Abstract
Understanding the photophysical properties and stability of near-infrared fluorescent proteins (NIR FPs) based on bacterial phytochromes is of great importance for the design of efficient fluorescent probes for use in cells and in vivo. Previously, the natural ligand of NIR FPs biliverdin (BV) has been revealed to be capable of covalent binding to the inherent cysteine residue in the PAS domain (Cys15), and to the cysteine residue introduced into the GAF domain (Cys256), as well as simultaneously with these two residues. Here, based on the spectroscopic analysis of several NIR FPs with both cysteine residues in PAS and GAF domains, we show that the covalent binding of BV simultaneously with two domains is the reason for the higher quantum yield of BV fluorescence in these proteins as a result of rigid fixation of the chromophore in their chromophore-binding pocket. We demonstrate that since the attachment sites are located in different regions of the polypeptide chain forming a figure-of-eight knot, their binding to BV leads to shielding of many sites of proteolytic degradation due to additional stabilization of the entire protein structure. This makes NIR FPs with both cysteine residues in PAS and GAF domains less susceptible to cleavage by intracellular proteases.
1. Introduction
Genetically encoded protein fluorescent markers are routinely used to study biological processes by selectively labeling cellular targets (proteins, organelles) involved in these processes [1,2]. An indisputable advantage of near-infrared (NIR) biomarkers is the possibility of their use in experiments on living model animals since the absorption and fluorescence spectra of these proteins fall within the “transparency window” of biological tissues—the spectral region in which cell components no longer absorb and water does not yet absorb [3].
Most of the currently available NIR biomarkers are developed from NIR fluorescent proteins (NIR FPs), which consist of chromophore-binding domains composed of two full-length bacterial phytochrome domains, PAS (Per-ARNT-Sim) and GAF (cGMP PDE/AC/FhlA) [1,4]. The choice of bacterial phytochromes for the development of NIR FPs is largely due to the fact that the natural ligand of bacterial phytochromes is biliverdin (BV), a product of endogenous synthesis from heme, which is always present in cells [5].
When BV interacts with NIR FP, it incorporates into the so-called chromophore-binding pocket formed by the PAS and GAF domains and covalently binds to inherent Cys15 in the N-terminal region of the PAS domain (CysPAS). However, these NIR FPs have a very low fluorescence quantum yield. To increase the fluorescence quantum yield, a mutant form of NIR FP was created that has a cysteine residue in the GAF domain (CysGAF), in a position equivalent to the attachment site of the plant and cyanobacteria photoreceptor (Cys256). The spectral properties of dimeric NIR FPs with all possible combinations of CysPAS and CysGAF have been studied [6,7]. It has been shown that the highest fluorescence quantum yield among BV-bearing NIR FPs exhibits those of them containing both cysteine residues, CysPAS and CysGAF. Crystallography of the monomeric protein miRFP670, containing both CysPAS and CysGAF, has revealed the covalent attachment of BV to these two residues simultaneously [8]. This type of BV attachment is believed to be relevant for the highest fluorescence quantum yield of BV-containing NIR FPs with CysPAS and CysGAF compared to other BV-containing NIR FPs.
To date, this assumption has been confirmed only by analyzing the Raman spectra of the iRFP682 (by the band shift at 1656 cm−1) [9]. We were surprised that the covalent binding of BV to two cysteine residues of NIR FPs was not confirmed directly by mass spectroscopy. In this regard, we decided to try to obtain the samples of several NIR FPs, in which BV is covalently bound simultaneously to CysPAS and CysGAF, and to more accurately determine the spectral characteristics of these proteins. These NIR FPs include a monomeric protein BphP1-FP, and dimeric proteins iRFP713/V256C and iRFP670 derived from bacterial phytochromes RpBphP1, RpBphP2, and RpBphP6 from Rhodopseudomonas palustris, respectively. In this work, we analyzed the resistance of NIR FPs containing two cysteine residues to proteases and revealed the reasons for their high resistance to proteolytic cleavage.
2. Results and Discussion
2.1. Preparation of NIR FPs Enriched in CysGAF-BV-CysPAS
Monomeric [8] and dimeric (Figure 1) NIR FPs with two cysteine residues in the PAS and GAF domains can bind their chromophore, BV, in two different ways: covalently through the C32 atom of BV to CysGAF (BV-CysGAF) and covalently simultaneously through the C31 atom of BV to CysGAF and the C32 atom of BV to CysPAS (CysGAF-BV-CysPAS). The ratio of NIR FP molecules, in which the mode of BV binging differs, is approximately equimolar. The presence of two types of protein molecules in approximately equal amounts is evidenced by two bands of the same total intensity on the gels visualizing the separation of these proteins under denaturing conditions (Figure 1). At the same time, the band of higher electrophoretic mobility corresponds to NIR FP monomers containing BV covalently bound to two cysteine residues simultaneously. The covalent attachment of BV simultaneously to the two cysteine residues in the PAS and GAF domains of the NIR FP is supposed to result in an inability to untie the unique figure-eight knot [10] encompassing these domains [8], which makes the denatured state of the NIR FP rather compact.
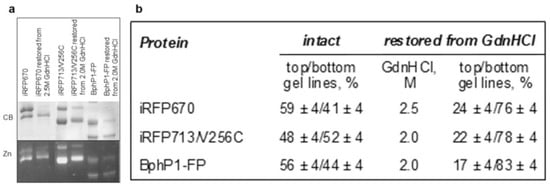
Figure 1.
