Integrative Analysis of miRNAs and Their Targets Involved in Ray Floret Growth in Gerbera hybrida
Abstract
1. Introduction
2. Results
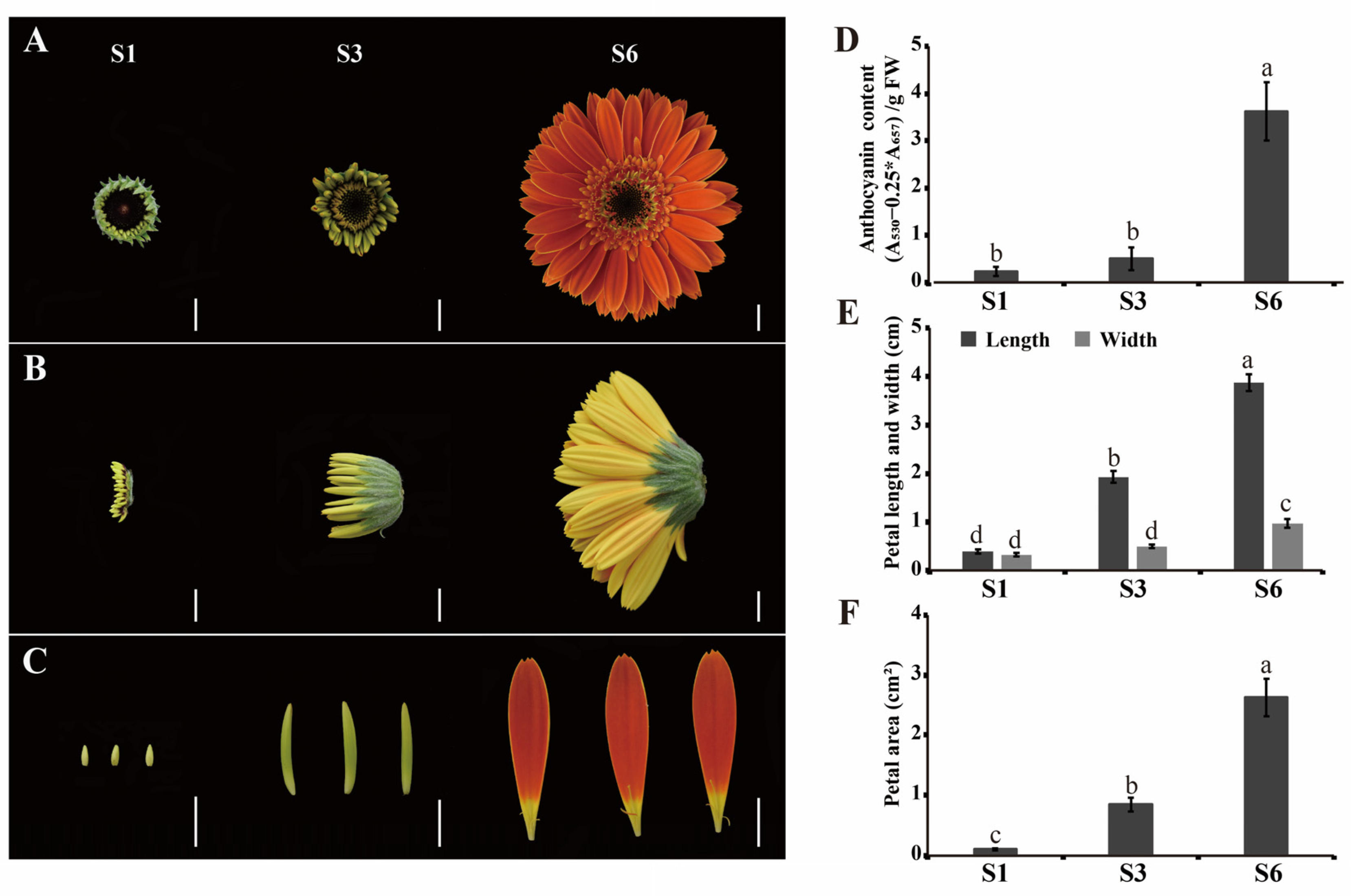
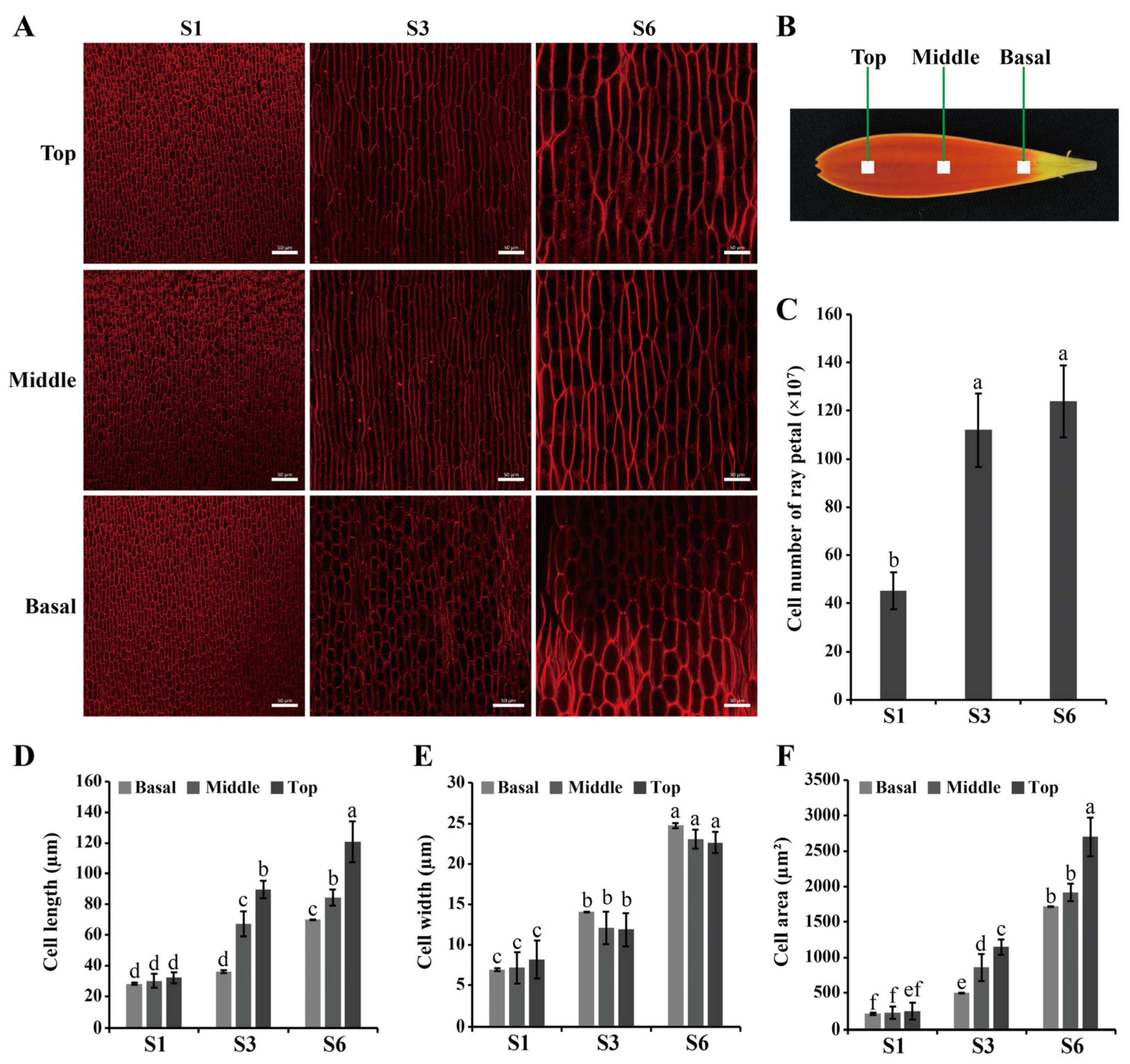
2.1. Phenotypic Characterization of Ray Florets during Inflorescence Opening
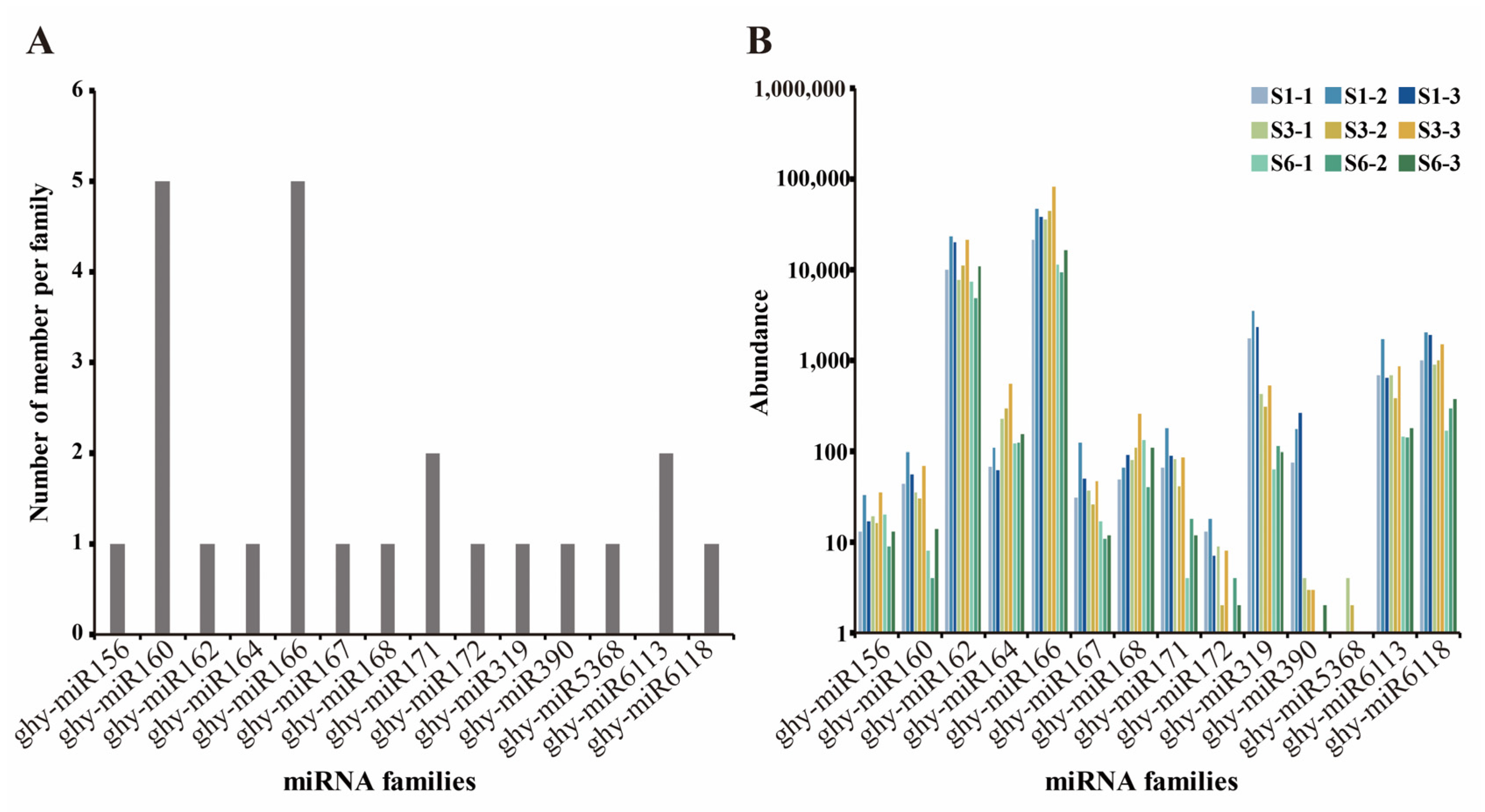
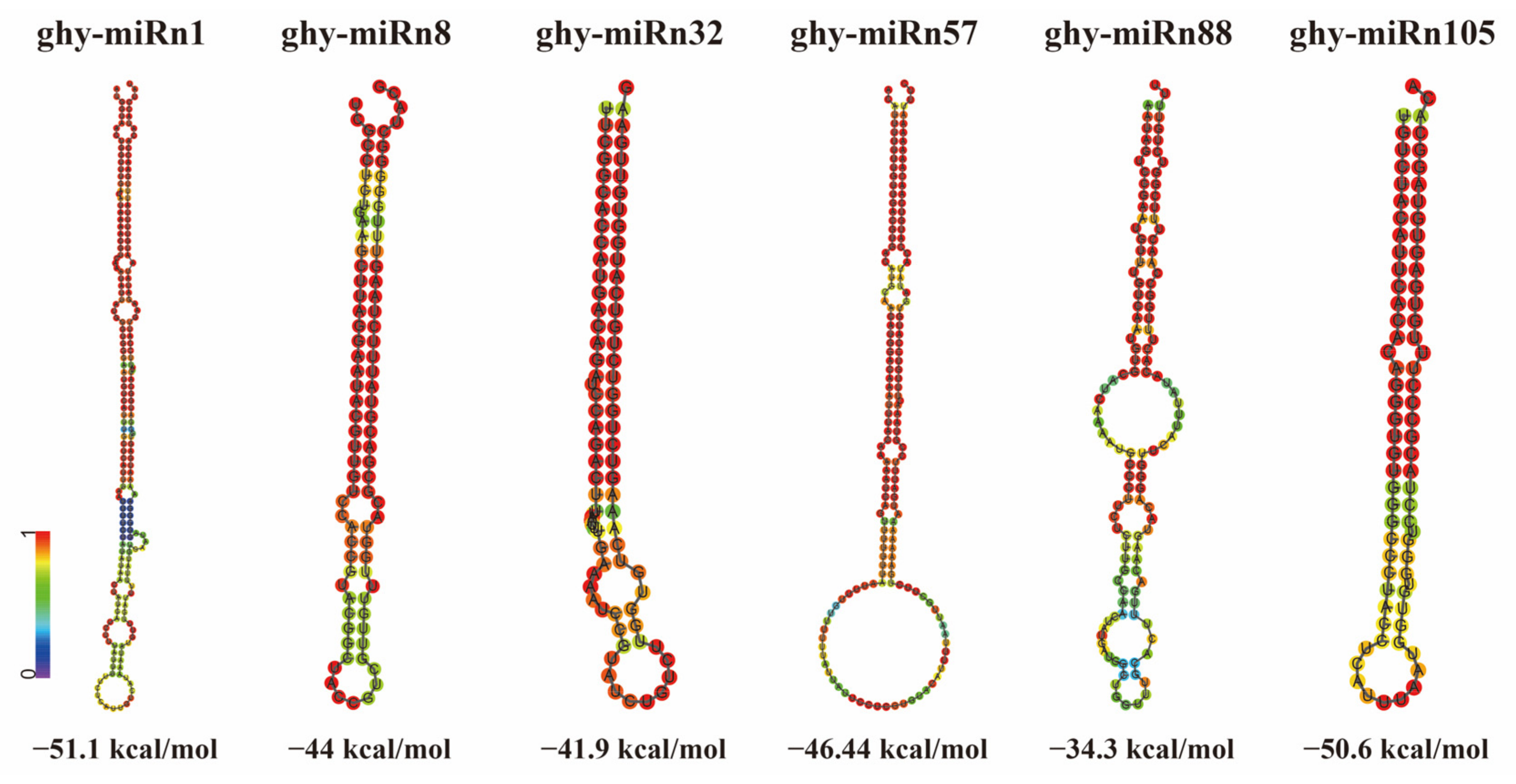
2.2. Identification of Conserved and Novel miRNAs in Gerbera
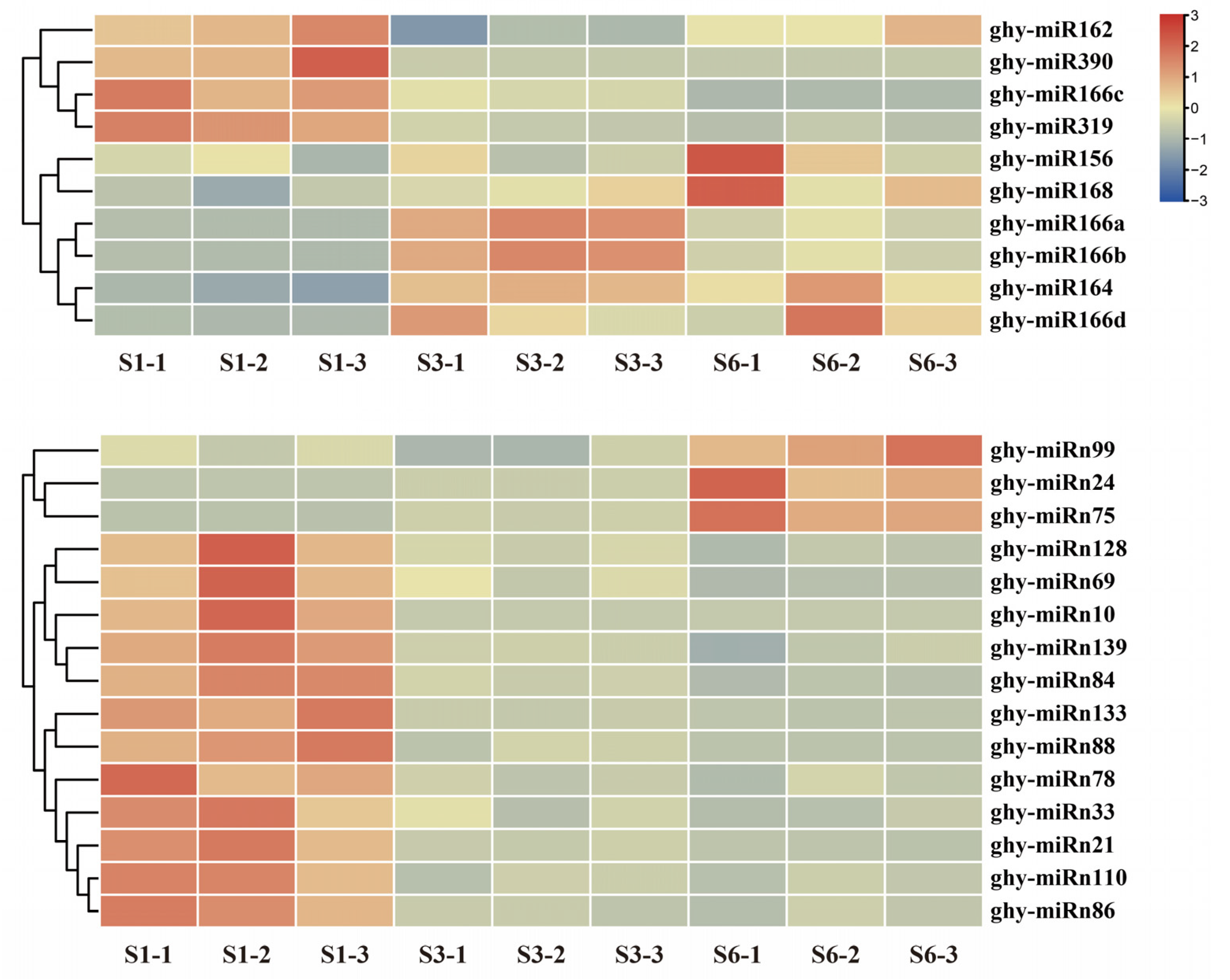
2.3. Differential Expression of miRNAs during the Growth of Ray Floret
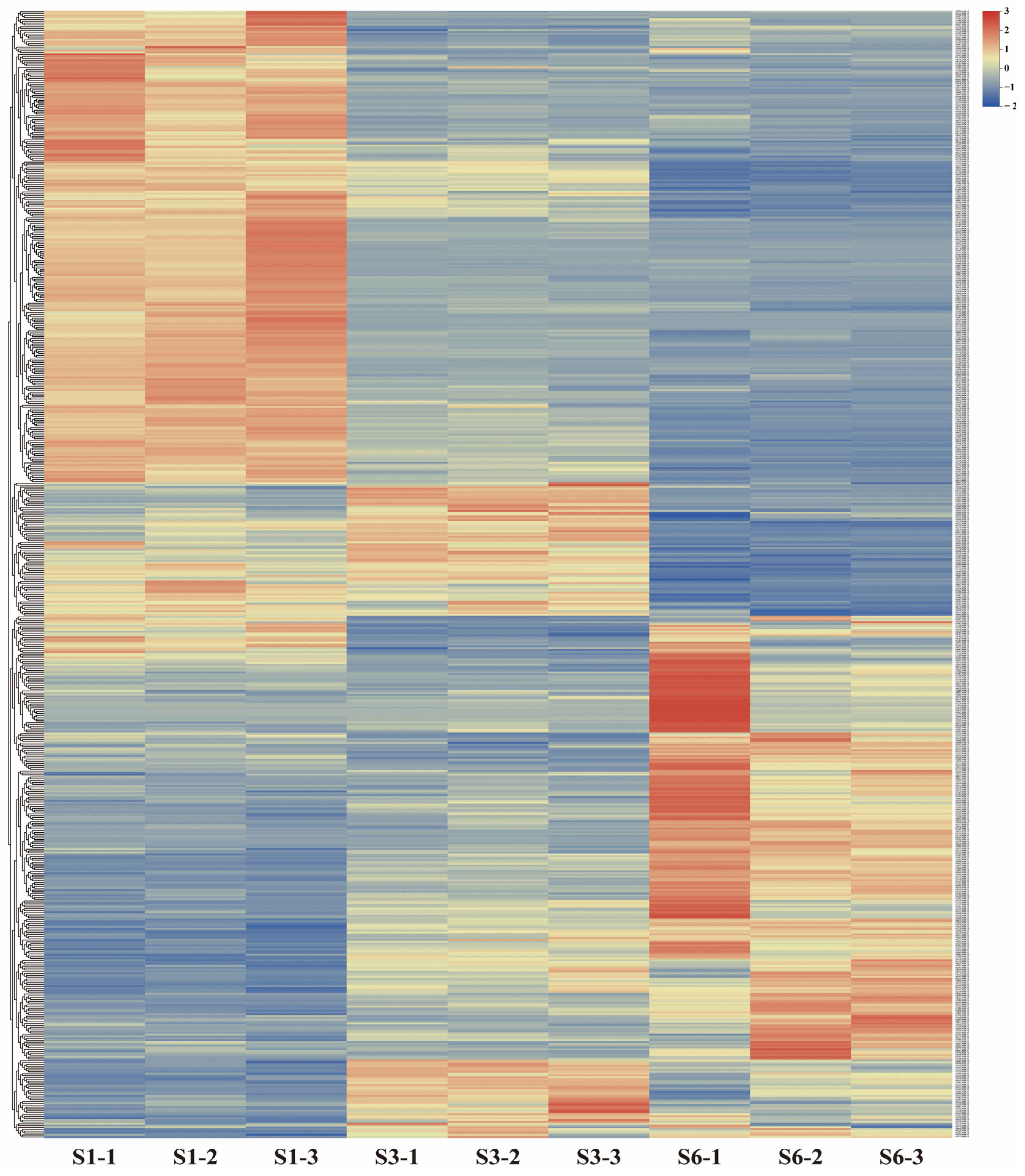
2.4. Expression Profiles of miRNA Target Genes in Gerbera
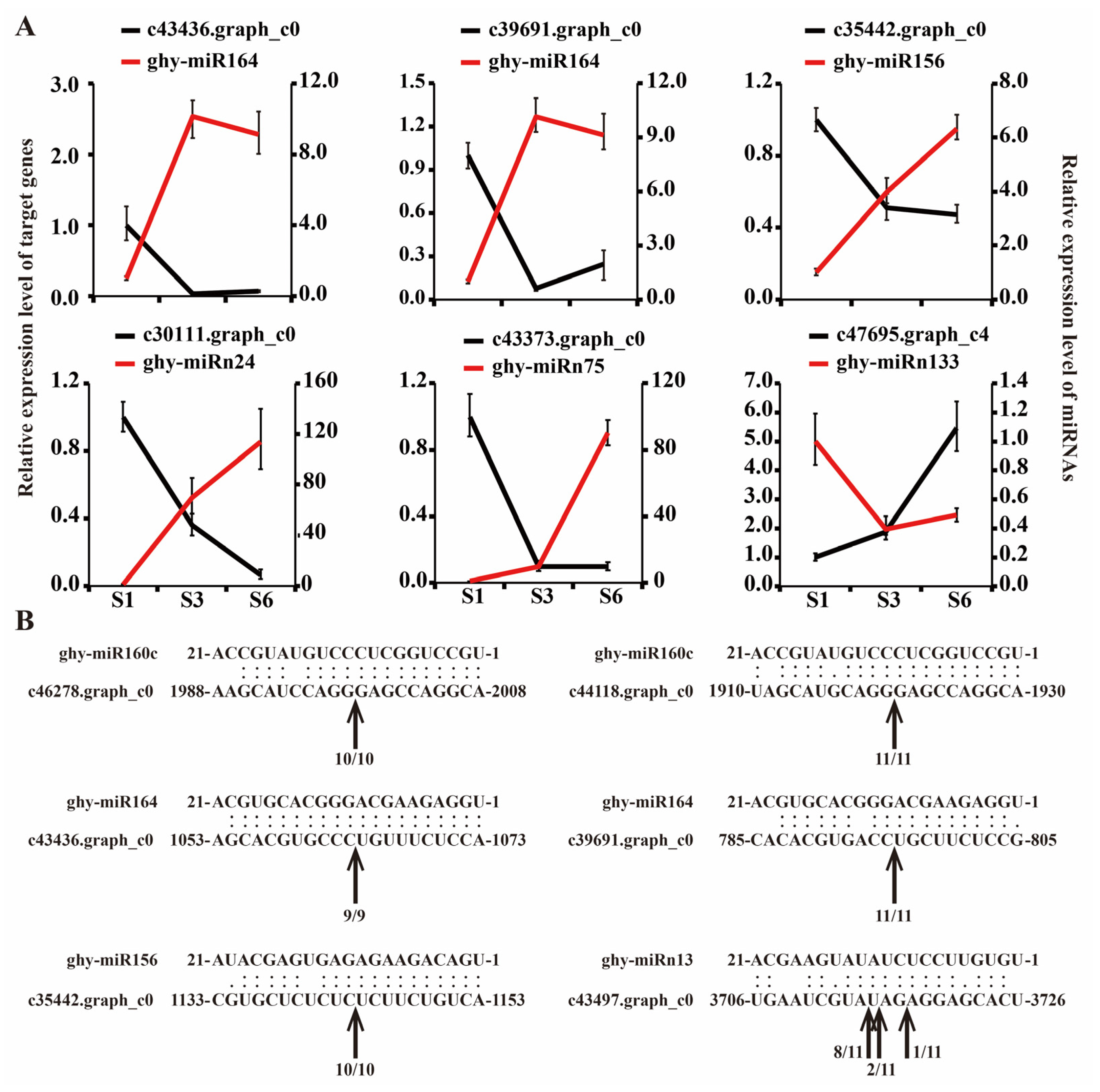
2.5. Validation of miRNA and Target Gene Expression
3. Discussion
3.1. Conserved and Novel miRNAs in Gerbera Ray Petals
3.2. Correlation Analysis of Differentially Expressed miRNAs and miRNA Target Genes
4. Materials and Methods
4.1. Plant Materials
4.2. Length, Width and Size of Petals and Cells
4.3. Measurement of Anthocyanin Content
4.4. Library Construction and Sequencing
4.5. Bioinformatics of miRNAs
4.6. Analysis of Differentially Expressed miRNAs (DEMs)
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis of Differentially Expressed miRNAs
4.8. Prediction and Annotation of miRNA Target Genes
4.9. Validation of miRNA Targets
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodwillie, C.; Sargent, R.D.; Eckert, C.G.; Elle, E.; Geber, M.A.; Johnston, M.O.; Kalisz, S.; Moeller, D.A.; Ree, R.H.; Vallejo-Marin, M.; et al. Correlated evolution of mating system and floral display traits in flowering plants and its implications for the distribution of mating system variation. New Phytol. 2010, 185, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Hermann, K.; Kuhlemeier, C. The genetic architecture of natural variation in flower morphology. Curr. Opin. Plant Biol. 2011, 14, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2010, 33, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Fenster, C.B.; Cheely, G.; Dudash, M.R.; Reynolds, R.J. Nectar reward and advertisement in hummingbird-pollinated Silene virginica (Caryophyllaceae). Am. J. Bot. 2006, 93, 1800–1807. [Google Scholar] [CrossRef]
- Huang, T.; Irish, V.F. Gene networks controlling petal organogenesis. J. Exp. Bot. 2015, 67, 61–68. [Google Scholar] [CrossRef]
- Moyroud, E.; Glover, B.J. The Evolution of Diverse Floral Morphologies. Curr. Biol. 2017, 27, R941–R951. [Google Scholar] [CrossRef]
- Huang, R.; Huang, T.; Irish, V.F. Do epigenetic timers control petal development? Front. Plant Sci. 2021, 12, 709360. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and Comparison of Small RNAs from Plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef]
