Alterations of Phenylpropanoid Biosynthesis Lead to the Natural Formation of Pinkish-Skinned and White-Fleshed Strawberry (Fragaria × ananassa)
Abstract
:1. Introduction
2. Results
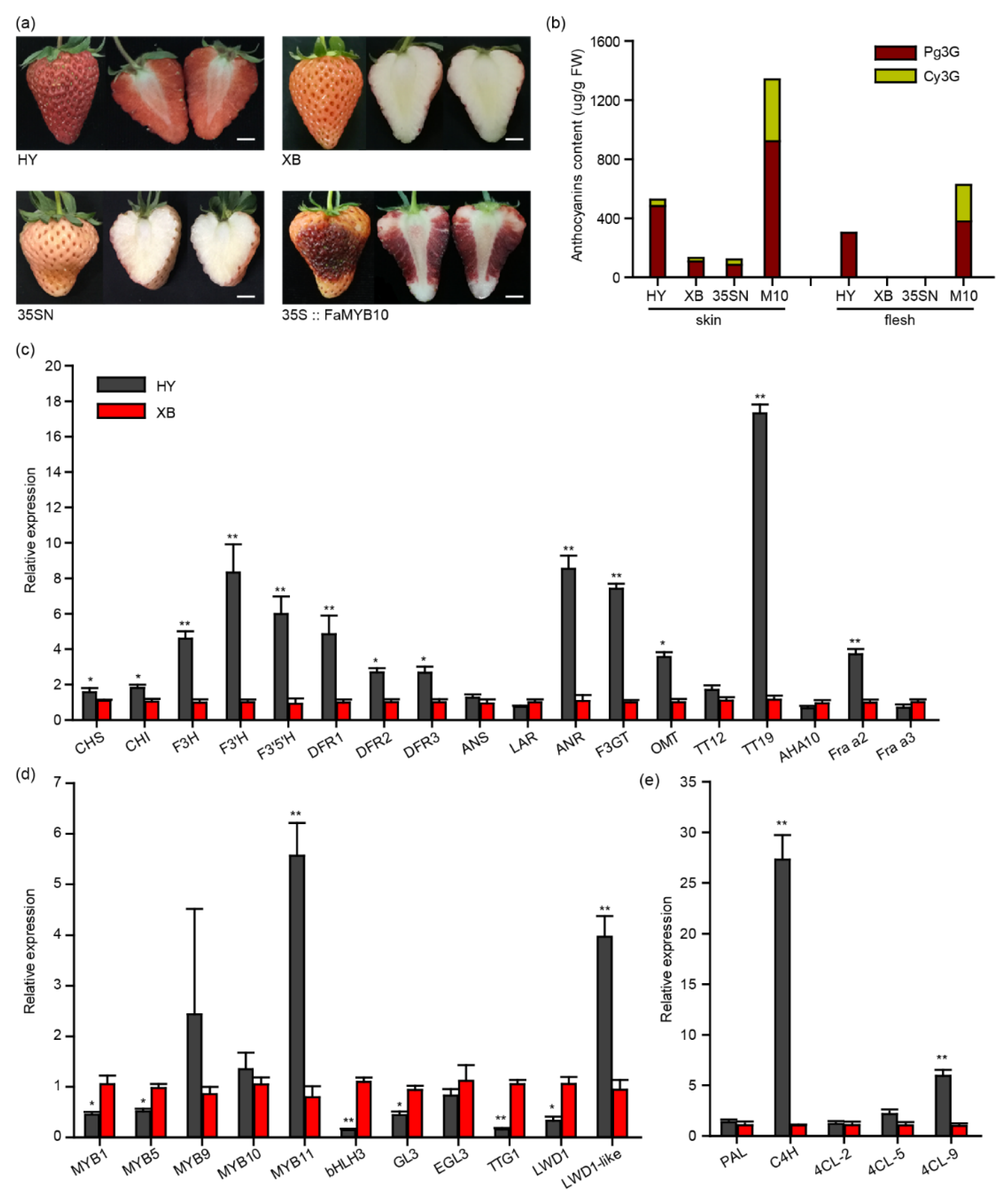
2.1. Variations in Anthocyanin-Related Genes in the Flesh of Benihoppe and Xiaobai Strawberries
2.2. Sequence Characteristics of the FaMYB10, FaC4H, and FaTT19 in Benihoppe and Xiaobai
2.3. Detection of Various Flavonoid Metabolites in Strawberry Flesh by Targeted Metabolomics
2.4. Detection of Other Metabolites in Strawberry Flesh by Quasi-Targeted Metabonomics
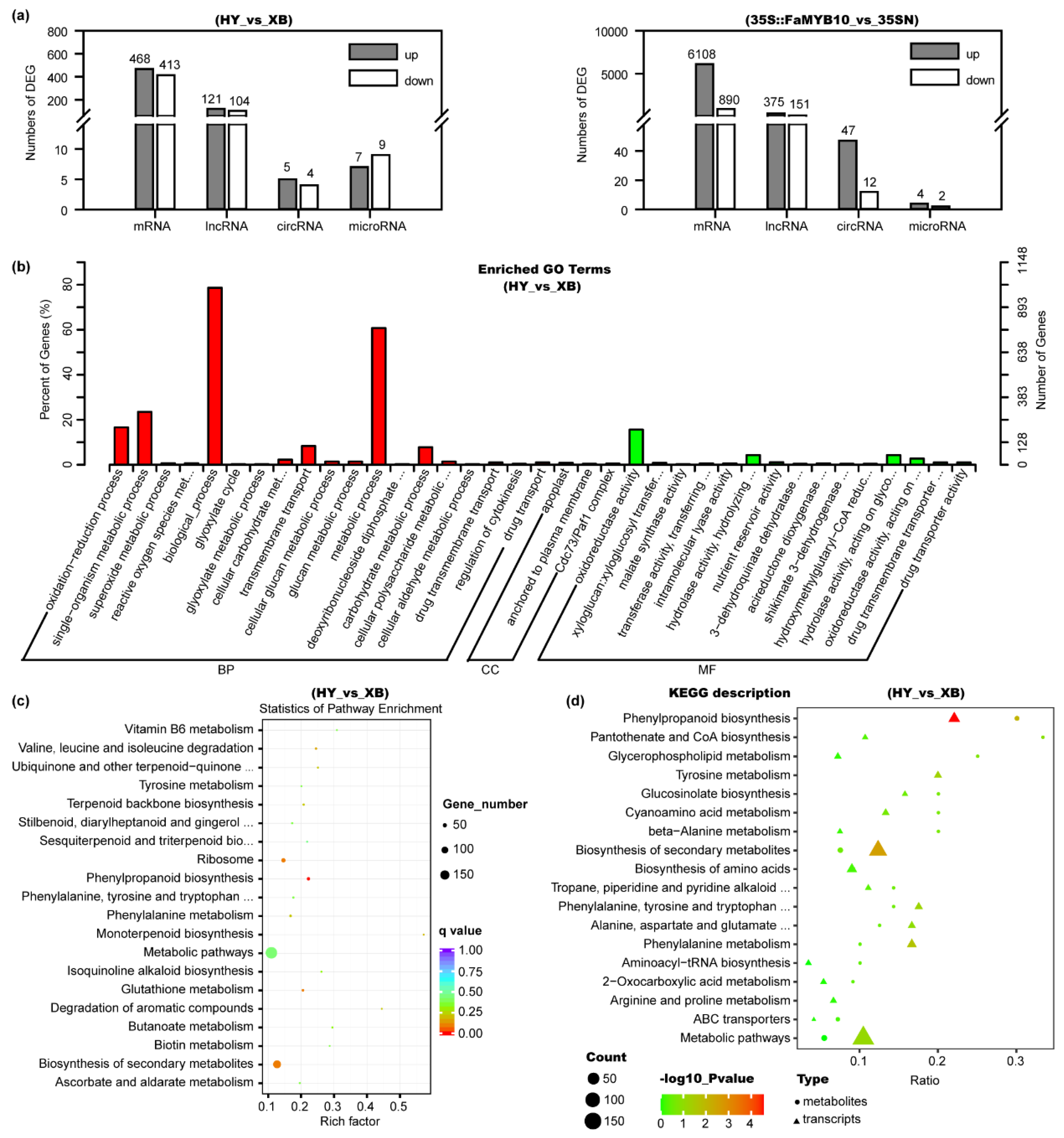
2.5. Whole Transcriptomic Sequencing of Red- and White-Fleshed Strawberries
2.6. Analysis of the Differentially Expressed mRNAs, lncRNAs, circRNAs, and microRNAs
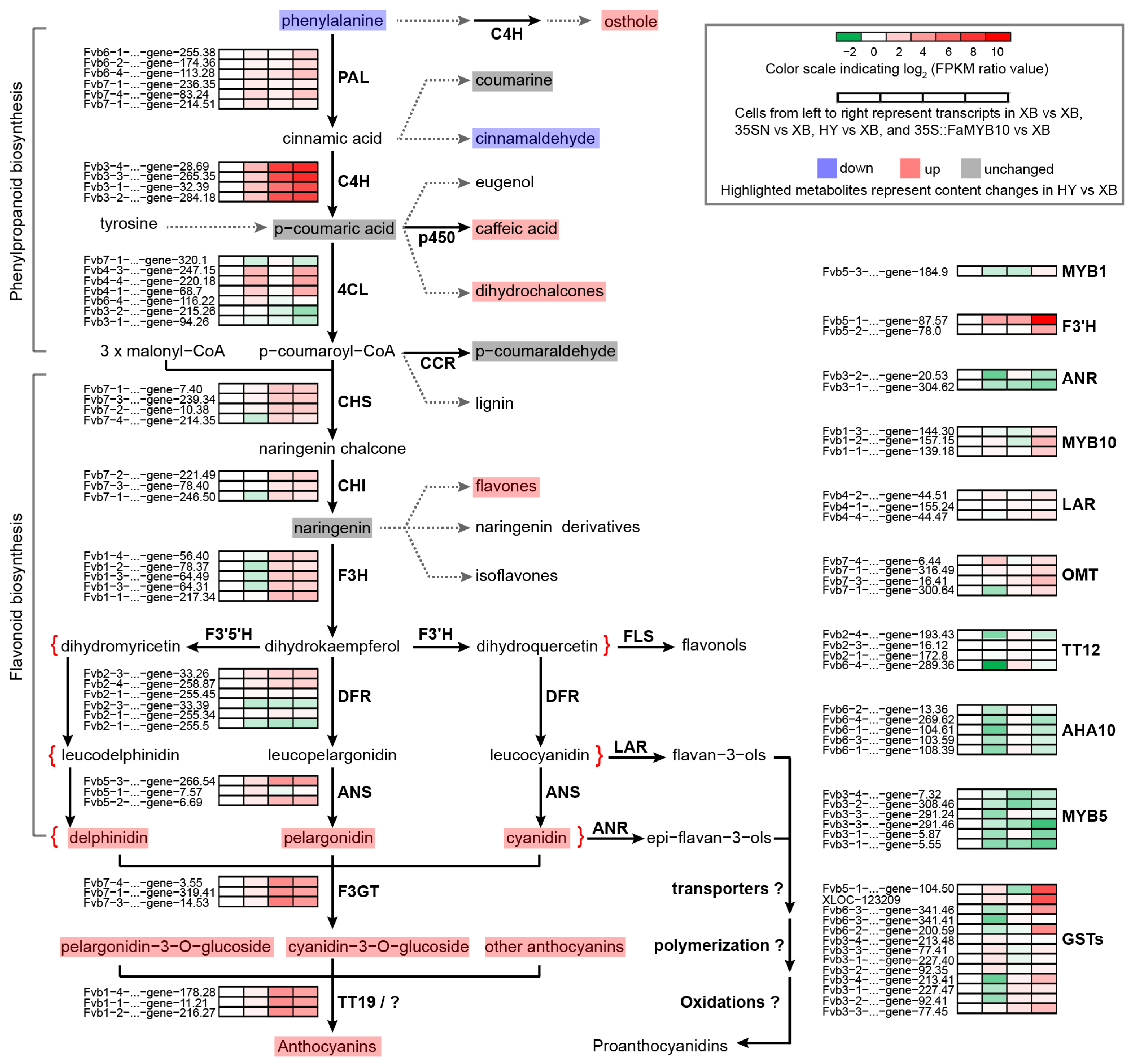
2.7. Combined Analysis of Metabolomics and Transcriptomics
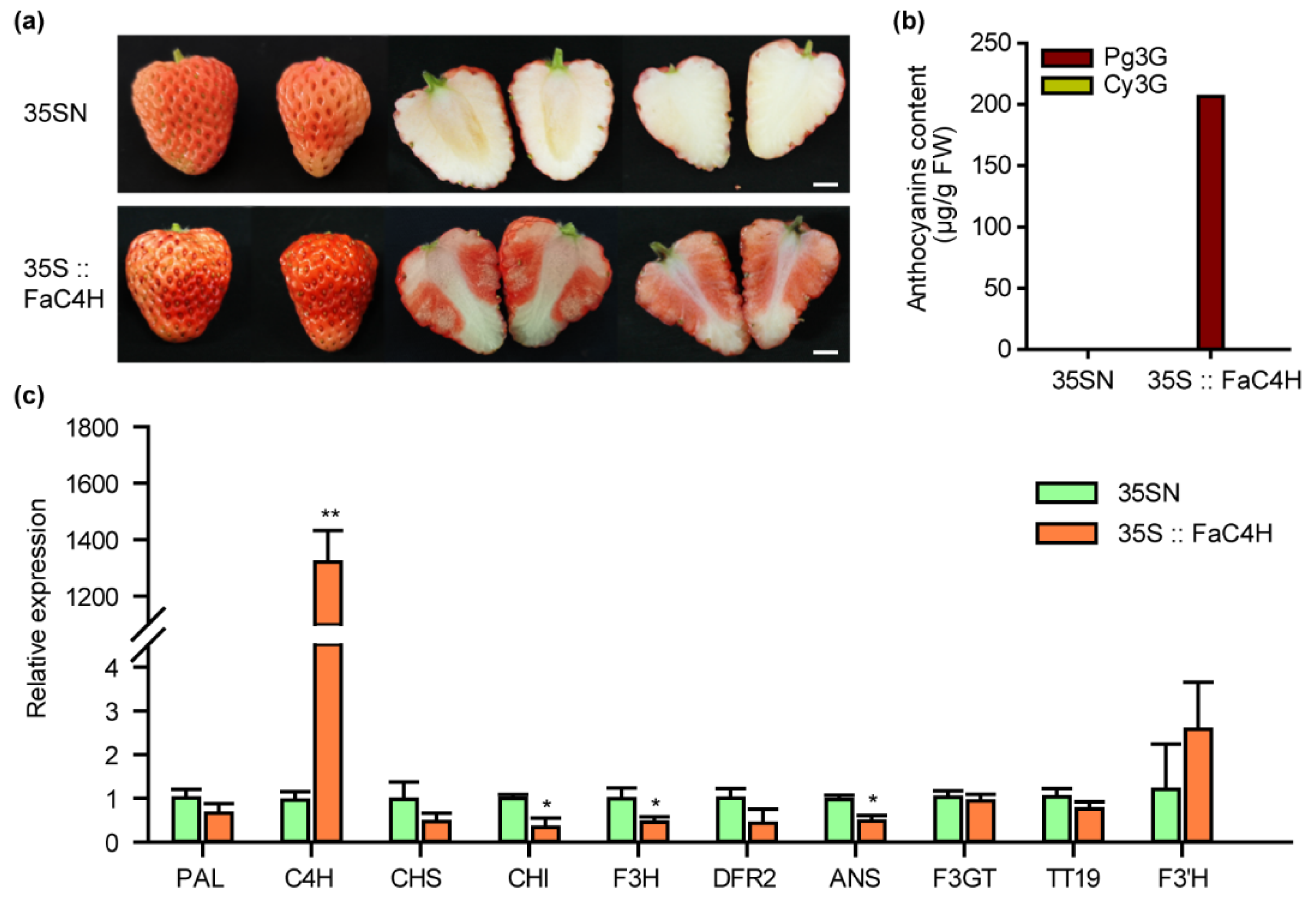
2.8. Overexpression of FaC4H Restored Anthocyanin Accumulation in Xiaobai’s Flesh
2.9. Methylation Detection of FaC4H and FaF3′H Promoters
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Anthocyanins and PAs Detection
4.3. RT-qPCR
4.4. Transient Overexpression Assays in Strawberry Fruits
4.5. Total RNA Sequencing and Analysis
4.6. Metabolomics Detection
4.7. DNA Extraction and Bisulfite Sequencing Assays
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almeida, J.R.M.; D’Amico, E.; Preuss, A.; Carbone, F.; de Vos, C.H.R.; Deiml, B.; Mourgues, F.; Perrotta, G.; Fischer, T.C.; Bovy, A.G.; et al. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch. Biochem. Biophys. 2007, 465, 61–71. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. N. Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Pillet, J.; Yu, H.W.; Chambers, A.H.; Whitaker, V.M.; Folta, K.M. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 2015, 66, 4455–4467. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Ibdah, M.; Martens, S.; Gang, D.R. Biosynthetic pathway and metabolic engineering of plant dihydrochalcones. J. Agric. Food Chem. 2018, 66, 2273–2280. [Google Scholar] [CrossRef]
- Sun, M.; Sun, M.; Zhang, J. Osthole: An overview of its sources, biological activities, and modification development. Med. Chem. Res. 2021, 30, 1767–1794. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- da Silva, F.L.; Escribano-Bailón, M.T.; Alonso, J.J.P.; Rivas-Gonzalo, J.C.; Santos-Buelga, C. Anthocyanin pigments in strawberry. LWT-Food Sci. Technol. 2007, 40, 374–382. [Google Scholar] [CrossRef]
- Fischer, T.C.; Mirbeth, B.; Rentsch, J.; Sutter, C.; Ring, L.; Flachowsky, H.; Habegger, R.; Hoffmann, T.; Hanke, M.V.; Schwab, W. Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria × ananassa). N. Phytol. 2014, 201, 440–451. [Google Scholar] [CrossRef]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 651. [Google Scholar] [CrossRef] [Green Version]
- Medina-Puche, L.; Cumplido-Laso, G.; Amil-Ruiz, F.; Hoffmann, T.; Ring, L.; Rodríguez-Franco, A.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J.; Blanco-Portales, R. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 2014, 65, 401–417. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.R.; Kim, H.T.; Shanmugam, A.; Nath, U.K.; Goswami, G.; Song, J.Y.; Park, J.I.; Nou, I.S. Expression profiling of regulatory and biosynthetic genes in contrastingly anthocyanin rich strawberry (Fragaria × ananassa) cultivars reveals key genetic determinants of fruit color. Int. J. Mol. Sci. 2018, 19, 656. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, H.; Yang, Y.; Li, M.; Zhang, Y.; Liu, J.; Dong, J.; Li, J.; Butelli, E.; Xue, Z.; et al. The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnol. J. 2020, 18, 1169–1184. [Google Scholar] [CrossRef] [Green Version]