The content of two types of molecules containing BV-CysGAF or CysGAF-BV-CysPAS in NIR FPs with two cysteine residues in the PAS and GAF domains in their original state and treated with GdnHCl at moderate concentrations followed by restoration. (a) The SDS PAGE of NIR FP samples followed by staining with Coomassie blue (CB) and detection of zinc-induced fluorescence (Zn). (b) The proportion of NIR FP monomers that contain BV-CysGAF or CysGAF-BV-CysPAS.
To analyze the effect of covalent BV binding simultaneously with two cysteine residues on the spectral properties of NIR FPs, we obtained protein samples with increased content of CysGAF-BV-CysPAS. This was achieved using the procedure proposed by Buhrke for iRFP682 [9], which was modified by taking into account the results of our studies of the unfolding–refolding processes and the structural stability of NIR FPs [7,11]. We have previously shown that dimeric and monomeric NIR FPs containing both cysteine residues in the PAS and GAF domains are more resistant to chemical denaturing agents compared to those NIR FPs with cysteine residues capable of covalent binding the chromophore either in the PAS domain or in the GAF domain or missing [7,11]. The stabilization of NIR FPs in the presence of two cysteine residues is likely attributed to the binding of two regions of the polypeptide chain at the formation of CysGAF-BV-CysPAS, which fastens both domains of the proteins together [12]. With this in mind, we chose the conditions (denaturing agent concentration) for the incubation of NIR FPs with two cysteine residues under which their monomers containing BV-CysGAF would predominantly denature, while monomers containing CysGAF-BV-CysPAS would retain its intact structure. We propose another rather simple approach for the subsequent separation of denatured and native NIR FP molecules instead of the gel filtration [9], based on the fact that the covalent attachment of a chromophore leads to irreversible denaturation of NIR FPs [13]. According to this procedure, after incubation of the analyzed NIR FPs in the presence of 2–2.5 M GdnHCl (Figure 1b), the denaturant is removed from the solution by the method of equilibrium microdialysis. The change in the composition of the buffer stimulates the aggregation of denatured NIR FP molecules due to the accumulation of misfolded forms of the protein. Indeed, the samples of NIR FPs analyzed in this work treated in this way showed the presence of a clearly visible precipitate. This fraction of aggregates in the samples of NIR FPs is easily removed by centrifugation.
After the above-described processing of NIR FPs and their separation by SDS PAGE, the assessment of the total intensity of the bands corresponding to molecules containing BV-CysGAF and CysGAF-BV-CysPAS indicated an increase in the content of the latter molecules from 50 to 80% (Figure 1). Thus, according to the procedure described above, NIR FP samples were obtained, the monomers of which contained mainly CysGAF-BV-CysPAS (Figure 1).
2.2. Spectral Properties of NIR FPs Containing Predominantly CysGAF-BV-CysPAS
The spectral properties of NIR FPs enriched in CysGAF-BV-CysPAS were characterized by absorption, fluorescence spectroscopy, and circular dichroism (CD). The absorption and fluorescence spectra of NIR FPs enriched in CysGAF-BV-CysPAS almost coincided with the spectra of the original NIR FPs (Figure 2). However, the absorption spectra of dimeric NIR FPs containing predominantly CysGAF-BV-CysPAS were narrower compared to the spectra of the original proteins. This confirms the data obtained earlier by X-ray diffraction analysis on the structure of BV-CysGAF and CysGAF-BV-CysPAS and the identity of a system of conjugated π-bonds in them, which determines the spectral similarity of differently bound chromophore [8].
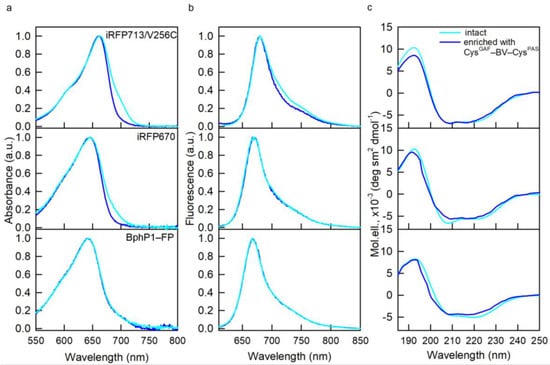
Figure 2.
Spectral properties of NIR FPs enriched with CysGAF-BV-CysPAS (curves in blue). (a) Absorption spectra of the chromophore. (b) chromophore fluorescence spectra (λex = 590 nm). (c) CD spectra in the spectral far-UV region. The spectral characteristics of the original NIR FPs containing both cysteine residues in the PAS and GAF domains are also shown (cyan curves).
According to the data of far-UV CD, NIR FPs containing predominantly CysGAF-BV-CysPAS retain the secondary structure inherent in the original NIR FPs (Figure 2). The tryptophan fluorescence spectra of NIR FPs containing predominantly CysGAF-BV-CysPAS were blue-shifted by 1–3 nm relative to the spectra of the original proteins (Table 1). The blue shift of the tryptophan spectrum indicates an increase in the rigidity of the microenvironment of protein tryptophan residues and a decrease in their accessibility to the solvent [14,15]. These data testify to some compaction of the structure of NIR FPs containing predominantly CysGAF-BV-CysPAS; however, there was observed no significant change in the spatial structure of these proteins compared to NIR FPs with equimolar content of CysGAF-BV-CysPAS and BV-CysGAF.

Table 1.
Parameters of the chromophore fluorescence of NIR FPs in the holoform, containing CysPAS and CysGAF, original and restored after GdnHCl-induced denaturation.
