|---|---|---|---|
| NEAT1 | miR-140 | Human | 2015 |
| Gm15290 | miR-27b | Mouse | 2017 |
| MEG3 | miR-140-5p | Human | 2020 |
| H19 | miR-188 | Mouse | 2018 |
| TINCR | miR-31-5p | Human | 2018 |
| GAS5 | miR-18a | Human | 2018 |
| GAS5 | miR-21a-5p | Mouse | 2018 |
| H19 | miR-30a | Human | 2019 |
| lnc PGC1β-OT1 | miR-148a-3p | Mouse | 2019 |
| LncRNA HCG11 | miR-204-5p | Human | 2020 |
| lncRNA RP11-142A22.4 | miR-587 | Human | 2020 |
| lncRNA-Adi | miR-449a | Human | 2020 |
| lncNEF | miR-155 | Human | 2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Wu, S.; He, Y.; Li, X.; Zhu, Y.; Lin, X.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. LncRNA-Mediated Adipogenesis in Different Adipocytes. Int. J. Mol. Sci. 2022, 23, 7488. https://doi.org/10.3390/ijms23137488
Zhang P, Wu S, He Y, Li X, Zhu Y, Lin X, Chen L, Zhao Y, Niu L, Zhang S, et al. LncRNA-Mediated Adipogenesis in Different Adipocytes. International Journal of Molecular Sciences. 2022; 23(13):7488. https://doi.org/10.3390/ijms23137488
Chicago/Turabian StyleZhang, Peiwen, Shuang Wu, Yuxu He, Xinrong Li, Yan Zhu, Xutao Lin, Lei Chen, Ye Zhao, Lili Niu, Shunhua Zhang, and et al. 2022. "LncRNA-Mediated Adipogenesis in Different Adipocytes" International Journal of Molecular Sciences 23, no. 13: 7488. https://doi.org/10.3390/ijms23137488
APA StyleZhang, P., Wu, S., He, Y., Li, X., Zhu, Y., Lin, X., Chen, L., Zhao, Y., Niu, L., Zhang, S., Li, X., Zhu, L., & Shen, L. (2022). LncRNA-Mediated Adipogenesis in Different Adipocytes. International Journal of Molecular Sciences, 23(13), 7488. https://doi.org/10.3390/ijms23137488