- Xie, Z.; Allen, E.; Fahlgren, N.; Calamar, A.; Givan, S.A.; Carrington, J.C. Expression of Arabidopsis MIRNA Genes. Plant Physiol. 2005, 138, 2145–2154. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zheng, B.; Yu, Y.; Won, S.Y.; Mo, B.; Chen, X. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011, 30, 814–822. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Pashkovskiy, P.P.; Ryazansky, S.S. Biogenesis, evolution, and functions of plant microRNAs. Biochem. (Mosc.) 2013, 78, 627–637. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Carrington, J.C.; Ambros, V. Role of MicroRNAs in Plant and Animal Development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef]
- Gou, J.-Y.; Felippes, F.F.; Liu, C.-J.; Weigel, D.; Wang, J.-W. Negative Regulation of Anthocyanin Biosynthesis in Arabidopsis by a miR156-Targeted SPL Transcription Factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Pei, H.; Ma, N.; Chen, J.; Zheng, Y.; Tian, J.; Li, J.; Zhang, S.; Fei, Z.; Gao, J. Integrative Analysis of miRNA and mRNA Profiles in Response to Ethylene in Rose Petals during Flower Opening. PLoS ONE 2013, 8, e64290. [Google Scholar] [CrossRef]
- Hong, Y.; Jackson, S. Floral induction and flower formation-the role and potential applications of miRNAs. Plant Biotechnol. J. 2015, 13, 282–292. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef]
- Takada, S.; Hibara, K.; Ishida, T.; Tasaka, M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar] [CrossRef]
- Laufs, P.; Peaucelle, A.; Morin, H.; Traas, J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322. [Google Scholar] [CrossRef]
- Baker, C.C.; Sieber, P.; Wellmer, F.; Meyerowitz, E.M. The early extra petals1 Mutant Uncovers a Role for MicroRNA miR164c in Regulating Petal Number in Arabidopsis. Curr. Biol. 2005, 15, 303–315. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Nag, A.; King, S.; Jack, T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef]
- Schiessl, K.; Muiño, J.M.; Sablowski, R. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc. Natl. Acad. Sci. USA 2014, 111, 2830–2835. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic Organization, Differential Expression, and Interaction of SQUAMOSA Promoter-Binding-Like Transcription Factors and microRNA156 in Rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Tiwari, M.; Pandey, A.; Bhatia, C.; Sharma, A.; Trivedi, P.K. MicroRNA858 Is a Potential Regulator of Phenylpropanoid Pathway and Plant Development. Plant Physiol. 2016, 171, 944–959. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Sung, G.-H.; Spatafora, J.W.; Carrington, J. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004, 36, 1282–1290. [Google Scholar] [CrossRef]