The overlap of the CD spectra in the visible region for NIR FPs containing predominantly CysGAF-BV-CysPAS (Figure 3) and their initial forms points at the same 15Z-configuration of BV-CysGAF and CysGAF-BV-CysPAS, which is characteristic of the Pr-form of phytochromes [16,17,18]. Nevertheless, narrower CD spectra in the near UV region of NIR FPs containing predominantly CysGAF-BV-CysPAS compared to the spectra of the original proteins stand out (Figure 3). This implies a greater rigidity of the microenvironment of CysGAF-BV-CysPAS compared to BV-CysGAF, since the optical activity of NIR probes in this spectral range is linked to the interaction of the pyrrole rings of their chromophore with aromatic residues of the protein [13,17].
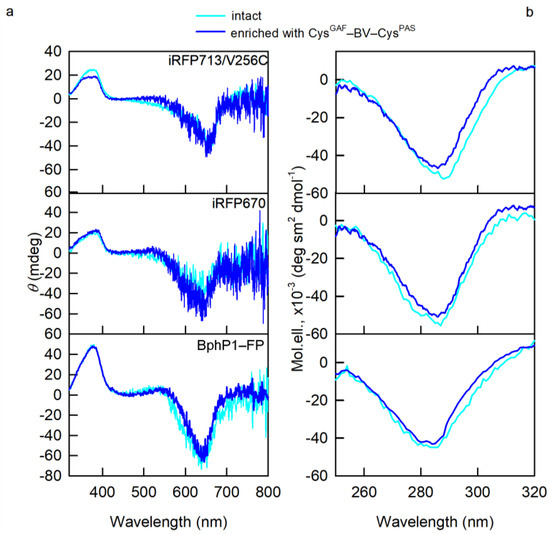
Figure 3.
Properties of BV environment in NIR FPs enriched with CysGAF-BV-CysPAS (curves in blue). (a) CD spectra in the visible spectral region (b) CD spectra in the near-UV spectral region. The spectral characteristics of the original NIR FPs containing both cysteine residues in the PAS and GAF domains are also shown (cyan curves).
The fluorescence quantum yield of NIR FPs containing predominantly CysGAF-BV-CysPAS exceeded by 1–2% this characteristic for NIR FPs containing both CysGAF-BV-CysPAS and BV-CysGAF in an equimolar ratio (Table 1). An increase in the fluorescence quantum yield of a NIR FP upon its saturation with CysGAF-BV-CysPAS also correlated with a higher fluorescence lifetime of the protein (Table 1) [19,20].
Taken together, the obtained data confirm that CysGAF-BV-CysPAS exhibits a higher fluorescence quantum yield compared to BV bound in NIR FPs differently. Our findings demonstrate also that the increased quantum yield of the former is associated with its rigid fixation in the GAF domain of the analyzed NIR FPs, which leads to a decrease in the efficiency of nonradiative deactivation of the chromophore excited state [20,21]. These data are consistent with the results of a study of the structural properties of the iRFP682 chromophore involved in covalent binding to both cysteine residues in the GAF and PAS domains of the protein using resonance Raman spectroscopy [9]. According to data of SDS PAGE (Figure 1), the content of two types of molecules with BV-CysGAF or CysGAF-BV-CysPAS may differ slightly for iRFP713/V256C, iRFP670, and BphP1-FP. This could be one of the reasons for the higher quantum yield of iRFP713/V256C compared to BphP1-FP and iRFP670. According to the results of our previous studies, the way BV binds to NIR FPs may depend on the amino acid composition of the GAF domain pocket [22]. The influence of the microenvironment of the BV chromophore in NIR FPs on the dynamics of the chromophore and, consequently, the efficiency of nonradiative deactivation of its excited state should not be ruled out [20,23]. In particular, it is known that the residue at position 204 (numbering of amino acids is given according to the RpBphP2 sequence) is directly involved in the hydrogen bond network with the chromophore and other amino acids in NIR FPs and, in addition, can affect the number of water molecules inside the pocket of the GAF domain, also impacting the proton wire proteins [21,24,25,26,27].
2.3. Effect of CysGAF-BV-CysPAS on NIR FPs Stability in the Cell
Obtaining NIR FPs with increased content of CysGAF-BV-CysPAS by selective denaturation of monomers with CysGAF-BV in the parent NIR FPs implies that the cross-linking of the PAS and GAF domains through simultaneous binding of cysteine residues in them to the chromophore indeed stabilizes the NIR FP structure. The stability of the biomarker in the cell, in addition to the stability of its structure, is affected by its resistance to the action of proteolytic enzymes. Therefore, we analyzed the influence of proteases, trypsin, and chymotrypsin, on the structure of NIR FPs containing both cysteine residues in the PAS and GAF domains using iRFP670 as an example. Protease-treated samples were analyzed using the Tricine-SDS polyacrylamide gel electrophoresis, used to separate peptides over a wide range of molecular weights [28]. The iRFP670 protein has 33 and 22 cleavage sites of trypsin and chymotrypsin, respectively (Figure S1). The peptide size of iRFP670 processed with proteases is calculated considering the presence of BV differently bound to protein monomers, CysGAF-BV and CysGAF-BV-CysPAS (Table S1).
We found that only bands corresponding to the target uncleaved protein were visualized on gels of iRFP670 in the holoform after its treatment with proteases (Figure S2, gel 1). The pre-incubation of the protein under denaturing conditions (5 M urea) to destabilize its structure [29,30] did not promote protein cleavage by proteases (Figure S2, gel 1). This indicates a high resistance of iRFP670 in the holoform to the action of proteases.
