BHLHE22 Expression Is Associated with a Proinflammatory Immune Microenvironment and Confers a Favorable Prognosis in Endometrial Cancer
Abstract
1. Introduction
2. Results
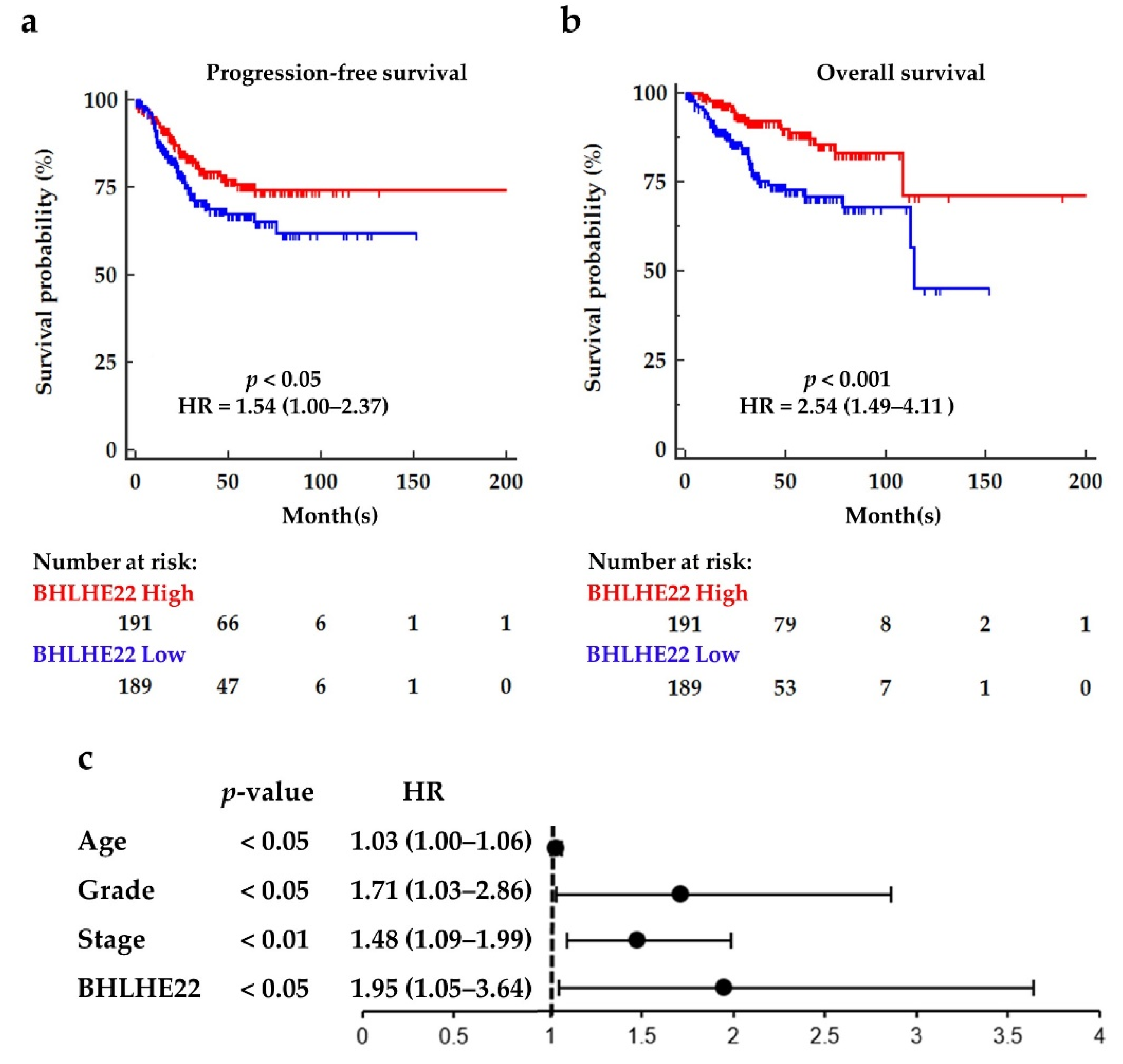
2.1. BHLHE22 Is Downregulated in EC Tissues and Correlated with Poor Survival
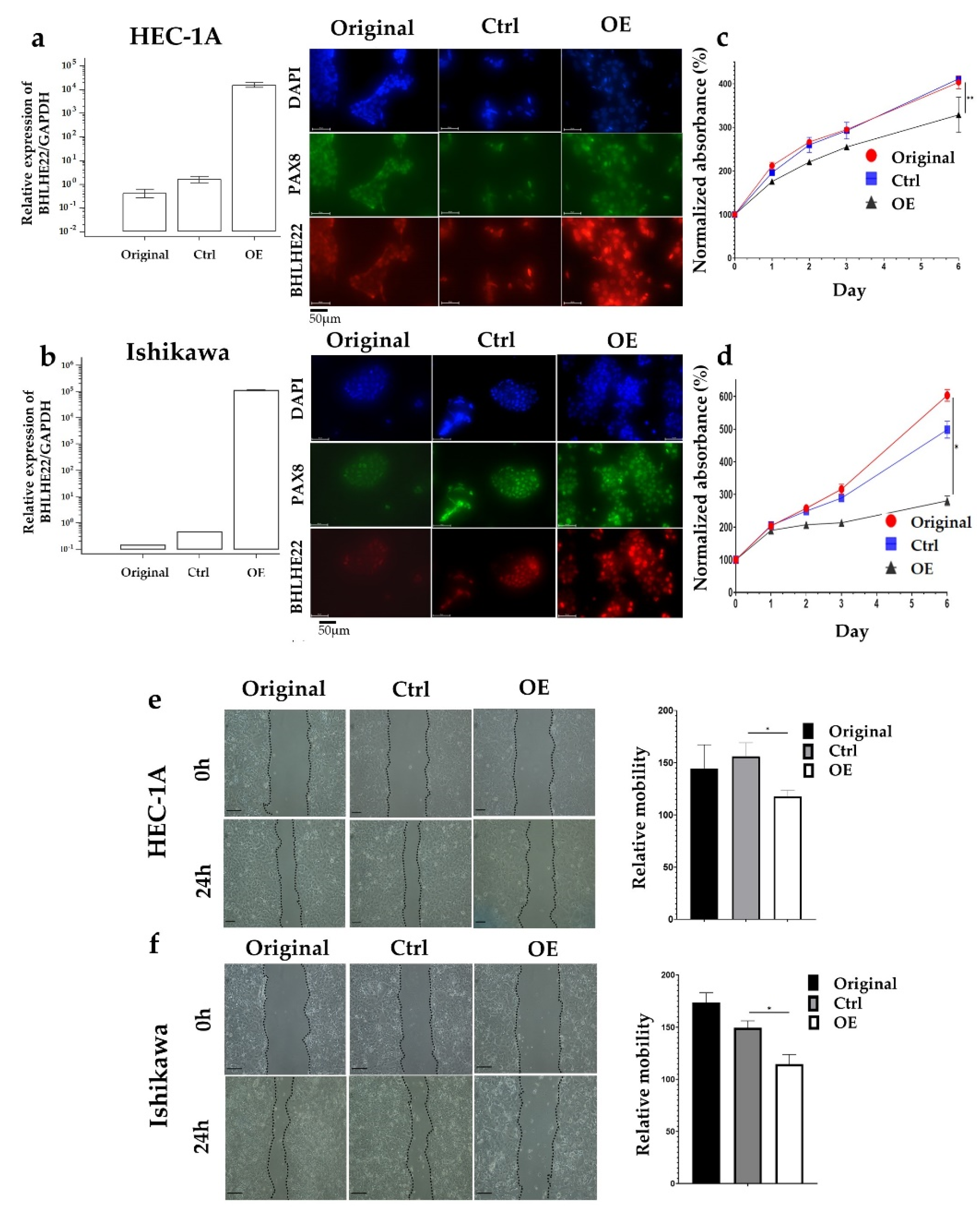
2.2. BHLHE22 Expression Inhibits EC Proliferation and Migration
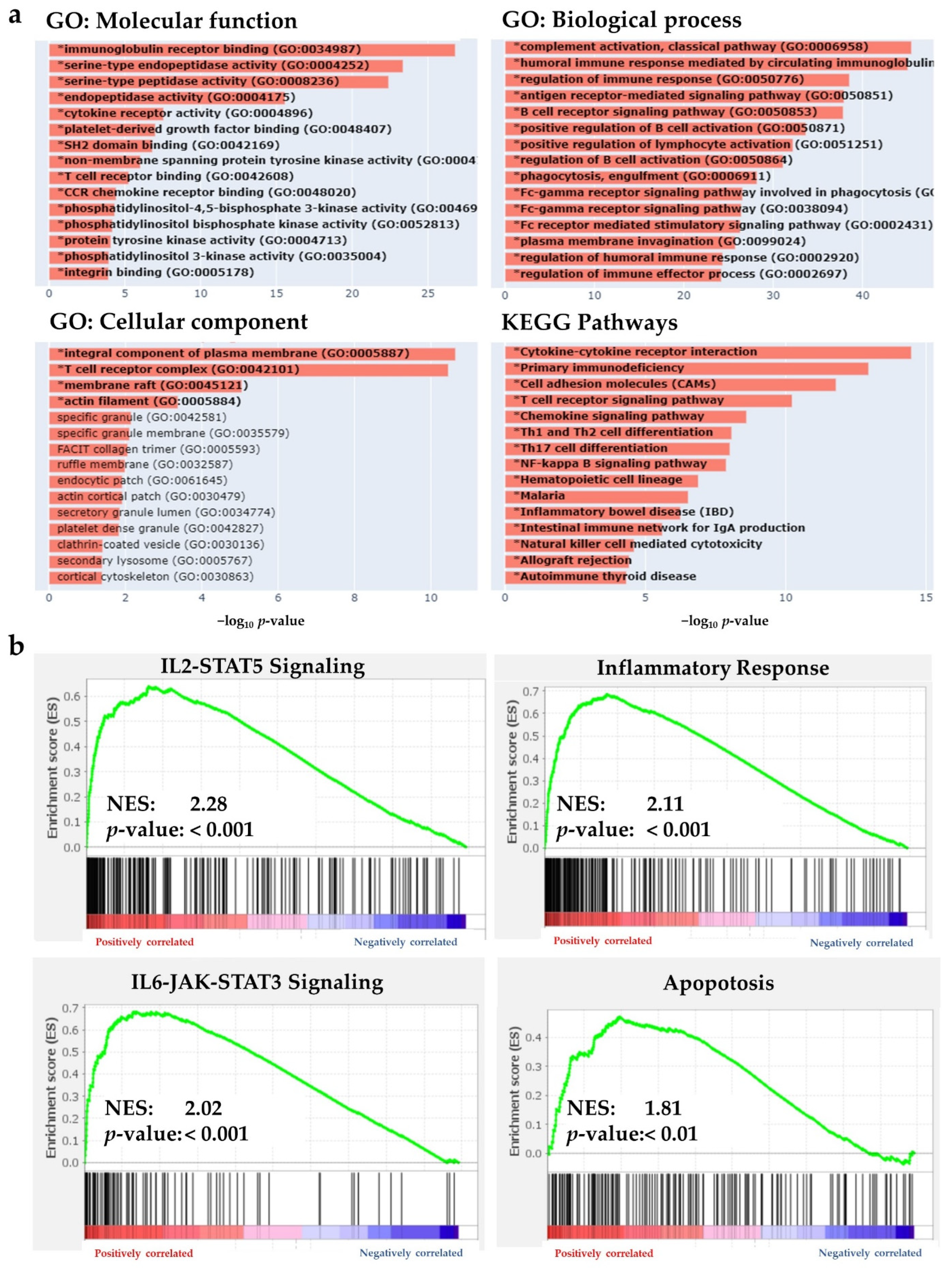
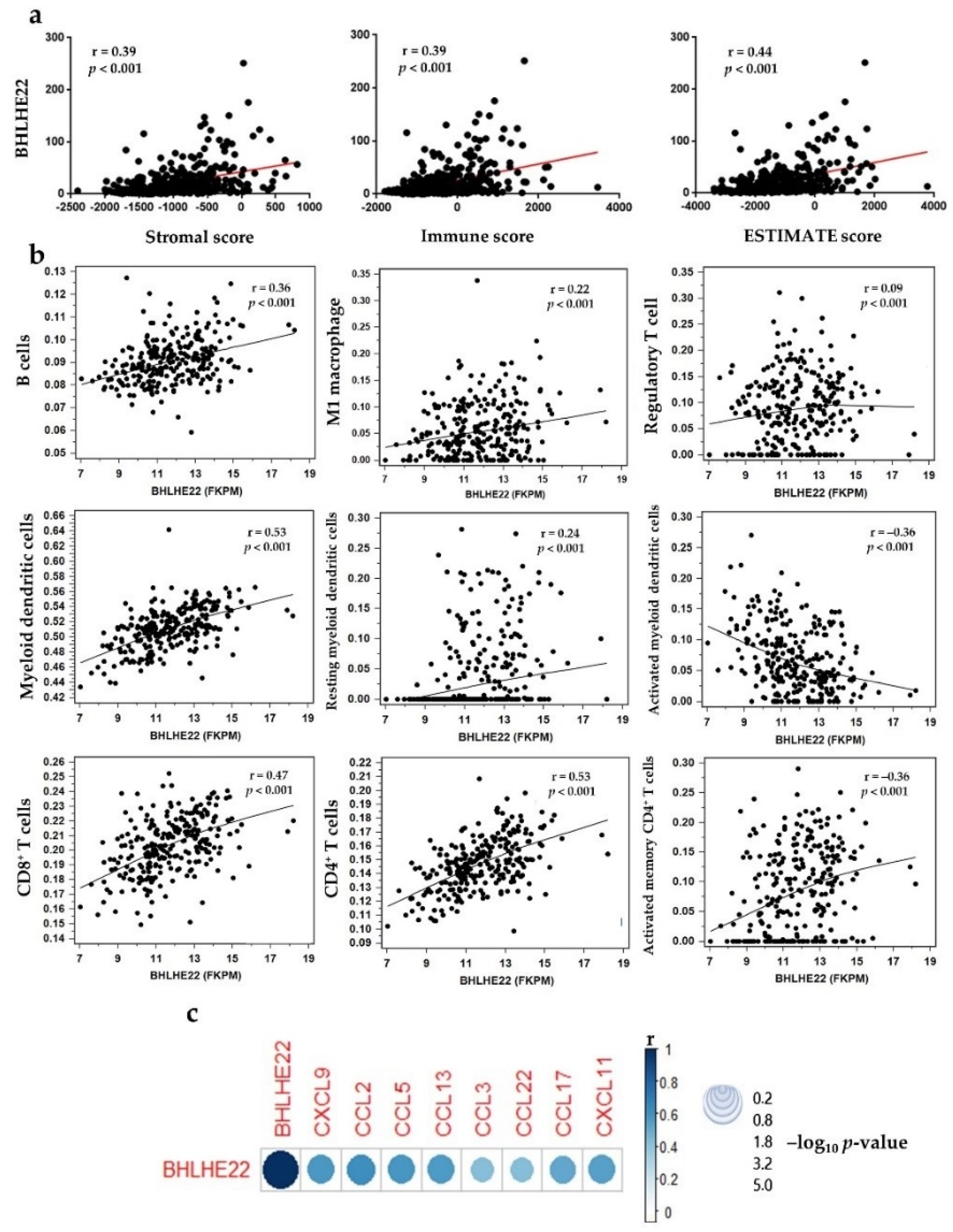
2.3. BHLHE22 Is Associated with Immune Function in EC
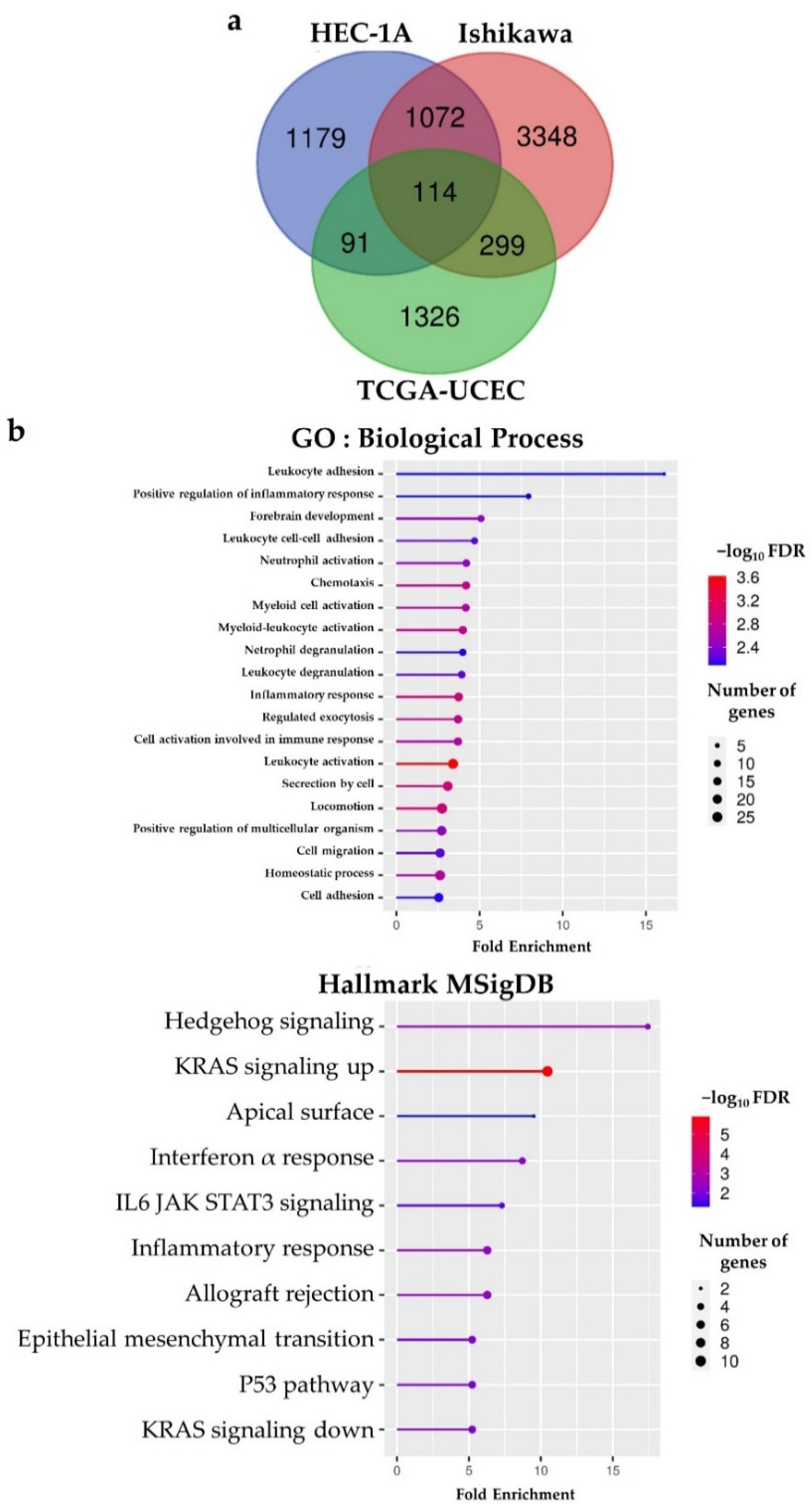
2.4. BHLHE22 Positively Correlated with Chemokines Expression and Proinflammatory Immune Cells in EC Microenvironment
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.2. Cell Lines Culture Condition and Generation of Stable Cell Lines
4.3. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (qPCR)
4.4. RNA Sequencing
4.5. Immunofluorescence (IF)
4.6. Cell Proliferation Assay
4.7. Wound Healing Assay
4.8. Data Sources
4.9. LinkedOmics Database and Gene Set Enrichment Analysis (GSEA)
4.10. TIMER and EPIC Database Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.; Gong, T.T.; Liu, F.H.; Jiang, Y.T.; Sun, H.; Ma, X.X.; Zhao, Y.H.; Wu, Q.J. Global, Regional, and National Burden of Endometrial Cancer, 1990–2017: Results From the Global Burden of Disease Study, 2017. Front. Oncol. 