Neurogranin and Neurofilament Light Chain as Preclinical Biomarkers in Scrapie
Abstract
:1. Introduction
2. Results
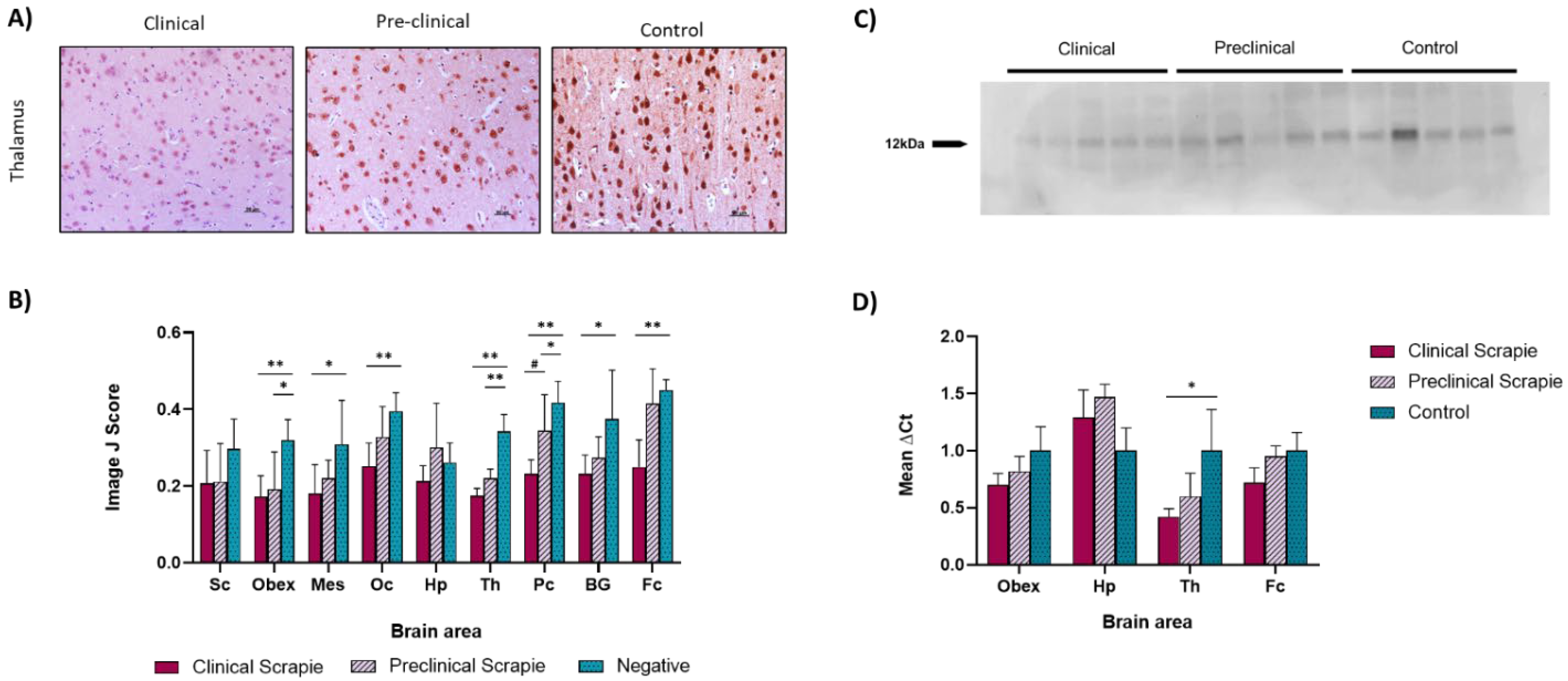
2.1. Ng Expression Is Downregulated in Scrapie-Infected Sheep
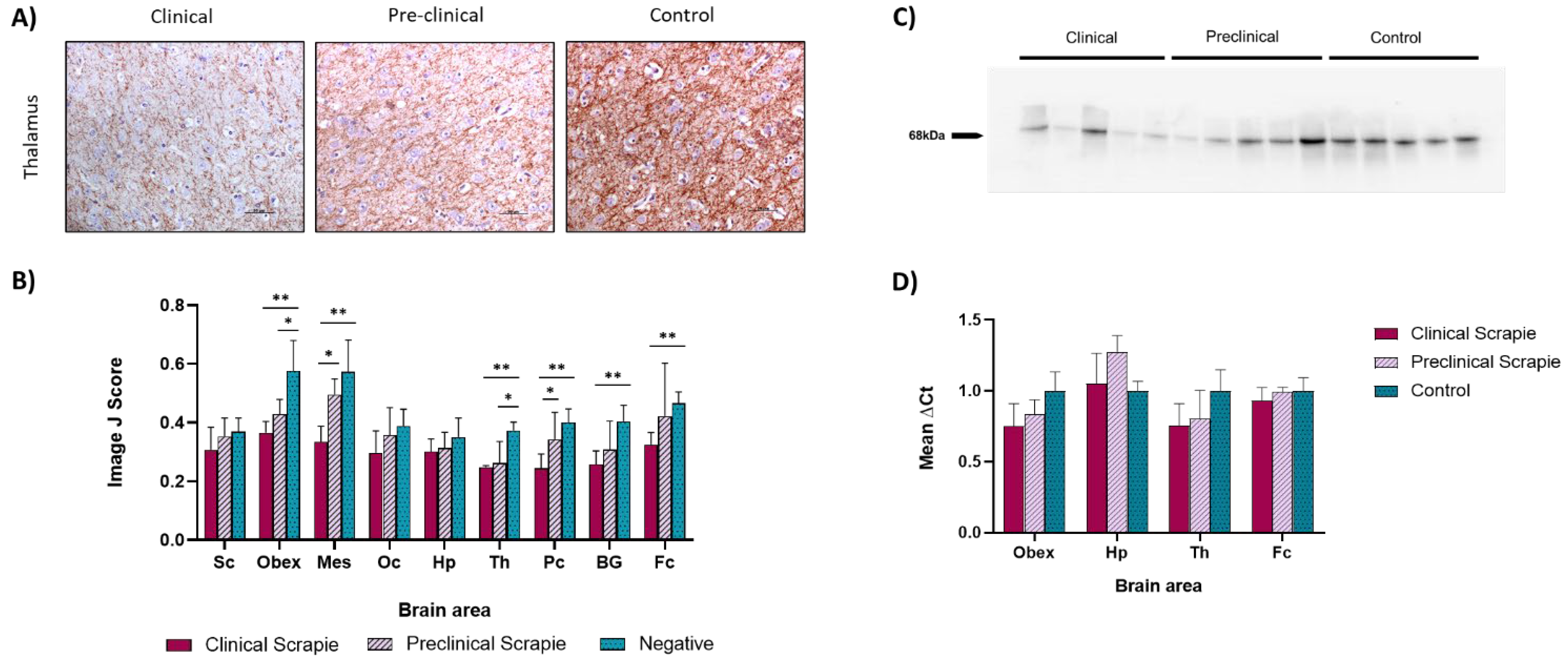
2.2. NfL Expression Is Downregulated in Most Brain Areas of Scrapie-Infected Sheep
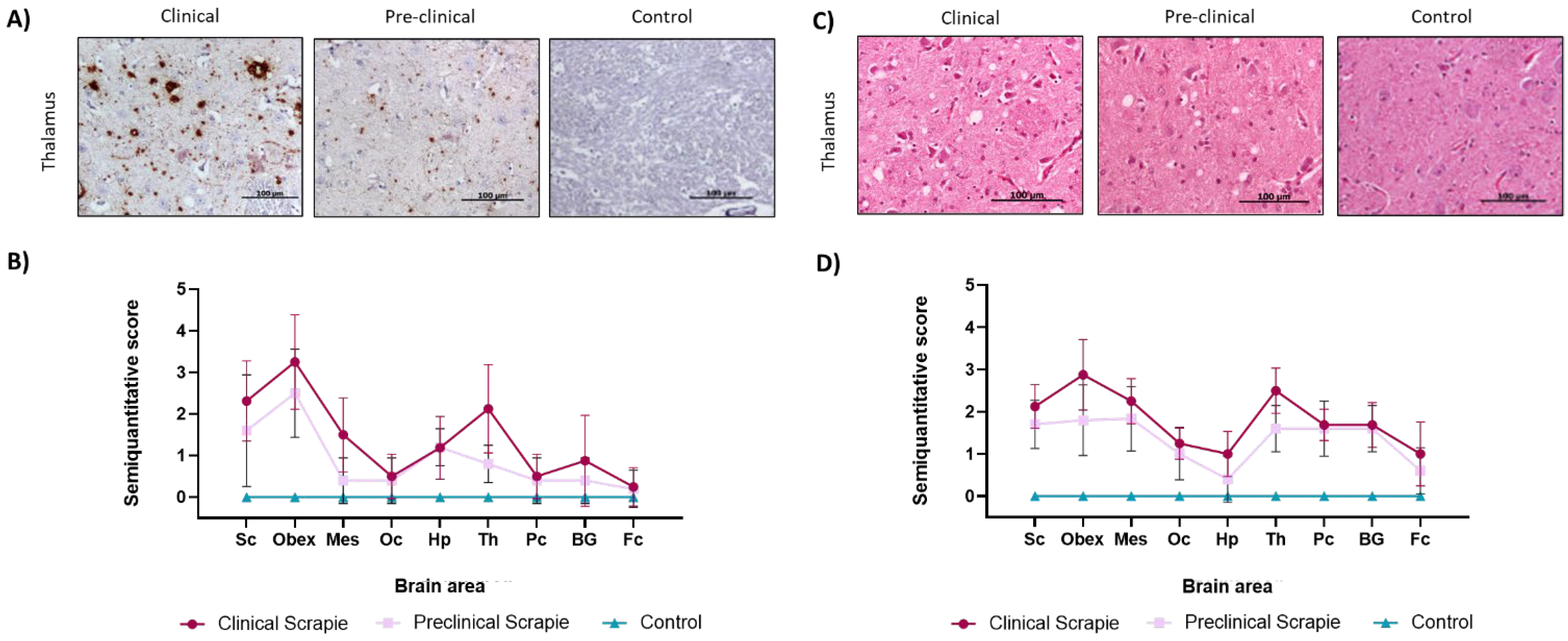
2.3. Scrapie-Related Neuropathological Features in Sheep and Correlation with Ng and NfL Protein Expression
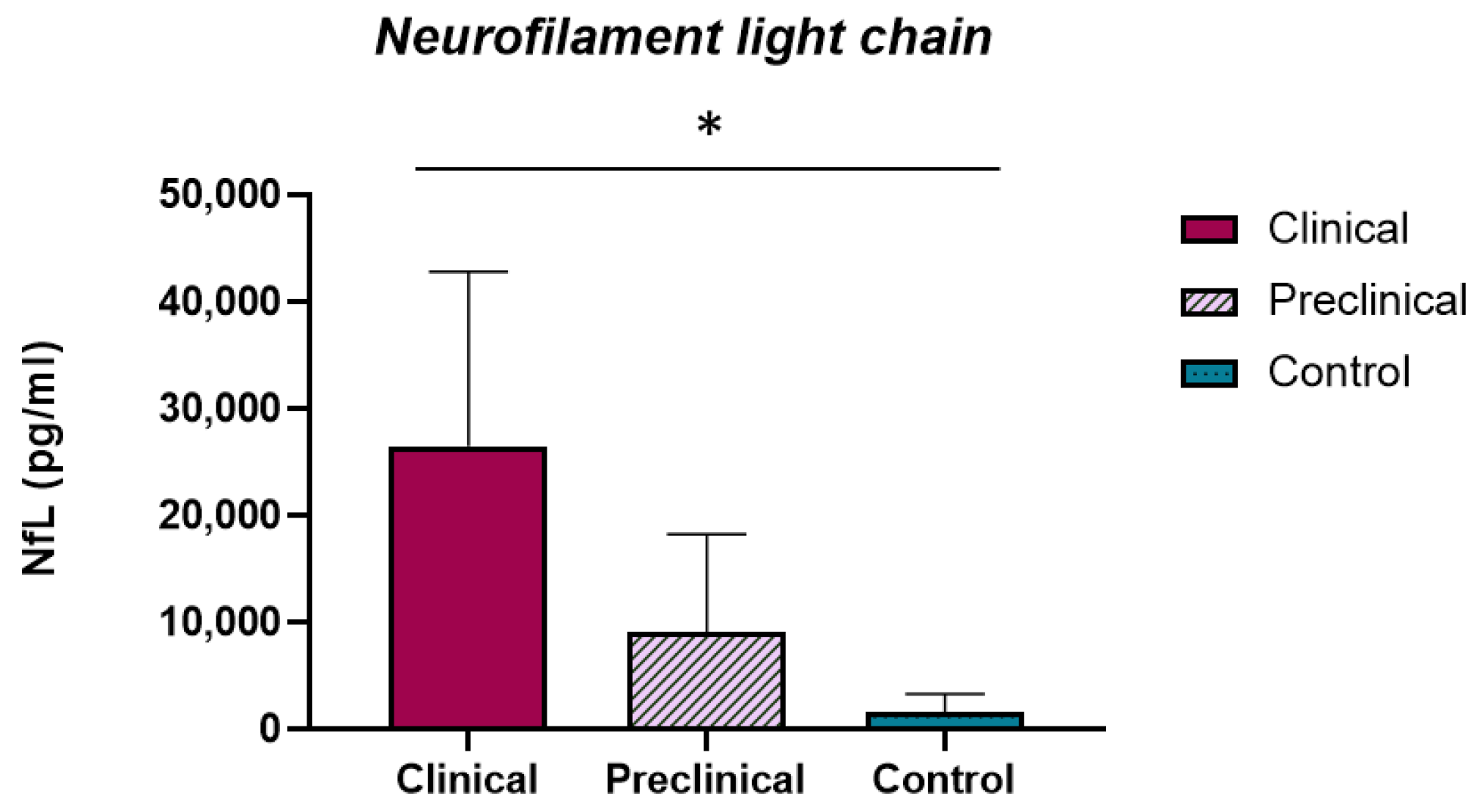
2.4. Ng and NfL Concentrations in Sheep CSF
3. Discussion
3.1. Ng Expression in Brain Tissue Was Affected by Prion-Related Pathogenesis
3.2. NfL Expression in Brain Tissue Was Reduced throughout the Course of the Disease
3.3. Ng and NfL Levels in Brain Tissue Are Negatively Correlated with Prion-Associated Lesions
3.4. NfL Levels Are Increased in the CSF of Scrapie-Affected Sheep
3.5. Conclusions
4. Materials and Methods
4.1. Animals and Sample Collection
4.2. Histological and Immunohistochemical Analyses
4.3. Cerebrospinal Fluid Analyses
4.4. Western Blot Analyses
4.5. Gene Expression Analyses
4.6. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prusiner, S. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.L.N.; McGill, I.S.; Done, S.H.; Bradley, R. Neuropathology of scrapie: A study of the distribution patterns of brain lesions in 222 cases of natural scrapie in sheep, 1982–1991. Vet. Rec. 1997, 140, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.; Appleby, B.; Brandel, J.-P.; Caughey, B.; Collins, S.; Geschwind, M.D.; Green, A.; Haïk, S.; Kovacs, G.G.; Ladogana, A.; et al. Biomarkers and diagnostic guidelines for sporadic Creutzfeldt-Jakob disease. Lancet Neurol. 2021, 20, 235–246. [Google Scholar] [CrossRef]
- Puoti, G.; Bizzi, A.; Forloni, G.; Safar, J.G.; Tagliavini, F.; Gambetti, P. Sporadic human prion diseases: Molecular insights and diagnosis. Lancet Neurol. 2012, 11, 618–628. [Google Scholar] [CrossRef]
- Otto, M.; Wiltfang, J.; Cepek, L.; Neumann, M.; Mollenhauer, B.; Steinacker, P.; Ciesielczyk, B.; Schulz–Schaeffer, W.; Kretzschmar, H.A.; Poser, S. Tau protein and 14-3-3 protein in the differential diagnosis of Creutzfeldt–Jakob disease. Neurology 2002, 58, 192–197. [Google Scholar] [CrossRef]