Partial cleavage of iRFP670 by chymotrypsin, but not trypsin, in the presence of ProteaseMAX™ Surfactant (Trypsin Enhancer) was observed [31,32,33] (Figure S2, gel 2 and 3). It is believed that this chemical increases the efficiency of several proteolytic enzymes by destabilizing the structure of the analyzed proteins and solubilizing them [31]. We found the temperatures of iRFP670 incubation in the presence of chymotrypsin should be lowered to avoid autolysis of the protease (Figure S2, gel 4). In the solution of iRFP670 cleaved with chymotrypsin, peptides were found mainly with a molecular weight of about 14–15 kDa and higher (Figure 4). These peptides are modified with a covalently attached chromophore as evidenced by Zn-induced fluorescence of the corresponding electrophoretic bands (Figure 4). The analyzed solution contained trace amounts of peptides with a molecular weight of about 12 kDa and 7–10 kDa with a covalently attached chromophore in their composition (Figure 4).
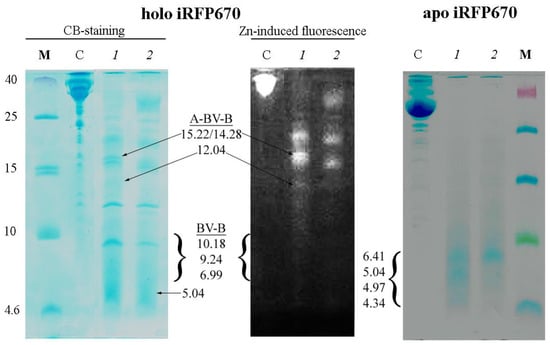
Figure 4.
Cleavage of iRFP670 in the holoform (left panels) and the apoform (right panel) with chymotrypsin. Tricine-SDS PAGE of protein samples followed by staining with Coomassie blue (CB) and detection of zinc-induced fluorescence (Zn). The following samples were loaded into the wells: M—marker peptides, C—control samples of the original protein, 1 and 2—iRFP670 samples cleaved at 4 and 20 °C. The expected size of the detected peptides and the nature of BV binding to them are indicated (see also Table 2).
To clarify the reasons for partial proteolysis of iRFP670, we analyzed the location of chymotrypsin cleavage sites in the protein using crystallographic data on the structure of phytochromes (Table 2, Figure 5). We discovered potential steric inaccessibility for the action of chymotrypsin at some cleavage sites in the protein sequence. Thus, the inaccessibility of the Y213 cleavage site is likely associated with its location near one of the intersections of the protein polypeptide chain that forms the knot (Figure 5). The F247 cleavage site is located near another intersection of the polypeptide chain in a loop through which the N-terminal segment of the protein is pulled (Figure 5). These two cleavage sites are located N-terminally from the CysGAF residue. According to the calculation, the solution of iRFP670 not hydrolyzed by chymotrypsin at Y213 and F247 sites should contain peptides with a covalently attached chromophore with a molecular weight of 6.99 (peptide B-BV) and 12.04 kDa (peptide B-BV-peptide A) (Table 2). The protease access to the F260 cleavage site located at the C-terminus of CysGAF may be limited by the covalent binding of BV in the protein (Figure 5), which will lead to the appearance in the analyzed solution of peptides with a covalently attached chromophore of a size of 9.24 (peptide B-BV) and 14.28 kDa (peptide B-BV-peptide A) (Table 2). The W281 cleavage site which is C-terminal to CysGAF is buried deep within the protein globule in its GAF domain (Figure 5). In the case of non-hydrolysis of the iRFP670 polypeptide chain at all of the sites listed, including Y213, F247, F260, and W281, the size of the anticipated protein fragments will be 10.18 kDa (peptide B-BV) and 15.22 kDa (peptide B-BV-peptide A) (Table 2). At the cleavage of iRFP670 monomers containing CysGAF-BV, a peptide with a CysPAS residue without a covalently attached chromophore and with a molecular weight of about 5.04 kDa is also expected (Table S1).

Table 2.
Estimated size of BV-containing peptides obtained by partial and complete cleavage of iRFP670 in the holoform with chymotrypsin (cleavage after residues F, Y, and W).
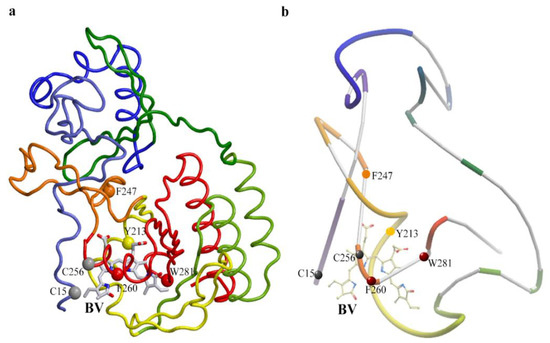
Figure 5.
Location of proteolytic cleavage sites in the iRFP670 structure that is potentially inaccessible to chymotrypsin. (a) Spatial structure of the protein. (b) The schematic representation of the structure of the protein shows the knot topology. The figure was created based on X-ray data of PAS-GAF domains of RpBphP2 protein from Rhodopseudomonas palustris (PDB: 4e04 file [34]) using the graphic software VMD (Visual Molecular Dynamics) [35], Raster3D [36] and ICM Pro [37]. Chymotrypsin cleavage sites Y213, F247, F260, and W281 are shown as van der Waals spheres (CA atoms of residues are shown). BV chromophore is displayed by sticks.