2019, 9, 1440. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- McAlpine, J.N.; Temkin, S.M.; Mackay, H.J. Endometrial cancer: Not your grandmother’s cancer. Cancer 2016, 122, 2787–2798. [Google Scholar] [CrossRef]
- Temkin, S.M.; Kohn, E.C.; Penberthy, L.; Cronin, K.A.; Rubinsak, L.; Dickie, L.A.; Minasian, L.; Noone, A.M. Hysterectomy-corrected rates of endometrial cancer among women younger than age 50 in the United States. Cancer Causes Control 2018, 29, 427–433. [Google Scholar] [CrossRef]
- Surveillance, Epidemiology, and End Results (SEER) Program. Cancer Stat Facts: Uterine Cancer. Available online: https://seer.cancer.gov/statfacts/html/corp.html (accessed on 7 April 2021).
- Connor, E.V.; Rose, P.G. Management Strategies for Recurrent Endometrial Cancer. Expert Rev. Anticancer Ther. 2018, 18, 873–885. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Lu, K.H.; Broaddus, R.R. Endometrial Cancer. N. Engl. J. Med. 2020, 383, 2053–2064. [Google Scholar] [CrossRef]
- Levine, D.A. The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Howitt, B.E.; Shukla, S.A.; Sholl, L.M.; Ritterhouse, L.L.; Watkins, J.C.; Rodig, S.; Stover, E.; Strickland, K.C.; D’Andrea, A.D.; Wu, C.J.; et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol. 2015, 1, 1319–1323. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Bang, Y.J.; Berton-Rigaud, D.; Elez, E.; Pishvaian, M.J.; Rugo, H.S.; Puzanov, I.; Mehnert, J.M.; Aung, K.L.; Lopez, J.; et al. Safety and Antitumor Activity of Pembrolizumab in Advanced Programmed Death Ligand 1-Positive Endometrial Cancer: Results From the KEYNOTE-028 Study. J. Clin. Oncol. 2017, 35, 2535–2541. [Google Scholar] [CrossRef] [PubMed]
- Oaknin, A.; Gilbert, L.; Tinker, A.V.; Brown, J.; Mathews, C.; Press, J.; Sabatier, R.; O’Malley, D.M.; Samouelian, V.; Boni, V.; et al. Safety and antitumor activity of dostarlimab in patients with advanced or recurrent DNA mismatch repair deficient/microsatellite instability-high (dMMR/MSI-H) or proficient/stable (MMRp/MSS) endometrial cancer: Interim results from GARNET-a phase I, single-arm study. J. Immunother. Cancer 2022, 10, e003777. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.L.; Su, P.H.; Liao, Y.P.; Wu, T.I.; Hsu, Y.T.; Lin, W.Y.; Wang, H.C.; Weng, Y.C.; Ou, Y.C.; Huang, T.H.; et al. Integrated Epigenomics Analysis Reveals a DNA Methylation Panel for Endometrial Cancer Detection Using Cervical Scrapings. Clin. Cancer Res. 2017, 23, 263–272. [Google Scholar] [CrossRef]