- Cramm, M.; Schmitz, M.; Karch, A.; Mitrova, E.; Kuhn, F.; Schroeder, B.; Raeber, A.; Varges, D.; Kim, Y.-S.; Satoh, K.; et al. Stability and Reproducibility Underscore Utility of RT-QuIC for Diagnosis of Creutzfeldt-Jakob Disease. Mol. Neurobiol. 2015, 53, 1896–1904. [Google Scholar] [CrossRef] [Green Version]
- González, L.; Dagleish, M.P.; Martin, S.; Dexter, G.; Steele, P.; Finlayson, J.; Jeffrey, M. Diagnosis of preclinical scrapie in live sheep by the immunohistochemical examination of rectal biopsies. Veter. Rec. 2008, 162, 397–403. [Google Scholar] [CrossRef]
- Llorens, F.; Barrio, T.; Correia, A.; Villar-Piqué, A.; Thüne, K.; Lange, P.; Badiola, J.J.; Schmitz, M.; Lachmann, I.; Bolea, R.; et al. Cerebrospinal Fluid Prion Disease Biomarkers in Pre-clinical and Clinical Naturally Occurring Scrapie. Mol. Neurobiol. 2018, 55, 8586–8591. [Google Scholar] [CrossRef]
- Xiang, Y.; Xin, J.; Le, W.; Yang, Y. Neurogranin: A Potential Biomarker of Neurological and Mental Diseases. Front. Aging Neurosci. 2020, 12, 584743. [Google Scholar] [CrossRef]
- Nazir, F.H.; Camporesi, E.; Brinkmalm, G.; Lashley, T.; Toomey, C.E.; Kvartsberg, H.; Zetterberg, H.; Blennow, K.; Becker, B. Molecular forms of neurogranin in cerebrospinal fluid. J. Neurochem. 2020, 157, 816–833. [Google Scholar] [CrossRef]
- Alvarez-Castelao, B.; Schuman, E.M. The Regulation of Synaptic Protein Turnover. J. Biol. Chem. 2015, 290, 28623–28630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Represa, A.; Deloulme, J.C.; Sensenbrenner, M.; Ben-Ari, Y.; Baudier, J. Neurogranin: Immunocytochemical localization of a brain-specific protein kinase C substrate. J. Neurosci. 1990, 10, 3782–3792. [Google Scholar] [CrossRef] [PubMed]
- Prichard, L.; Deloulme, J.C.; Storm, D.R. Interactions between Neurogranin and Calmodulin in Vivo. J. Biol. Chem. 1999, 274, 7689–7694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez-González, I.; Cuesta, S.N.; Algaba, A.; Díez-Guerra, F.J. Neurogranin binds to phosphatidic acid and associates to cellular membranes. Biochem. J. 2007, 404, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Korley, F.K.; Dai, M.; Everett, A.D. Serum neurogranin measurement as a biomarker of acute traumatic brain injury. Clin. Biochem. 2015, 48, 843–848. [Google Scholar] [CrossRef] [Green Version]