- Fahlgren, N.; Howell, M.D.; Kasschau, K.D.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.; Law, T.F.; Grant, S.R.; Dangl, J.L.; et al. High-Throughput Sequencing of Arabidopsis microRNAs: Evidence for Frequent Birth and Death of MIRNA Genes. PLoS ONE 2007, 2, e219. [Google Scholar] [CrossRef]
- Ma, Z.R.; Coruh, C.; Axtell, M.J. Arabidopsis lyrata Small RNAs: Transient MIRNA and Small Interfering RNA Loci within the Arabidopsis Genus. Plant Cell 2010, 22, 1090–1103. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Funk, V.A.; Susanna, A.; Stuessy, T.; Bayer, R. Systematics, Evolution, and Biogeography of the Compositae; IAPT, International Association for Plant Taxonomy: Washington, DC, USA, 2009; pp. 293–313. [Google Scholar]
- Heiden, G.; Iganci, J.R.V.; Macias, L. Baccharis sect. Caulopterae (Asteraceae, Astereae) no Rio Grande do Sul, Brasil. Rodriguésia 2009, 60, 943–983. [Google Scholar] [CrossRef]
- Lopes, D.C.D.X.P.; de Oliveira, T.B.; Viçosa, A.L.; Valverde, S.S.; Ricci Júnior, E. Anti-inflammatory activity of the compositae family and its therapeutic potential. Planta Med. 2021, 87, 71–100. [Google Scholar] [CrossRef]
- Laitinen, R.A.; Immanen, J.; Auvinen, P.; Rudd, S.; Alatalo, E.; Paulin, L.; Ainasoja, M.; Kotilainen, M.; Koskela, S.; Teeri, T.H.; et al. Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae). Genome Res. 2005, 15, 475–486. [Google Scholar] [CrossRef][Green Version]
- Teeri, T.H.; Elomaa, P.; Kotilainen, M.; Albert, V.A. Mining plant diversity: Gerbera as a model system for plant developmental and biosynthetic research. BioEssays 2006, 28, 756–767. [Google Scholar] [CrossRef]
- Kotilainen, M.; Elomaa, P.; Uimari, A.; Albert, V.A.; Yu, D.; Teeri, T.H. GRCD1, an AGL2-Like MADS Box Gene, Participates in the C Function during Stamen Development in Gerbera hybrida. Plant Cell 2000, 12, 1893. [Google Scholar] [CrossRef]
- Uimari, A.; Kotilainen, M.; Elomaa, P.; Yu, D.; Albert, V.A.; Teeri, T.H. Integration of reproductive meristem fates by a SEPALLATA -like MADS-box gene. Proc. Natl. Acad. Sci. USA 2004, 101, 15817–15822. [Google Scholar] [CrossRef]
- Broholm, S.K.; Tähtiharju, S.; Laitinen, R.A.E.; Albert, V.A.; Teeri, T.H.; Elomaa, P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 9117–9122. [Google Scholar] [CrossRef]
- Ruokolainen, S.; Ng, Y.P.; Broholm, S.K.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Characterization of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproductive transition. BMC Plant Biol. 2010, 10, 128. [Google Scholar] [CrossRef]