- Liew, P.L.; Huang, R.L.; Wu, T.I.; Liao, C.C.; Chen, C.W.; Su, P.H.; Wang, H.C.; Weng, Y.C.; Lai, H.C. Combined genetic mutations and DNA-methylated genes as biomarkers for endometrial cancer detection from cervical scrapings. Clin. Epigenetics 2019, 11, 170. [Google Scholar] [CrossRef]
- Xu, Z.P.; Dutra, A.; Stellrecht, C.M.; Wu, C.; Piatigorsky, J.; Saunders, G.F. Functional and structural characterization of the human gene BHLHB5, encoding a basic helix-loop-helix transcription factor. Genomics 2002, 80, 311–318. [Google Scholar] [CrossRef]
- Ross, S.E.; McCord, A.E.; Jung, C.; Atan, D.; Mok, S.I.; Hemberg, M.; Kim, T.K.; Salogiannis, J.; Hu, L.; Cohen, S.; et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron 2012, 73, 292–303. [Google Scholar] [CrossRef]
- Li, Q.; Kannan, A.; DeMayo, F.J.; Lydon, J.P.; Cooke, P.S.; Yamagishi, H.; Srivastava, D.; Bagchi, M.K.; Bagchi, I.C. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 2011, 331, 912–916. [Google Scholar] [CrossRef]
- Fukuda, T.; Shirane, A.; Wada-Hiraike, O.; Oda, K.; Tanikawa, M.; Sakuabashi, A.; Hirano, M.; Fu, H.; Morita, Y.; Miyamoto, Y.; et al. HAND2-mediated proteolysis negatively regulates the function of estrogen receptor α. Mol. Med. Rep. 2015, 12, 5538–5544. [Google Scholar] [CrossRef][Green Version]