- Bereczki, E.; Bogstedt, A.; Höglund, K.; Tsitsi, P.; Brodin, L.; Ballard, C.; Svenningsson, P.; Aarsland, D. Synaptic proteins in CSF relate to Parkinson’s disease stage markers. Npj Park. Dis. 2017, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- De Vos, A.; Bjerke, M.; Brouns, R.; De Roeck, N.; Jacobs, D.; Van den Abbeele, L.; Guldolf, K.; Zetterberg, H.; Blennow, K.; Engelborghs, S.; et al. Neurogranin and tau in cerebrospinal fluid and plasma of patients with acute ischemic stroke. BMC Neurol. 2017, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Blennow, K.; Díaz-Lucena, D.; Zetterberg, H.; Villar-Pique, A.; Karch, A.; Vidal, E.; Hermann, P.; Schmitz, M.; Abizanda, I.; Zerr, I.; et al. CSF neurogranin as a neuronal damage marker in CJD: A comparative study with AD. J. Neurol. Neurosurg. Psychiatry 2019, 90, 846–853. [Google Scholar] [CrossRef]
- Mons, N.; Enderlin, V.; Jaffard, R.; Higueret, P. Selective age-related changes in the PKC-sensitive, calmodulin-binding protein, neurogranin, in the mouse brain. J. Neurochem. 2008, 79, 859–867. [Google Scholar] [CrossRef]
- Pak, J.H.; Huang, F.L.; Li, J.; Balschun, D.; Reymann, K.G.; Chiang, C.; Westphal, H.; Huang, K.-P. Involvement of neurogranin in the modulation of calcium/calmodulin-dependent protein kinase II, synaptic plasticity, and spatial learning: A study with knockout mice. Proc. Natl. Acad. Sci. USA 2000, 97, 11232–11237. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Cherry, T.; Bies, C.E.; Florence, M.A.; Gerges, N.Z. Neurogranin enhances synaptic strength through its interaction with calmodulin. EMBO J. 2009, 28, 3027–3039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, S.G.; Kang, M.; Kim, Y.-S.; Kim, N.-H.; Nam, D.W.; Song, E.J.; Mook-Jung, I.; Moon, M. Intrahippocampal injection of a lentiviral vector expressing neurogranin enhances cognitive function in 5XFAD mice. Exp. Mol. Med. 2018, 50, e461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, A.; Rao, M.V.; Veeranna; Nixon, R.A. Neurofilaments and Neurofilament Proteins in Health and Disease. Cold Spring Harb. Perspect. Biol. 2017, 9, a018309. [Google Scholar] [CrossRef] [Green Version]