- Ruokolainen, S.; Ng, Y.P.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol. 2010, 10, 129. [Google Scholar] [CrossRef]
- Ruokolainen, S.; Ng, Y.P.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 2011, 107, 1491–1499. [Google Scholar] [CrossRef]
- Tähtiharju, S.; Rijpkema, A.S.; Vetterli, A.; Albert, V.A.; Teeri, T.; Elomaa, P. Evolution and Diversification of the CYC/TB1 Gene Family in Asteraceae—A Comparative Study in Gerbera (Mutisieae) and Sunflower (Heliantheae). Mol. Biol. Evol. 2011, 29, 1155–1166. [Google Scholar] [CrossRef]
- Zhang, L.L.; Li, L.; Wu, J.; Peng, J.; Zhang, L.; Wang, X. Cell Expansion and microtubule behavior in ray floret petals of Gerbera hybrida: Responses to light and gibberellic acid. Photochem. Photobiol. Sci. 2012, 11, 279–288. [Google Scholar] [CrossRef]
- Juntheikki-Palovaara, I.; Tähtiharju, S.; Lan, T.; Broholm, S.K.; Rijpkema, A.S.; Ruonala, R.; Kale, L.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida(Asteraceae). Plant J. 2014, 79, 783–796. [Google Scholar] [CrossRef]
- Li, L.; Zhang, W.; Zhang, L.; Li, N.; Peng, J.; Wang, Y.; Zhong, C.; Yang, Y.; Sun, S.; Liang, S.; et al. Transcriptomic insights into antagonistic effects of gibberellin and abscisic acid on petal growth in Gerbera hybrida. Front. Plant Sci. 2015, 6, 168. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, T.; Broholm, S.K.; Tähtiharju, S.; Mouhu, K.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Evolutionary co-option of floral meristem identity genes for patterning of the flower-like asteraceae inflorescence. Plant Physiol. 2016, 172, 284–296. [Google Scholar] [PubMed]
- Zhao, Y.; Broholm, S.K.; Wang, F.; Rijpkema, A.S.; Lan, T.; Albert, V.A.; Teeri, T.H.; Elomaa, P. TCP and MADS-Box Transcription Factor Networks Regulate Heteromorphic Flower Type Identity in Gerbera hybrida. Plant Physiol. 2020, 184, 1455–1468. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Jin, X.; Yao, W.; Kong, L.; Huang, G.; Tao, Y.; Li, L.; Wang, X.; Wang, Y. A Mini Zinc-Finger Protein (MIF) from Gerbera hybrida Activates the GASA Protein Family Gene, GEG, to Inhibit Ray Petal Elongation. Front. Plant Sci. 2017, 8, 1649. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Han, M.; Yao, W.; Wang, Y. Transcriptome analysis reveals the regulation of brassinosteroids on petal growth in Gerbera hybrida. PeerJ 2017, 5, e3382. [Google Scholar] [CrossRef]
- Huang, G.; Han, M.; Jian, L.; Chen, Y.; Sun, S.; Wang, X.; Wang, Y. An ETHYLENE INSENSITIVE3-LIKE1 Protein Directly Targets the GEG Promoter and Mediates Ethylene-Induced Ray Petal Elongation in Gerbera hybrida. Front. Plant Sci. 2020, 10, 1737. [Google Scholar] [CrossRef]