- Murata, H.; Tanaka, S.; Tsuzuki-Nakao, T.; Kido, T.; Kakita-Kobayashi, M.; Kida, N.; Hisamatsu, Y.; Tsubokura, H.; Hashimoto, Y.; Kitada, M.; et al. The transcription factor HAND2 up-regulates transcription of the IL15 gene in human endometrial stromal cells. J. Biol. Chem. 2020, 295, 9596–9605. [Google Scholar] [CrossRef]
- Buell-Gutbrod, R.; Cavallo, A.; Lee, N.; Montag, A.; Gwin, K. Heart and Neural Crest Derivatives Expressed Transcript 2 (HAND2): A novel biomarker for the identification of atypical hyperplasia and Type I endometrial carcinoma. Int. J. Gynecol. Pathol. 2015, 34, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.E.; Jarjour, N.N.; Lin, C.C.; Edelson, B.T. Transcription Factor Bhlhe40 in Immunity and Autoimmunity. Trends Immunol. 2020, 41, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, B.; Son, Y.M.; Wang, Z.; Jiang, L.; Xiang, M.; Ye, Z.; Beckermann, K.E.; Wu, Y.; Jenkins, J.W.; et al. The Transcription Factor Bhlhe40 Programs Mitochondrial Regulation of Resident CD8(+) T Cell Fitness and Functionality. Immunity 2019, 51, 491–507.e497. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013, 4, 2612. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Song, Y.; Gu, Y.; Hu, X.; Wang, M.; He, Q.; Li, Y. Endometrial Tumors with MSI-H and dMMR Share a Similar Tumor Immune Microenvironment. OncoTargets Ther. 2021, 14, 4485–4497. [Google Scholar] [CrossRef]