- Gaetani, L.; Blennow, K.; Calabresi, P.; Di Filippo, M.; Parnetti, L.; Zetterberg, H. Neurofilament light chain as a biomarker in neurological disorders. J. Neurol. Neurosurg. Psychiatry 2019, 90, 870–881. [Google Scholar] [CrossRef]
- van Eijk, J.J.J.; van Everbroeck, B.; Abdo, W.F.; Kremer, B.P.H.; Verbeek, M.M. CSF Neurofilament Proteins Levels are Elevated in Sporadic Creutzfeldt-Jakob Disease. J. Alzheimer’s Dis. 2010, 21, 569–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanata, E.; Golanska, E.; Villar-Piqué, A.; Karsanidou, A.; Dafou, D.; Xanthopoulos, K.; Schmitz, M.; Ferrer, I.; Karch, A.; Sikorska, B.; et al. Cerebrospinal fluid neurofilament light in suspected sporadic Creutzfeldt-Jakob disease. J. Clin. Neurosci. 2018, 60, 124–127. [Google Scholar] [CrossRef] [Green Version]
- Steinacker, P.; Blennow, K.; Halbgebauer, S.; Shi, S.; Ruf, V.; Oeckl, P.; Giese, A.; Kuhle, J.; Slivarichova, D.; Zetterberg, H.; et al. Neurofilaments in blood and CSF for diagnosis and prediction of onset in Creutzfeldt-Jakob disease. Sci. Rep. 2016, 6, 38737. [Google Scholar] [CrossRef]
- Zetterberg, H.; Bozzetta, E.; Favole, A.; Corona, C.; Cavarretta, M.C.; Ingravalle, F.; Blennow, K.; Pocchiari, M.; Meloni, D. Neurofilaments in blood is a new promising preclinical biomarker for the screening of natural scrapie in sheep. PLoS ONE 2019, 14, e0226697. [Google Scholar] [CrossRef] [Green Version]
- Soto, C.; Satani, N. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol. Med. 2011, 17, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Hilton, K.J.; Cunningham, C.; Reynolds, R.A.; Perry, V.H. Early Hippocampal Synaptic Loss Precedes Neuronal Loss and Associates with Early Behavioural Deficits in Three Distinct Strains of Prion Disease. PLoS ONE 2013, 8, e68062. [Google Scholar] [CrossRef] [Green Version]