Considering the structural analysis performed, the presence on the gel of intense Zn-positive bands corresponding to peptides of 14–15 kDa, barely pronounced Zn-positive bands corresponding to peptides of 12 kDa and 7–10 kDa, as well as Zn-negative band corresponding to the peptide of 5 kDa can be interpreted in favor of only a marginal cleavage of the protein monomer with CysGAF-BV-CysPAS, and almost complete cleavage of the monomer with CysGAF-BV (Figure 4). Our data are in good agreement with the literature on the presence of two types of chromophore binding in dimeric and monomeric NIR FPs with two cysteine residues CysPAS and CysGAF [8,12]. The large size of chromophore-bearing peptides obtained by cleavage of iRFP670 with chymotrypsin also is in good agreement with the assumption of simultaneous covalent attachment of BV to cysteine residues in both domains in NIR FPs.
In the solution of iRFP670 in the apoform (free of the chromophore) treated with chymotrypsin in the presence of ProteaseMAX™ Surfactant, short peptides with a size between 4.6 and 10 kDa were detected (Figure 4). According to the analysis of the amino acid sequence of iRFP670, only peptide A with a molecular mass of 5.04 kDa falls into this size range (Table S2). Complete hydrolysis by the protease of the rest of the apoprotein polypeptide chain is expected to yield peptides with a molecular mass of no more than 4.34 kDa (Table S2). In the case of incomplete hydrolysis of the apoprotein iRFP670, when cleavage sites Y213 and F247, or only F247, located at the intersecting sections of the polypeptide chain in the knot, are omitted, peptides with a molecular weight of 6.41 and 4.97 kDa (peptide B variants) should be formed, respectively (Table S2). This situation is likely realized in our case (Figure 4).
Our findings also highlight the possible function of the knot in the polypeptide chain of the chromophore-binding domain of phytochromes in increasing the resistance of these proteins to proteolytic degradation. Our data also show that the simultaneous covalent binding of BV to both domains of the NIR FPs with two cysteine residues CysPAS and CysGAF enhances significantly the stabilizing effect of the knot, making these proteins almost insensitive to the action of proteases.
3. Conclusions
In this work, we analyzed the effect of the covalent binding of BV to two cysteine residues in the PAS and GAF domains in NIR FPs on the structural and spectral properties of these proteins. Previously, the allosteric influence of protein monomers in dimeric NIR FPs on each other’s structure has been shown to govern the nature of BV binding in them [7]. Indeed, dimeric NIR FPs, in which only one of the cysteine residues in the PAS or GAF domains is present, have BV covalently bound to protein only in one monomer, while the chromophore in the second protein monomer is incorporated into the pocket of the GAF domain, but is not fixed by a covalent bond. The reason for this is that the covalent binding of BV in one monomer of the dimeric NIR FP bearing one cysteine residue may prevent the subsequent formation of a covalent bond between BV and another protein monomer. This explains the low molecular brightness of NIR FPs with one cysteine residue [7]. In dimeric NIR FPs with two cysteine residues, inhibition of covalent binding of the chromophore is not observed; BV in such proteins is covalently bound to both monomers. The absence of BV not covalently bound to the protein in NIR FPs with both cysteine residues CysPAS and CysGAF contributes to their higher molecular brightness compared to NIR FPs bearing one of these two cysteine residues or without them [7].
The analysis in the current work of several NIR FPs with two cysteine residues CysPAS and CysGAF corroborated with earlier studies of individual NIR FPs of this group [8,9], indicates that the quantum yield of their fluorescence is additionally affected by the covalent binding of BV simultaneously with two domains in these proteins, which leads to rigid fixation of the chromophore in their chromophore-binding pocket. It should be noted that, according to the literature, the kinetics of covalent binding of the chromophore depends on the number of cysteine residues in NIR FPs: the rate of covalent attachment of BV to CysGAF is significantly higher in proteins containing both CysGAF and CysPAS [38]. In that work, it has been suggested that as a result of rapid covalent fixation of BV in NIR FPs with two cysteine residues, they avoid the formation of non-fluorescent complexes with protoporphyrin in the cell, which increases their brightness [38]. Thus, several molecular mechanisms are responsible for the higher brightness of dimeric NIR FPs with two cysteine residues CysPAS and CysGAF compared to other BV-containing NIR FPs.
Our data indicate that the covalent binding of BV simultaneously to two regions of the polypeptide chain, which additionally forms a figure-of-eight knot, in NIR FPs with two cysteine residues CysPAS and CysGAF leads to the screening of many sites of proteolytic degradation in them. As a result, covalent binding of BV in NIR FPs simultaneously with two cysteine residues CysPAS and CysGAF not only stabilizes their structure but also increases their resistance to proteolytic degradation in the cell, which is important for their use as fluorescent tags in the cell.
4. Materials and Methods
4.1. Materials
Guanidine hydrochloride (GdnHCl), buffer components, and trypsin from bovine pancreas were purchased from Sigma (St. Louis, MO, USA) and used without further purification. Chymotrypsin and ProteaseMAX™ Surfactant (Trypsin Enhancer) were purchased from Promega (Madison, WI, USA).