- Aviña-Padilla, K.; Ramírez-Rafael, J.A.; Herrera-Oropeza, G.E.; Muley, V.Y.; Valdivia, D.I.; Díaz-Valenzuela, E.; García-García, A.; Varela-Echavarría, A.; Hernández-Rosales, M. Evolutionary Perspective and Expression Analysis of Intronless Genes Highlight the Conservation of Their Regulatory Role. Front. Genet. 2021, 12, 654256. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shen, Y.; Wang, L.; Qiao, J.; Zhao, Y.; Wang, Q. A novel 12-gene prognostic signature in breast cancer based on the tumor microenvironment. Ann. Transl. Med. 2022, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Dennis, D.J.; Han, S.; Schuurmans, C. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019, 1705, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Li, B.L.; Wan, X.P. Prognostic significance of immune landscape in tumour microenvironment of endometrial cancer. J. Cell Mol. Med. 2020, 24, 7767–7777. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.J.; Nakamura, T.; Miyazaki, D.; Ohbayashi, M.; Dawson, M.; Toda, M. Chemokines: Roles in leukocyte development, trafficking, and effector function. J. Allergy Clin. Immunol. 2003, 111, 1185–1199; quiz 1200. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pagès, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Wang, W.; Kryczek, I.; Dostál, L.; Lin, H.; Tan, L.; Zhao, L.; Lu, F.; Wei, S.; Maj, T.; Peng, D.; et al. Effector T Cells Abrogate Stroma-Mediated Chemoresistance in Ovarian Cancer. Cell 2016, 165, 1092–1105. [Google Scholar] [CrossRef]
- Mollica Poeta, V.; Massara, M.; Capucetti, A.; Bonecchi, R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front. Immunol. 2019, 10, 379. [Google Scholar] [CrossRef]
- Mughees, M.; Kaushal, J.B.; Sharma, G.; Wajid, S.; Batra, S.K.; Siddiqui, J.A. Chemokines and cytokines: Axis and allies in prostate cancer pathogenesis. Semin. Cancer Biol. 2022, in press. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Grywalska, E.; Sobstyl, M.; Putowski, L.; Roliński, J. Current Possibilities of Gynecologic Cancer Treatment with the Use of Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2019, 20, 4705. [Google Scholar] [CrossRef]
- Whelan, S.; Ophir, E.; Kotturi, M.F.; Levy, O.; Ganguly, S.; Leung, L.; Vaknin, I.; Kumar, S.; Dassa, L.; Hansen, K.; et al. PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8(+) T-cell Function. Cancer Immunol. Res. 2019, 7, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darmawi; Chen, L.-Y.; Su, P.-H.; Liew, P.-L.; Wang, H.