- Gossner, A.G.; Foster, J.D.; Fazakerley, J.K.; Hunter, N.; Hopkins, J. Gene expression analysis in distinct regions of the central nervous system during the development of SSBP/1 sheep scrapie. Veter- Microbiol. 2011, 147, 42–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaetani, L.; Höglund, K.; Parnetti, L.; Pujol-Calderon, F.; Becker, B.; Eusebi, P.; Sarchielli, P.; Calabresi, P.; Di Filippo, M.; Zetterberg, H.; et al. A new enzyme-linked immunosorbent assay for neurofilament light in cerebrospinal fluid: Analytical validation and clinical evaluation. Alzheimer’s Res. Ther. 2018, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Kvartsberg, H.; Lashley, T.; Murray, C.E.; Brinkmalm, G.; Cullen, N.C.; Höglund, K.; Zetterberg, H.; Blennow, K.; Portelius, E. The intact postsynaptic protein neurogranin is reduced in brain tissue from patients with familial and sporadic Alzheimer’s disease. Acta Neuropathol. 2019, 137, 89–102. [Google Scholar] [CrossRef] [Green Version]
- Castilla, J.; Hetz, C.; Soto, C. Molecular Mechanisms of Neurotoxicity of Pathological Prion Protein. Curr. Mol. Med. 2004, 4, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Benestad, S.L.; Austbø, L.; Tranulis, M.A.; Espenes, A.; Olsaker, I. Healthy goats naturally devoid of prion protein. Veter. Res. 2012, 43, 87. [Google Scholar] [CrossRef] [Green Version]
- Mallucci, G.R.; Ratté, S.; Asante, E.A.; Linehan, J.; Gowland, I.; Jefferys, J.G.R.; Collinge, J. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 2002, 21, 202–210. [Google Scholar] [CrossRef]
- Westergard, L.; Christensen, H.M.; Harris, D.A. The cellular prion protein (PrPC): Its physiological function and role in disease. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2007, 1772, 629–644. [Google Scholar] [CrossRef] [Green Version]
- Herms, J.; Tings, T.; Gall, S.; Madlung, A.; Giese, A.; Siebert, H.; Schürmann, P.; Windl, O.; Brose, N.; Kretzschmar, H. Evidence of Presynaptic Location and Function of the Prion Protein. J. Neurosci. 1999, 19, 8866–8875. [Google Scholar] [CrossRef]
- Moya, K.L.; Sales, N.; Hassig, R.; Creminon, C.; Grassi, J.; Di Giamberardino, L. Immunolocalization of the cellular prion protein in normal brain. Microsc. Res. Tech. 2000, 50, 58–65. [Google Scholar] [CrossRef]
- Spielhaupter, C.; Schätzl, H.M. PrPC Directly Interacts with Proteins Involved in Signaling Pathways. J. Biol. Chem. 2001, 276, 44604–44612. [Google Scholar] [CrossRef] [Green Version]