- Aharoni, A.; De Vos, C.H.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Y.; Ma, Y.; Yang, X.; Yin, X.; Zhang, Y.; Xiao, Y.; Liu, W.; Li, Y.; Li, S.; et al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 2020, 18, 2267–2279. [Google Scholar] [CrossRef] [Green Version]
- Gupta, O.P.; Karkute, S.G.; Banerjee, S.; Meena, N.L.; Dahuja, A. Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Front. Plant Sci. 2017, 8, 374. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Hazra, S.; Chattopadhyay, S. Identification of conserved miRNAs and their putative target genes in Podophyllum hexandrum (Himalayan Mayapple). Plant Gene 2016, 6, 82–89. [Google Scholar] [CrossRef] [Green Version]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Gao, Q.; Luo, H.; Li, Y.; Liu, Z.; Kang, C. Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol. J. 2020, 18, 1550–1561. [Google Scholar] [CrossRef] [Green Version]
- Ou, C.; Zhang, X.; Wang, F.; Zhang, L.; Zhang, Y.; Fang, M.; Wang, J.; Wang, J.; Jiang, S.; Zhang, Z. A 14 nucleotide deletion mutation in the coding region of the PpBBX24 gene is associated with the red skin of “Zaosu Red” pear (Pyrus pyrifolia White Pear Group): A deletion in the PpBBX24 gene is associated with the red skin of pear. Hortic. Res. 2020, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, Y.; Dou, Y.; Li, W.; Wang, S.; Shi, W.; Sun, Y.; Zhang, Z. Single nucleotide mutation in FvMYB10 may lead to the yellow fruit in Fragaria vesca. Mol. Breed. 2017, 37, 35. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef]