-C.; Weng, Y.-C.; Huang, R.-L.; Lai, H.-C. BHLHE22 Expression Is Associated with a Proinflammatory Immune Microenvironment and Confers a Favorable Prognosis in Endometrial Cancer. Int. J. Mol. Sci. 2022, 23, 7158. https://doi.org/10.3390/ijms23137158
Darmawi, Chen L-Y, Su P-H, Liew P-L, Wang H-C, Weng Y-C, Huang R-L, Lai H-C. BHLHE22 Expression Is Associated with a Proinflammatory Immune Microenvironment and Confers a Favorable Prognosis in Endometrial Cancer. International Journal of Molecular Sciences. 2022; 23(13):7158. https://doi.org/10.3390/ijms23137158
Chicago/Turabian StyleDarmawi, Lin-Yu Chen, Po-Hsuan Su, Phui-Ly Liew, Hui-Chen Wang, Yu-Chun Weng, Rui-Lan Huang, and Hung-Cheng Lai. 2022. "BHLHE22 Expression Is Associated with a Proinflammatory Immune Microenvironment and Confers a Favorable Prognosis in Endometrial Cancer" International Journal of Molecular Sciences 23, no. 13: 7158. https://doi.org/10.3390/ijms23137158
APA StyleDarmawi, Chen, L.-Y., Su, P.-H., Liew, P.-L., Wang, H.-C., Weng, Y.-C., Huang, R.-L., & Lai, H.-C. (2022). BHLHE22 Expression Is Associated with a Proinflammatory Immune Microenvironment and Confers a Favorable Prognosis in Endometrial Cancer. International Journal of Molecular Sciences, 23(13), 7158. https://doi.org/10.3390/ijms23137158