- Zhang, T.T.; Zhao, Y.; Juntheikki, I.; Mouhu, K.; Broholm, S.K.; Rijpkema, A.S.; Kins, L.; Lan, T.; Albert, V.A.; Teeri, T.H.; et al. Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 2017, 216, 939–954. [Google Scholar] [CrossRef]
- Ren, G.; Li, L.; Huang, Y.; Wang, Y.; Zhang, W.; Zheng, R.; Zhong, C.; Wang, X. GhWIP2, a WIP zinc finger protein, suppresses cell expansion in Gerbera hybrida by mediating crosstalk between gibberellin, abscisic acid, and auxin. New Phytol. 2018, 219, 728–742. [Google Scholar] [CrossRef]
- Ortiz, J.P.A.; Leblanc, O.; Rohr, C.; Grisolia, M.; Siena, L.A.; Podio, M.; Colono, C.; Azzaro, C.; Pessino, S.C. Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genom. 2019, 20, 487. [Google Scholar] [CrossRef]
- Mallory, A.C.; Dugas, D.; Bartel, D.P.; Bartel, B. MicroRNA Regulation of NAC-Domain Targets Is Required for Proper Formation and Separation of Adjacent Embryonic, Vegetative, and Floral Organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Liu, L.; Lee, E.; Han, X.; Rim, Y.; Chu, H.; Kim, S.-W.; Sack, F.; Kim, J.-Y. The Arabidopsis Callose Synthase Gene GSL8 Is Required for Cytokinesis and Cell Patterning. Plant Physiol. 2009, 150, 105–113. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 Regulates Arabidopsis Petal Growth by Interacting with the bHLH Transcription Factor BIGPETALp. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef]
- Damodharan, S.; Corem, S.; Gupta, S.K.; Arazi, T. Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 2018, 96, 855–868. [Google Scholar] [CrossRef]
- Dai, X.; Lu, Q.; Wang, J.; Wang, L.; Xiang, F.; Liu, Z. MiR160 and its target genes ARF10, ARF16 and ARF17 modulate hypocotyl elongation in a light, BRZ, or PAC-dependent manner in arabidopsis: miR160 promotes hypocotyl elongation. Plant Sci. 2021, 303, 110686. [Google Scholar] [CrossRef]
- Haley, B.; Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004, 11, 599–606. [Google Scholar] [CrossRef]
- Jiang, A.; Guo, Z.; Pan, J.; Yang, Y.; Zhuang, Y.; Zuo, D.; Hao, C.; Gao, Z.; Xin, P.; Chu, J.; et al. The PIF1-miR408-PLANTACYANIN repression cascade regulates light-dependent seed germination. Plant Cell 2021, 33, 1506–1529. [Google Scholar] [CrossRef]
- Sun, Z.-C.; Zhang, L.-S.; Wang, Z.-J. Genome-wide analysis of miRNAs in Carya cathayensis. BMC Plant Biol. 2017, 17, 228. [Google Scholar] [CrossRef]
- Shi, Y.; Xia, H.; Cheng, X.; Zhang, L. Genome-wide miRNA analysis and integrated network for flavonoid biosynthesis in Osmanthus fragrans. BMC Genom. 2021, 22, 141. [Google Scholar] [CrossRef]
- Liu, G.; Liu, J.; Pei, W.; Li, X.; Wang, N.; Ma, J.; Zang, X.; Zhang, J.; Yu, S.; Wu, M.; et al. Analysis of the MIR160 gene family and the role of MIR160a_A05 in regulating fiber length in cotton. Planta 2019, 250, 2147–2158. [Google Scholar] [CrossRef]