4.2. Protein Expression and Purification
The target NIR FP genes, containing N-terminal polyhistidine tag, were amplified and cloned into a pBAD/His-B vector (Invitrogen, Waltham, MA, USA) using BglII and EcoRI sites expression in LMG194 host cells (Invitrogen, Waltham, MA, USA). NIR FPs are expressed and purified as previously described [7,39]. Briefly, NIR FPs in apo, (i.e., in the absence of a chromophore) and holoform, (i.e., in a complex with biliverdin) were obtained by expressing only the target protein gene and together with the hemoxygenase (HO) gene from the pWA23h-HO vector, respectively. The HO enzyme provides BV synthesis. The RM medium (48 mM Na2HPO4, 22 mM KH2PO4, 19 mM NH4Cl, 8.5 mM NaCl, 2% casamino acids, 1 mM MgCl2, 1 mM thiamine) supplemented with ampicillin was used for the expression of NIR FPs in the apoforms. Ampicillin and kanamycin were included in the culture medium for the expression of NIR FPs in holoform. Arabinose and rhamnose, respectively, were used for induction of the NIR FPs and HO expression. Cell lysates were purified with sequential affinity chromatography on a His Gravitrap column (GE Healthcare, Chicago, IL, USA) and ion-exchange chromatography on a MonoQ column (GE Healthcare, Chicago, IL, USA). The purity of the proteins was tested by SDS-PAGE using 15% polyacrylamide gels [40]. The protein solutions were dialyzed and stored in a 20 mM Tris/HCl solution, pH 8.0.
4.3. Spectral Measurements
Absorption spectra of NIR FPs were acquired using a U-3900H spectrophotometer (Hitachi, Tokyo, Japan) with microcells 101.016-QS 5 × 5 mm (Hellma, Müllheim, Germany).
The fluorescence spectra were measured using a Cary Eclipse spectrofluorimeter (Varian, Melbourne, Australia) with cells 10 × 10 × 4 mm (Starna, Atascadero, CA, USA). The raw fluorescence intensity of BV was corrected for the secondary inner filter effect [41]. The extinction coefficient of NIR FPs was determined by comparing the absorbance values at the far-red peak with the absorbance value at the Soret peak at about 390 nm. The extinction coefficient of tested proteins at the Soret band was supposed to be equal to that of free BV (39,900 M−1·cm−1). The fluorescence quantum yield of NIR FPs was calculated using iRFP713 as standard.
Tryptophan fluorescence spectra of NIR FPs were recorded using 295 nm excitation wavelengths and corrected for instrument sensitivity. The position of these spectra was determined by recording the parameter A = I320/I365, i.e., the ratio of fluorescence intensities on two slopes of the protein tryptophan fluorescence spectrum [42]. With this approach, a tiny spectral shift of 1 nm can be reliably detected.
Chromophore fluorescence decay curves NIR FPs were collected using a spectrometer FluoTime 300 (PicoQuant, Berlin, Germany) with the Laser Diode Head LDH-C-375 (λex = 375 nm) or LDH-C-660 (λex = 660 nm). The fitting of fluorescence decay curves was performed using the standard convolute-and-compare nonlinear least-squares procedure [43] with minimization according to Marquardt [44].
CD spectra were obtained using a Jasco-810 spectropolarimeter (Jasco, Tokyo, Japan). The far-UV CD spectra were recorded in a 1 mm path length cell from 250 to 190 nm, with a step size of 0.1 nm. The near-UV CD spectra were recorded in a 10 mm path length cell from 320 to 250 nm, with a step size of 0.1 nm. The visible CD spectra were scanned from 800 to 320 nm, with a step size of 0.1 nm, using a 10 mm path length cell. An average of three scans was obtained for all spectra. The CD spectra of the appropriate buffer solution were recorded and subtracted from the protein spectra.
4.4. Biochemical Measurements
The chromophore binding with NIR FPs was assayed by zinc-induced fluorescence and staining with Coomassie blue of protein samples separated by SDS-PAGE [45]. The content of NIR FPs with covalently attached BV was evaluated in ImageJ on the total intensity of the bands visualized with CB staining.
Protease-treated samples were analyzed by the 16/6% Tricine-SDS polyacrylamide gel electrophoresis, used to separate peptides over a wide range of molecular weights [28]. The peptides with a known molecular weight of up to 40 kDa were used as markers. Protein samples were incubated with a protease in a ratio of 20:1 overnight at 37 °C for trypsin and 50 °C for chymotrypsin (an optimum temperature for enzyme activity). Some samples were pre-incubated with urea at a concentration of 5 M at 50 °C for 30 min before adding the protease. Alternatively, samples were pre-incubated with 0.2, 0.1, and 0.025% ProteaseMAX™ Surfactant at 37 °C for 7 h without urea or with 2 M urea added. The samples thus prepared were mixed with proteases in a ratio of 20:1 and incubated overnight at 4, 20, 37, or 50 °C.
4.5. Analysis of Protein 3D Structure
The X-ray data of the PAS-GAF domains of the bacterial phytochrome RpBphP2 (4E04.ent file [34] from PDB [46]) were used to analyze the location of proteolytic cleavage sites in NIR FPs.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/ijms23137347/s1.