- Collinge, J.; Whittington, M.A.; Sidle, K.C.L.; Smith, C.J.; Palmer, M.S.; Clarke, A.R.; Jefferys, J.G.R. Prion protein is necessary for normal synaptic function. Nature 1994, 370, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Otero, A.; Betancor, M.; Eraña, H.; Fernández Borges, N.; Lucas, J.J.; Badiola, J.J.; Castilla, J.; Bolea, R. Prion-Associated Neurodegeneration Causes Both Endoplasmic Reticulum Stress and Proteasome Impairment in a Murine Model of Spontaneous Disease. Int. J. Mol. Sci. 2021, 22, 465. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-B.; Shi, Q.; Xu, Y.; Xie, W.-L.; Zhang, J.; Tian, C.; Guo, Y.; Wang, K.; Zhang, B.-Y.; Chen, C.; et al. Protein Disulfide Isomerase Regulates Endoplasmic Reticulum Stress and the Apoptotic Process during Prion Infection and PrP Mutant-Induced Cytotoxicity. PLoS ONE 2012, 7, e38221. [Google Scholar] [CrossRef]
- Nakagawa, T.; Zhu, H.; Morishima, N.; Li, E.; Xu, J.; Yankner, B.A.; Yuan, J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 2000, 403, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, I.M. Calcineurin in memory and bidirectional plasticity. Biochem. Biophys. Res. Commun. 2003, 311, 1195–1208. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, E.; Beekes, M.; Diringer, H. Evidence for an alternative direct route of access for the scrapie agent to the brain by-passing the spinal cord. J. Gen. Virol. 1997, 78, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.A.; Striebel, J.F.; Rangel, A.; Woods, T.; Phillips, K.; Peterson, K.E.; Race, B.; Chesebro, B. Prion Strain Differences in Accumulation of PrPSc on Neurons and Glia Are Associated with Similar Expression Profiles of Neuroinflammatory Genes: Comparison of Three Prion Strains. PLoS Pathog. 2016, 12, e1005551. [Google Scholar] [CrossRef]
- Makarava, N.; Chang, J.C.-Y.; Molesworth, K.; Baskakov, I.V. Region-specific glial homeostatic signature in prion diseases is replaced by a uniform neuroinflammation signature, common for brain regions and prion strains with different cell tropism. Neurobiol. Dis. 2020, 137, 104783. [Google Scholar] [CrossRef]
- Makarava, N.; Chang, J.C.-Y.; Baskakov, I.V. Region-Specific Sialylation Pattern of Prion Strains Provides Novel Insight into Prion Neurotropism. Int. J. Mol. Sci. 2020, 21, 828. [Google Scholar] [CrossRef] [Green Version]