- Ru, P.; Xu, L.; Ma, H.; Huang, H. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 2006, 16, 457–465. [Google Scholar] [CrossRef]
- Wu, M.-F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.; Clarke, T.H.; Reed, J.W.; van der Knaap, E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef]
- Liu, H.; Jia, S.; Shen, D.; Liu, J.; Li, J.; Zhao, H.; Han, S.; Wang, Y. Four AUXIN RESPONSE FACTOR genes downregulated by microRNA167 are associated with growth and development in Oryza sativa. Funct. Plant Biol. 2012, 39, 736–744. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, X.; Cai, W.; Huang, W.; Zhou, X.; Luo, Q.; Yang, H.; Wang, J.; Huang, J. Arabidopsis miR171-Targeted Scarecrow-Like Proteins Bind to GT cis-Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions. PLoS Genet. 2014, 10, e1004519. [Google Scholar] [CrossRef]
- Curaba, J.; Talbot, M.; Li, Z.; Helliwell, C. Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol. 2013, 13, 6. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Schmid, M.; Uhlenhaut, H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef]
- Jung, J.-H.; Seo, Y.-H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.-H.; Park, C.-M. The GIGANTEA -regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef]
- Wollmann, H.; Mica, E.; Todesco, M.; Long, J.A.; Weigel, D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 2010, 137, 3633–3642. [Google Scholar] [CrossRef]
- Vashisht, I.; Mishra, P.; Pal, T.; Chanumolu, S.; Singh, T.R.; Chauhan, R.S. Mining NGS transcriptomes for miRNAs and dissecting their role in regulating growth, development, and secondary metabolites production in different organs of a medicinal herb, Picrorhiza kurroa. Planta 2015, 241, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wan, L.; Bi, S.; Wan, X.; Li, Z.; Cao, J.; Tong, Z.; Xu, H.; He, F.; Li, X. Identification of Drought-Responsive MicroRNAs from Roots and Leaves of Alfalfa by High-Throughput Sequencing. Genes 2017, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Breuil-Broyer, S.; Morel, P.; de Almeida-Engler, J.; Coustham, V.; Negrutiu, I.; Trehin, C. High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J. 2004, 38, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; López-Giráldez, F.; Townsend, J.; Irish, V.F. RBE controls microRNA164 expression to effect floral organogenesis. Development 2012, 139, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Zhang, Y.; Wang, W.; Irish, V.F.; Huang, T. RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis. J. Exp. Bot. 2016, 67, 6473–6480. [Google Scholar] [CrossRef]