Author Contributions
Conceptualization and supervision: K.K.T., O.V.S. (Olesya V. Stepanenko) and I.M.K.; methodology: K.K.T., I.M.K., and O.V.S. (Olesya V. Stepanenko); investigation and visualization: O.V.S. (Olesya V. Stepanenko) and O.V.S. (Olga V. Stepanenko); writing—original draft preparation, review, and editing: O.V.S. (Olesya V. Stepanenko), O.V.S. (Olga V. Stepanenko), I.M.K. and K.K.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernov, K.G.; Redchuk, T.A.; Omelina, E.S.; Verkhusha, V.V. Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes. Chem. Rev. 2017, 117, 6423–6446. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001, 19, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Verkhusha, V.V. Near-Infrared Fluorescent Proteins: Multiplexing and Optogenetics across Scales. Trends Biotechnol. 2018, 36, 1230–1243. [Google Scholar] [CrossRef] [Green Version]
- Kapitulnik, J.; Maines, M.D. The role of bile pigments in health and disease: Effects on cell signaling, cytotoxicity, and cytoprotection. Front. Pharmacol. 2012, 3, 136. [Google Scholar] [CrossRef] [Green Version]
- Shcherbakova, D.M.; Baloban, M.; Pletnev, S.; Malashkevich, V.N.; Xiao, H.; Dauter, Z.; Verkhusha, V.V. Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins. Chem. Biol. 2015, 22, 540–1551. [Google Scholar] [CrossRef] [Green Version]
- Stepanenko, O.V.; Baloban, M.; Bublikov, G.S.; Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Kuznetsova, I.M.; Verkhusha, V.V. Allosteric effects of chromophore interaction with dimeric near-infrared fluorescent proteins engineered from bacterial phytochromes. Sci. Rep. 2016, 6, 18750. [Google Scholar] [CrossRef] [Green Version]
- Baloban, M.; Shcherbakova, D.M.; Pletnev, S.; Pletnev, V.Z.; Lagarias, J.C.; Verkhusha, V.V. Designing brighter near-infrared fluorescent proteins: Insights from structural and biochemical studies. Chem. Sci. 2017, 8, 4546–4557. [Google Scholar] [CrossRef] [Green Version]
- Buhrke, D.; Tavraz, N.N.; Shcherbakova, D.M.; Sauthof, L.; Moldenhauer, M.; Velazquez Escobar, F.; Verkhusha, V.V.; Hildebrandt, P.; Friedrich, T. Chromophore binding to two cysteines increases quantum yield of near-infrared fluorescent proteins. Sci. Rep. 2019, 9, 1866. [Google Scholar] [CrossRef]
- Jamroz, M.; Niemyska, W.; Rawdon, E.J.; Stasiak, A.; Millett, K.C.; Sulkowski, P.; Sulkowska, J.I. KnotProt: A database of proteins with knots and slipknots. Nucleic Acids Res. 2015, 43, D306–D314. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Stepanenko, O.V.; Bublikov, G.S.; Kuznetsova, I.M.; Verkhusha, V.V.; Turoverov, K.K. Stabilization of structure in near-infrared fluorescent proteins by binding of biliverdin chromophore. J. Mol. Struct. 2017, 1140, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Shcherbakova, D.M.; Verkhusha, V.V.; Turoverov, K.K. Interaction of Biliverdin Chromophore with Near-Infrared Fluorescent Protein BphP1-FP Engineered from Bacterial Phytochrome. Int. J. Mol. Sci. 2017, 18, 1009. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Bublikov, G.S.; Stepanenko, O.V.; Shcherbakova, D.M.; Verkhusha, V.V.; Turoverov, K.K.; Kuznetsova, I.M. A knot in the protein structure-probing the near-infrared fluorescent protein iRFP designed from a bacterial phytochrome. FEBS J. 2014, 281, 2284–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsova, I.M.; Yakusheva, T.A.; Turoverov, K.K. Contribution of separate tryptophan residues to intrinsic fluorescence of actin. Analysis of 3D structure. FEBS Lett. 1999, 452, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Turoverov, K.K.; Kuznetsova, I.M.; Zaitsev, V.N. The environment of the tryptophan residue in Pseudomonas aeruginosa azurin and its fluorescence properties. Biophys. Chem. 1985, 23, 79–89. [Google Scholar] [CrossRef]
- Bjorling, S.C.; Zhang, C.F.; Farrens, D.L.; Song, P.S.; Kliger, D.S. Time-resolved circular dichroism of native oat phytochrome photointermediates. J. Am. Chem. Soc. 1992, 114, 4581–4588. [Google Scholar] [CrossRef]
- Borucki, B.; Otto, H.; Rottwinkel, G.; Hughes, J.; Heyn, M.P.; Lamparter, T. Mechanism of Cph1 phytochrome assembly from stopped-flow kinetics and circular dichroism. Biochemistry 2003, 42, 13684–13697. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Shang, L.; Martin, S.S.; Lagarias, J.C. Distinct classes of red/far-red photochemistry within the phytochrome superfamily. Proc. Natl. Acad. Sci. USA 2009, 106, 6123–6127. [Google Scholar] [CrossRef] [Green Version]
- Toh, K.C.; Stojkovic, E.A.; Rupenyan, A.B.; van Stokkum, I.H.; Salumbides, M.; Groot, M.L.; Moffat, K.; Kennis, J.T. Primary reactions of bacteriophytochrome observed with ultrafast mid-infrared spectroscopy. J. Phys. Chem. A 2011, 115, 3778–3786. [Google Scholar] [CrossRef]
- Toh, K.C.; Stojkovic, E.A.; van Stokkum, I.H.; Moffat, K.; Kennis, J.T. Proton-transfer and hydrogen-bond interactions determine fluorescence quantum yield and photochemical efficiency of bacteriophytochrome. Proc. Natl. Acad. Sci. USA 2010, 107, 9170–9175. [Google Scholar] [CrossRef] [Green Version]