- Liberski, P.P. Axonal changes in experimental prion diseases recapitulate those following constriction of postganglionic branches of the superior cervical ganglion: A comparison 40 years later. Prion 2019, 13, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Strong, M.J.; Volkening, K.; Hammond, R.; Yang, W.; Strong, W.; Leystra-Lantz, C.; Shoesmith, C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007, 35, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Volkening, K.; Leystra-Lantz, C.; Yang, W.; Jaffee, H.; Strong, M.J. Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 2009, 1305, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Monleón, E.; Monzón, M.; Hortells, P.; Vargas, A.; Acín, C.; Badiola, J.J. Detection of PrPsc on Lymphoid Tissues from Naturally Affected Scrapie Animals: Comparison of Three Visualization Systems. J. Histochem. Cytochem. 2004, 52, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, H.; Dickinson, A.G. Scrapie in mice. J. Comp. Pathol. 1973, 83, 29–40. [Google Scholar] [CrossRef]
- Betancor, M.; Moreno-Martinez, L.; Lopez-Perez, O.; Otero, A.; Hernaiz, A.; Barrio, T.; Badiola, J.J.; Osta, R.; Bolea, R.; Martin-Burriel, I. Therapeutic Assay with the Non-toxic C-Terminal Fragment of Tetanus Toxin (TTC) in Transgenic Murine Models of Prion Disease. Mol. Neurobiol. 2021, 58, 5312–5326. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Lyahyai, J.; Serrano, C.; Ranera, B.; Badiola, J.J.; Zaragoza, P.; Martin-Burriel, I. Effect of scrapie on the stability of house-keeping genes. Anim. Biotechnol. 2010, 21, 1–13. [Google Scholar] [CrossRef]
| Ng | NfL | PrPSc | Spongiosis | ||
|---|---|---|---|---|---|
| Obex | Ng | - | 0.4918 * | −0.6006 ** | −0.7227 *** |
| NfL | 0.4918 * | - | −0.7755 **** | −0.7497 *** | |
| PrPSc | −0.6006 ** | −0.7755 **** | - | 0.8633 **** | |
| Spongiosis | −0.7227 *** | −0.7497 *** | 0.8633 **** | - | |
| Hp | Ng | - | 0.2959 n.s. | −0.2786 n.s. | −0.2238 n.s. |
| NfL | 0.2959 n.s. | - | −0.2145 n.s. | −0.2313 n.s. | |
| PrPSc | −0.2786 n.s. | −0.2145 n.s. | - | 0.7921 **** | |
| Spongiosis | −0.2238 n.s. | −0.2313 n.s. | 0.7921 **** | - | |
| Th | Ng | - | 0.6994 *** | −0.8124 **** | −0.8038 **** |