- Aydinoglu, F.; Lucas, S.J. Identification and expression profiles of putative leaf growth related microRNAs in maize (Zea mays L.) hybrid ADA313. Gene 2018, 690, 57–67. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Adenot, X.; Elmayan, T.; Lauressergues, D.; Boutet, S.; Bouché, N.; Gasciolli, V.; Vaucheret, H. DRB4-Dependent TAS3 trans-Acting siRNAs Control Leaf Morphology through AGO7. Curr. Biol. 2006, 16, 927–932. [Google Scholar] [CrossRef]
- Garcia, D.; Collier, S.A.; Byrne, M.E.; Martienssen, R.A. Specification of Leaf Polarity in Arabidopsis via the trans-Acting siRNA Pathway. Curr. Biol. 2006, 16, 933–938. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 Interaction and Dual Functionality in TAS3 trans-Acting siRNA Formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef]
- McConnell, J.; Barton, M. Leaf polarity and meristem formation in Arabidopsis. Development 1998, 125, 2935–2942. [Google Scholar] [CrossRef]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial Patterning of Arabidopsis Shoots by Class III HD-ZIP and KANADI Genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Juarez, M.T.; Kui, J.S.; Thomas, J.; Heller, B.A.; Timmermans, M.C.P. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004, 428, 84–88. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Walcher-Chevillet, C.L.; Kramer, E.M. Breaking the mold: Understanding the evolution and development of lateral organs in diverse plant models. Curr. Opin. Genet. Dev. 2016, 39, 79–84. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A New Class of Transcription Factors Mediates Brassinosteroid-Regulated Gene Expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- Vale, M.; Rodrigues, J.; Badim, H.; Gerós, H.; Conde, A. Exogenous application of non-mature miRNA-encoded miPEP164c inhibits proanthocyanidin synthesis and stimulates anthocyanin accumulation in grape berry cells. Front. Plant Sci. 2021, 12, 706679. [Google Scholar] [CrossRef]
- Schwinn, K.E.; Ngo, H.; Kenel, F.; Brummell, D.A.; Albert, N.W.; McCallum, J.A.; Pither-Joyce, M.; Crowhurst, R.N.; Eady, C.; Davies, K.M. The Onion (Allium cepa L.) R2R3-MYB Gene MYB1 Regulates Anthocyanin Biosynthesis. Front. Plant Sci. 2016, 7, 1865. [Google Scholar] [CrossRef]
- Elomaa, P.; Mehto, M.; Kotilainen, M.; Helariutta, Y.; Nevalainen, L.; Teeri, T.H. A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J. 1998, 16, 93–99. [Google Scholar] [CrossRef]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.; Teeri, T.H. Activation of Anthocyanin Biosynthesis in Gerbera hybrida (Asteraceae) Suggests Conserved Protein-Protein and Protein-Promoter Interactions between the Anciently Diverged Monocots and Eudicots. Plant Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef]