- Hawkins, C.; Caruana, J.; Schiksnis, E.; Liu, Z. Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep. 2016, 6, 29017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, J. A new variety of strawberry-Xiaobai (in Chinese). Shanxi Fruits 2014, 157, 46. [Google Scholar]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Alegre, S.; Pascual, J.; Trotta, A.; Angeleri, M.; Rahikainen, M.; Brosche, M.; Moffatt, B.; Kangasjärvi, S. Evolutionary conservation and post-translational control of S-adenosyl-L-homocysteine hydrolase in land plants. PLoS ONE 2020, 15, e0227466. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, L.; Chen, Q.; Li, Y.; Zhang, Y.; Luo, Y.; Zhang, Y.; Sun, B.; Wang, X.; Tang, H. Comparative Transcriptome Profiling Analysis of Red- and White-Fleshed Strawberry (Fragaria × ananassa) Provides New Insight into the Regulation of the Anthocyanin Pathway. Plant Cell Physiol. 2018, 59, 1844–1859. [Google Scholar] [CrossRef]
- Zhao, F.; Song, P.; Zhang, X.; Li, G.; Hu, P.; Aslam, A.; Zhao, X.; Zhou, H. Identification of candidate genes influencing anthocyanin biosynthesis during the development and ripening of red and white strawberry fruits via comparative transcriptome analysis. PeerJ 2021, 9, e10739. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The non-canonical aspects of microRNAs: Many roads to gene regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, A.; Bogdanovic, O. Enhancer DNA methylation: Implications for gene regulation. Essays Biochem. 2019, 63, 707–715. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, L.; Li, Y.; Chen, Q.; Ye, Y.; Zhang, Y.; Luo, Y.; Sun, B.; Wang, X.; Tang, H. Effect of red and blue light on anthocyanin accumulation and differential gene expression in strawberry (Fragaria × ananassa). Molecules 2018, 23, 820. [Google Scholar] [CrossRef] [Green Version]