- Toh, K.C.; Stojkovic, E.A.; van Stokkum, I.H.; Moffat, K.; Kennis, J.T. Fluorescence quantum yield and photochemistry of bacteriophytochrome constructs. Phys. Chem. Chem. Phys. 2011, 13, 11985–11997. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Stepanenko, O.V.; Turoverov, K.K.; Kuznetsova, I.M. Probing the allostery in dimeric near-infrared biomarkers derived from the bacterial phytochromes: The impact of the T204A substitution on the inter-monomer interaction. Int. J. Biol. Macromol. 2020, 162, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Buhrke, D.; Velazquez Escobar, F.; Sauthof, L.; Wilkening, S.; Herder, N.; Tavraz, N.N.; Willoweit, M.; Keidel, A.; Utesch, T.; Mroginski, M.A.; et al. The role of local and remote amino acid substitutions for optimizing fluorescence in bacteriophytochromes: A case study on iRFP. Sci. Rep. 2016, 6, 28444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auldridge, M.E.; Satyshur, K.A.; Anstrom, D.M.; Forest, K.T. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 2012, 287, 7000–7009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velazquez Escobar, F.; Hildebrandt, T.; Utesch, T.; Schmitt, F.J.; Seuffert, I.; Michael, N.; Schulz, C.; Mroginski, M.A.; Friedrich, T.; Hildebrandt, P. Structural parameters controlling the fluorescence properties of phytochromes. Biochemistry 2014, 53, 20–29. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Auldridge, M.E.; Lehtivuori, H.; Ihalainen, J.A.; Forest, K.T. Origins of fluorescence in evolved bacteriophytochromes. J. Biol. Chem. 2014, 289, 32144–32152. [Google Scholar] [CrossRef] [Green Version]
- Feliks, M.; Lafaye, C.; Shu, X.; Royant, A.; Field, M. Structural Determinants of Improved Fluorescence in a Family of Bacteriophytochrome-Based Infrared Fluorescent Proteins: Insights from Continuum Electrostatic Calculations and Molecular Dynamics Simulations. Biochemistry 2016, 55, 4263–4274. [Google Scholar] [CrossRef]
- Schagger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef]
- Proctor, V.A.; Cunningham, F.E. The chemistry of lysozyme and its use as a food preservative and a pharmaceutical. Crit. Rev. Food Sci. Nutr. 1988, 26, 359–395. [Google Scholar] [CrossRef]
- Mine, Y.; Ma, F.; Lauriau, S. Antimicrobial peptides released by enzymatic hydrolysis of hen egg white lysozyme. J. Agric. Food Chem. 2004, 52, 1088–1094. [Google Scholar] [CrossRef]
- Diaz-Gonzalez, F.; Milano, M.; Olguin-Araneda, V.; Pizarro-Cerda, J.; Castro-Cordova, P.; Tzeng, S.C.; Maier, C.S.; Sarker, M.R.; Paredes-Sabja, D. Protein composition of the outermost exosporium-like layer of Clostridium difficile 630 spores. J. Proteom. 2015, 123, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.B.; Pagana, J.; Gillespie, P.G. Twist-off purification of hair bundles. Methods Mol. Biol. 2009, 493, 241–255. [Google Scholar] [PubMed]
- Krey, J.F.; Sherman, N.E.; Jeffery, E.D.; Choi, D.; Barr-Gillespie, P.G. The proteome of mouse vestibular hair bundles over development. Sci. Data 2015, 2, 150047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellini, D.; Papiz, M.Z. Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. Acta Cryst. D Biol Cryst. 2012, 68 (Pt 8), 1058–1066. [Google Scholar] [CrossRef]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An introductory tutorial. Curr. Protoc. Bioinform. 2008, 24, 5.7.1–5.7.48. [Google Scholar] [CrossRef] [Green Version]
- Merritt, E.A.; Bacon, D.J. Raster3D: Photorealistic molecular graphics. Methods Enzym. 1997, 277, 505–524. [Google Scholar]
- Abagyan, R.; Totrov, M.; Kuznetsov, D. ICM-A New Method for Protein Modeling and Design: Applications to Docking and Structure Prediction from the Distorted Native Conformation. J. Comput. Chem. 1994, 15, 488–506. [Google Scholar] [CrossRef]
- Hontani, Y.; Baloban, M.; Escobar, F.V.; Jansen, S.A.; Shcherbakova, D.M.; Weissenborn, J.; Kloz, M.; Mroginski, M.A.; Verkhusha, V.V.; Kennis, J.T.M. Real-time observation of tetrapyrrole binding to an engineered bacterial phytochrome. Commun. Chem. 2021, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Stepanenko, O.V.; Shpironok, O.G.; Fonin, A.V.; Kuznetsova, I.M.; Turoverov, K.K. Near-Infrared Markers based on Bacterial Phytochromes with Phycocyanobilin as a Chromophore. Int. J. Mol. Sci. 2019, 20, 6067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Sulatskaya, A.I.; Kuznetsova, I.M.; Turoverov, K.K. Fluorescence of dyes in solutions with high absorbance. Inner filter effect correction. PLoS ONE 2014, 9, e103878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turoverov, K.K.; Kuznetsova, I.M. Intrinsic fluorescence of actin. J. Fluoresc. 2003, 13, 41–57. [Google Scholar] [CrossRef]
- O’Connor, D.V.; Phillips, D. Time-Correlated Single Photon Counting; Academic Press: New York, NY, USA, 1984; pp. 37–54. [Google Scholar]
- Marquardt, D.W. An algorithm for least-squares estimation of non linear parameters. J. Soc. Ind. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Berkelman, T.R.; Lagarias, J.C. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal. Biochem. 1986, 156, 194–201. [Google Scholar] [CrossRef]
- Dutta, S.; Burkhardt, K.; Swaminathan, G.J.; Kosada, T.; Henrick, K.; Nakamura, H.; Berman, H.M. Data deposition and annotation at the worldwide protein data bank. Methods Mol. Biol. 2008, 426, 81–101. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).