- Meng, X.; Wang, X. Regulation of flower Development and anthocyanin accumulation in Gerbera hybrida. J. Hortic. Sci. Biotech. 2004, 79, 131–137. [Google Scholar] [CrossRef]
- Rabino, I.; Mancinelli, A.L. Light, temperature, and anthocyanin production. Plant Physiol. 1986, 81, 922–924. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, P.; Zhao, Q.; Tang, Y.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Overexpression of a phosphate starvation response AP2/ERF gene from physic nut in Arabidopsis alters root morphological traits and phosphate starvation-induced anthocyanin accumulation. Front. Plant Sci. 2018, 9, 1186. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Q.; Li, L.; Peng, J.; Sun, S.; Wang, X. Transcriptome Analysis of Gerbera hybrida Ray Florets: Putative Genes Associated with Gibberellin Metabolism and Signal Transduction. PLoS ONE 2013, 8, e57715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Jiang, L.; Wang, J.; Gu, P.; Chen, M. MTide: An integrated tool for the identification of miRNA-target interaction in plants. Bioinformatics 2015, 31, 290–291. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Bhandari, M.; Abburi, V.; Natarajan, P.; Stommel, J.; Nimmakayala, P.; Reddy, U. Identification of miRNAs and Their Targets Involved in Flower and Fruit Development across Domesticated and Wild Capsicum Species. Int. J. Mol. Sci. 2021, 22, 4866. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Liao, B.; Lin, X.; Luo, Q.; Huang, X.; Wang, X.; Shan, Q.; Wang, Y. Integrative Analysis of miRNAs and Their Targets Involved in Ray Floret Growth in Gerbera hybrida. Int. J. Mol. Sci. 2022, 23, 7296. https://doi.org/10.3390/ijms23137296
Chen Y, Liao B, Lin X, Luo Q, Huang X, Wang X, Shan Q, Wang Y. Integrative Analysis of miRNAs and Their Targets Involved in Ray Floret Growth in Gerbera hybrida. International Journal of Molecular Sciences. 2022; 23(13):7296. https://doi.org/10.3390/ijms23137296
Chicago/Turabian StyleChen, Yanbo, Bingbing Liao, Xiaohui Lin, Qishan Luo, Xuanyan Huang, Xiaojing Wang, Qinli Shan, and Yaqin Wang. 2022. "Integrative Analysis of miRNAs and Their Targets Involved in Ray Floret Growth in Gerbera hybrida" International Journal of Molecular Sciences 23, no. 13: 7296. https://doi.org/10.3390/ijms23137296
APA StyleChen, Y., Liao, B., Lin, X., Luo, Q., Huang, X., Wang, X., Shan, Q., & Wang, Y. (2022). Integrative Analysis of miRNAs and Their Targets Involved in Ray Floret Growth in Gerbera hybrida. International Journal of Molecular Sciences, 23(13), 7296. https://doi.org/10.3390/ijms23137296