| NfL | 0.6994 *** | - | −0.8130 **** | −0.6530 ** | |
| PrPSc | −0.8124 **** | −0.8130 **** | - | 0.8112 **** | |
| Spongiosis | −0.8038 **** | −0.6530 ** | 0.8112 **** | - | |
| Fc | Ng | - | 0.2240 n.s. | −0.2587 n.s. | −0.5540 ** |
| NfL | 0.2240 n.s. | - | −0.1579 n.s. | −0.3233 n.s. | |
| PrPSc | −0.2587 n.s. | −0.1579 n.s. | - | 0.4971 * | |
| Spongiosis | −0.5540 ** | −0.3233 n.s. | 0.4971 * | - | |
| Gene | Forward (F) and Reverse (R) Primer Sequence (5′-3′) | Concentration (nM) | Accession Number |
|---|---|---|---|
| NRGN | F AACCTCCATCCCAGCCAAG | 400 | XM_027959688.2 |
| R CCCGGAAACTCGCCTGTA | 400 | ||
| NEFL | F CATTAGCGCTATGCAGGACA | 400 | XM_015093090.3 |
| R GCTGCAATCTCAATGTCCAA | 400 | ||
| GAPDH | F TCCATGACCACTTTGGCATCGT | 900 | AF_035421 |
| R GTCTTCTGGGTGGCAGTGA | 900 | ||
| SDHA | F CATCCACTACATGACGGAGCA | 300 | AY_970969 |
| R ATCTTGCCATCTTCAGTTCTGCTA | 300 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betancor, M.; Pérez-Lázaro, S.; Otero, A.; Marín, B.; Martín-Burriel, I.; Blennow, K.; Badiola, J.J.; Zetterberg, H.; Bolea, R. Neurogranin and Neurofilament Light Chain as Preclinical Biomarkers in Scrapie. Int. J. Mol. Sci. 2022, 23, 7182. https://doi.org/10.3390/ijms23137182
Betancor M, Pérez-Lázaro S, Otero A, Marín B, Martín-Burriel I, Blennow K, Badiola JJ, Zetterberg H, Bolea R. Neurogranin and Neurofilament Light Chain as Preclinical Biomarkers in Scrapie. International Journal of Molecular Sciences. 2022; 23(13):7182. https://doi.org/10.3390/ijms23137182
Chicago/Turabian StyleBetancor, Marina, Sonia Pérez-Lázaro, Alicia Otero, Belén Marín, Inmaculada Martín-Burriel, Kaj Blennow, Juan José Badiola, Henrik Zetterberg, and Rosa Bolea. 2022. "Neurogranin and Neurofilament Light Chain as Preclinical Biomarkers in Scrapie" International Journal of Molecular Sciences 23, no. 13: 7182. https://doi.org/10.3390/ijms23137182
APA StyleBetancor, M., Pérez-Lázaro, S., Otero, A., Marín, B., Martín-Burriel, I., Blennow, K., Badiola, J. J., Zetterberg, H., & Bolea, R. (2022). Neurogranin and Neurofilament Light Chain as Preclinical Biomarkers in Scrapie. International Journal of Molecular Sciences, 23(13), 7182. https://doi.org/10.3390/ijms23137182