- Skirycz, A.; Jozefczuk, S.; Stobiecki, M.; Muth, D.; Zanor, M.I.; Witt, I.; Mueller-Roeber, B. Transcription factor AtDOF4; 2 affects phenylpropanoid metabolism in Arabidopsis thaliana. N. Phytol. 2007, 175, 425–438. [Google Scholar] [CrossRef]
- Yang, Y.; He, Z.; Bing, Q.; Duan, X.; Chen, S.; Zeng, M.; Liu, X. Two Dof transcription factors promote flavonoid synthesis in kumquat fruit by activating C-glucosyltransferase. Plant Sci. 2022, 318, 111234. [Google Scholar] [CrossRef]
- Noguero, M.; Atif, R.M.; Ochatt, S. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef]
- Medina-Puche, L.; Molina-Hidalgo, F.J.; Boersma, M.; Schuurink, R.C.; López-Vidriero, I.; Solano, R.; Franco-Zorrilla, J.M.; Caballero, J.L.; Blanco-Portales, R.; Muñoz-Blanco, J. An R2R3-MYB transcription factor regulates eugenol production in ripe strawberry fruit receptacles. Plant Physiol. 2015, 168, 598–614. [Google Scholar] [CrossRef] [Green Version]
- Molina-Hidalgo, F.J.; Medina-Puche, L.; Cañete-Gómez, C.; Franco-Zorrilla, J.M.; López-Vidriero, I.; Solano, R.; Caballero, J.L.; Rodríguez-Franco, A.; Blanco-Portales, R.; Muñoz-Blanco, J.; et al. The fruit-specific transcription factor FaDOF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 2017, 68, 4529–4543. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, P.; Salvatierra, A.; Moya-León, M.A.; Herrera, R. Isolation of genes differentially expressed during development and ripening of Fragaria chiloensis fruit by suppression subtractive hybridization. J. Plant Physiol. 2010, 167, 1179–1187. [Google Scholar] [CrossRef]
- Li, J.; Ma, N.; An, Y.; Wang, L. FcMADS9 of fig regulates anthocyanin biosynthesis. Sci. Hortic. 2021, 278, 109820. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Mao, Z.; Liu, W.; Ding, L.; Zhang, X.; Yang, Y.; Wu, S.; Chen, X.; Wang, Y. Transcription factor McWRKY71 induced by ozone stress regulates anthocyanin and proanthocyanidin biosynthesis in Malus crabapple. Ecotoxicol. Environ. Saf. 2022, 232, 113274. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mao, W.; Liu, W.; Dai, H.; Liu, Y.; Ma, Y.; Zhang, Z. Deep sequencing discovery of novel and conserved microRNAs in wild type and a white-flesh mutant strawberry. Planta 2013, 238, 695–713. [Google Scholar] [CrossRef] [PubMed]
- Thill, J.; Miosic, S.; Gotame, T.P.; Mikulic-Petkovsek, M.; Gosch, C.; Veberic, R.; Preuss, A.; Schwab, W.; Stampar, F.; Stich, K.; et al. Differential expression of flavonoid 3′-hydroxylase during fruit development establishes the different B-ring hydroxylation patterns of flavonoids in Fragaria × ananassa and Fragaria vesca. Plant Physiol. Biochem. 2013, 72, 72–78. [Google Scholar] [CrossRef]
- Prior, R.L.; Fan, E.; Ji, H.; Howell, A.; Nio, C.; Payne, M.J.; Reed, J. Multi-laboratory validation of a standard method for quantifying proanthocyanidins in cranberry powders. J. Sci. Food Agric. 2010, 90, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, H.W.; Wang, X.R.; Xie, X.L.; Yue, X.Y.; Tang, H.R. An alternative cetyltrimethylammonium bromide-based protocol for RNA isolation from blackberry (Rubus L.). Genet. Mol. Res. 2012, 11, 1773–1782. [Google Scholar] [CrossRef]
- Ring, L.; Yeh, S.Y.; Hücherig, S.; Hoffmann, T.; Blanco-Portales, R.; Fouche, M.; Villatoro, C.; Denoyes, B.; Monfort, A.; Caballero, J.L.; et al. Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol. 2013, 163, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Spolaore, S.; Trainotti, L.; Casadoro, G. A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J. Exp. Bot. 2001, 52, 845–850. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Chen, F.Z.; You, L.J.; Yang, F.; Wang, L.N.; Guo, X.Q.; Gao, F.; Hua, C.; Tan, C.; Fang, L.; Shan, R.Q.; et al. CNGBdb: China national genebank database. Yi Chuan 2020, 42, 799–809. [Google Scholar] [CrossRef] [PubMed]

| No. | Compounds | Q1 1 (Da) | RT 2 (min) | Relative Quantification 3 | Fold Change | ||||
|---|---|---|---|---|---|---|---|---|---|
| XB | 35SN | HY | 35S::FaMYB10 | HY/XB | 35S::FaMYB10/35SN | ||||
| 1 | Pelargonidin 3-O-glucoside | 449.1 | 1.647 | (3.1 ± 1.0) × 104 | (3.0 ± 0.5) × 105 | (6.2 ± 0.4) × 106 | (9.9 ± 2.5) × 106 | 199.0166 | 33.5122 |
| 2 | Pelargonidin chloride | 287.0 | 3.773 | (1.3 ± 0.4) × 104 | (4.3 ± 0.4) × 104 | (5.0 ± 0.8) × 105 | (5.9 ± 0.6) × 105 | 38.6196 | 13.9370 |
| 3 | Pelargonidin 3,5-di-O-glucoside | 611.1 | 1.544 | 0 | (4.0 ± 1.4) × 103 | (2.8 ± 0.7) × 104 | (8.2 ± 1.5) × 104 | NA 4 | 20.3768 |
| 4 | Cyanidin O-rutinoside | 595.2 | 1.525 | (1.8 ± 0.2) × 104 | (5.3 ± 0.4) × 104 | (1.4 ± 0.1) × 105 | (6.6 ± 0.8) × 106 | 7.3731 | 123.0654 |
| 5 | Cyanidin O-acetylhexoside | 489.1 | 3.739 | (5.9 ± 3.1) × 104 | (9.3 ± 1.2) × 105 | (2.9 ± 0.2) × 106 | (4.4 ± 1.1) × 106 | 48.5049 | 4.7248 |
| 6 | Cyanidin O-syringic acid | 465.1 | 1.555 | (1.2 ± 0.2) × 104 | (3.5 ± 0.5) × 104 | (4.4 ± 0.3) × 105 | (4.0 ± 1.0) × 106 | 37.5528 | 115.2296 |
| 7 | Cyanidin-3-O-galactoside chloride | 465.1 | 1.560 | (1.1 ± 0.2) × 104 | (3.7 ± 0.6) × 104 | (3.9 ± 0.2) × 105 | (3.9 ± 1.0) × 106 | 34.0469 | 105.2074 |
| 8 | Cyanidin 3-O-malonylhexoside | 535.1 | 3.739 | (2.4 ± 1.0) × 104 | (3.8 ± 0.3) × 105 | (1.0 ± 0.1) × 106 | (2.0 ± 0.5) × 106 | 44.2875 | 5.1313 |
| 9 | Cyanidin 3-O-glucoside | 465.1 | 1.614 | 0 | 0 | (4.7 ± 0.2) × 104 | (5.8 ± 1.6) × 105 | NA | NA |
| 10 | Cyanidin 3-O-rutinoside chloride | 611.1 | 1.561 | 0 | 0 | (1.5 ± 0.2) × 104 | (9.0 ± 1.5) × 104 | NA | NA |
| 11 | Cyanidin chloride | 303.1 | 3.424 | 0 | 0 | (2.4 ± 0.2) × 104 | (6.8 ± 1.1) × 104 | NA | NA |
| 12 | Delphinidin O-malonylhexoside | 551.1 | 3.661 | 0 | (1.8 ± 0.2) × 104 | (8.7 ± 0.8) × 104 | (1.6 ± 0.3) × 106 | NA | 85.3311 |
| 13 | Delphinidin 3-β-d-Glucoside | 481.1 | 1.483 | (2.7 ± 1.0) × 103 | (3.5 ± 0.2) × 103 | (8.8 ± 2.5) × 103 | (2.8 ± 0.6) × 104 | 3.2300 | 7.8831 |
| 14 | Peonidin chloride | 317.1 | 3.812 | 0 | (2.2 ± 0.4) × 104 | (1.1 ± 0.4) × 105 | (1.1 ± 0.3) × 105 | NA | 5.1384 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Yue, M.; Liu, Y.; Ye, Y.; Zhang, Y.; Lin, Y.; Wang, X.; Chen, Q.; Tang, H. Alterations of Phenylpropanoid Biosynthesis Lead to the Natural Formation of Pinkish-Skinned and White-Fleshed Strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 2022, 23, 7375. https://doi.org/10.3390/ijms23137375
Jiang L, Yue M, Liu Y, Ye Y, Zhang Y, Lin Y, Wang X, Chen Q, Tang H. Alterations of Phenylpropanoid Biosynthesis Lead to the Natural Formation of Pinkish-Skinned and White-Fleshed Strawberry (Fragaria × ananassa). International Journal of Molecular Sciences. 2022; 23(13):7375. https://doi.org/10.3390/ijms23137375
Chicago/Turabian StyleJiang, Leiyu, Maolan Yue, Yongqiang Liu, Yuyun Ye, Yunting Zhang, Yuanxiu Lin, Xiaorong Wang, Qing Chen, and Haoru Tang. 2022. "Alterations of Phenylpropanoid Biosynthesis Lead to the Natural Formation of Pinkish-Skinned and White-Fleshed Strawberry (Fragaria × ananassa)" International Journal of Molecular Sciences 23, no. 13: 7375. https://doi.org/10.3390/ijms23137375
APA StyleJiang, L., Yue, M., Liu, Y., Ye, Y., Zhang, Y., Lin, Y., Wang, X., Chen, Q., & Tang, H. (2022). Alterations of Phenylpropanoid Biosynthesis Lead to the Natural Formation of Pinkish-Skinned and White-Fleshed Strawberry (Fragaria × ananassa). International Journal of Molecular Sciences, 23(13), 7375. https://doi.org/10.3390/ijms23137375






