Dynamics of Protein Phosphorylation during Arabidopsis Seed Germination
Abstract
:1. Introduction
2. Results
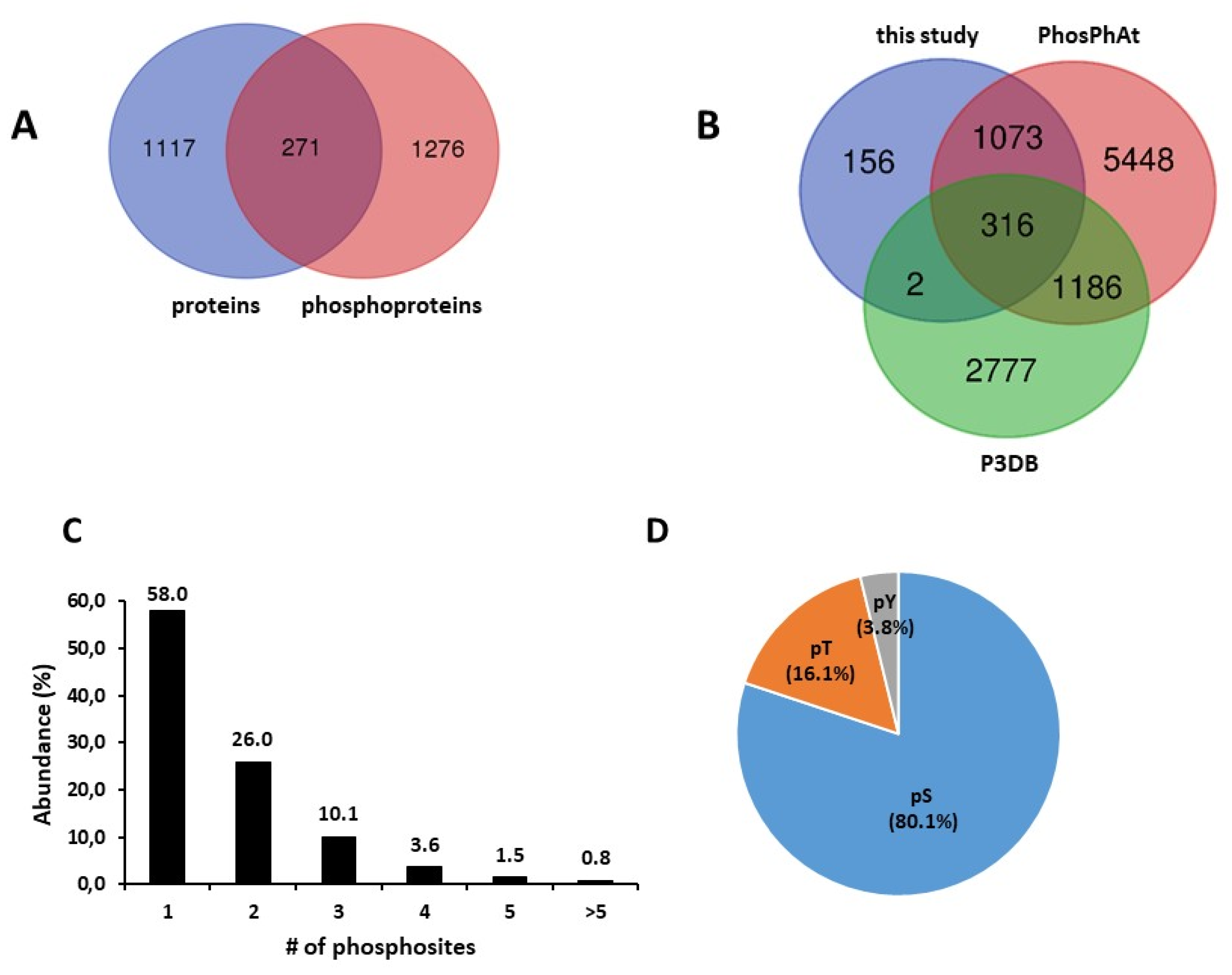
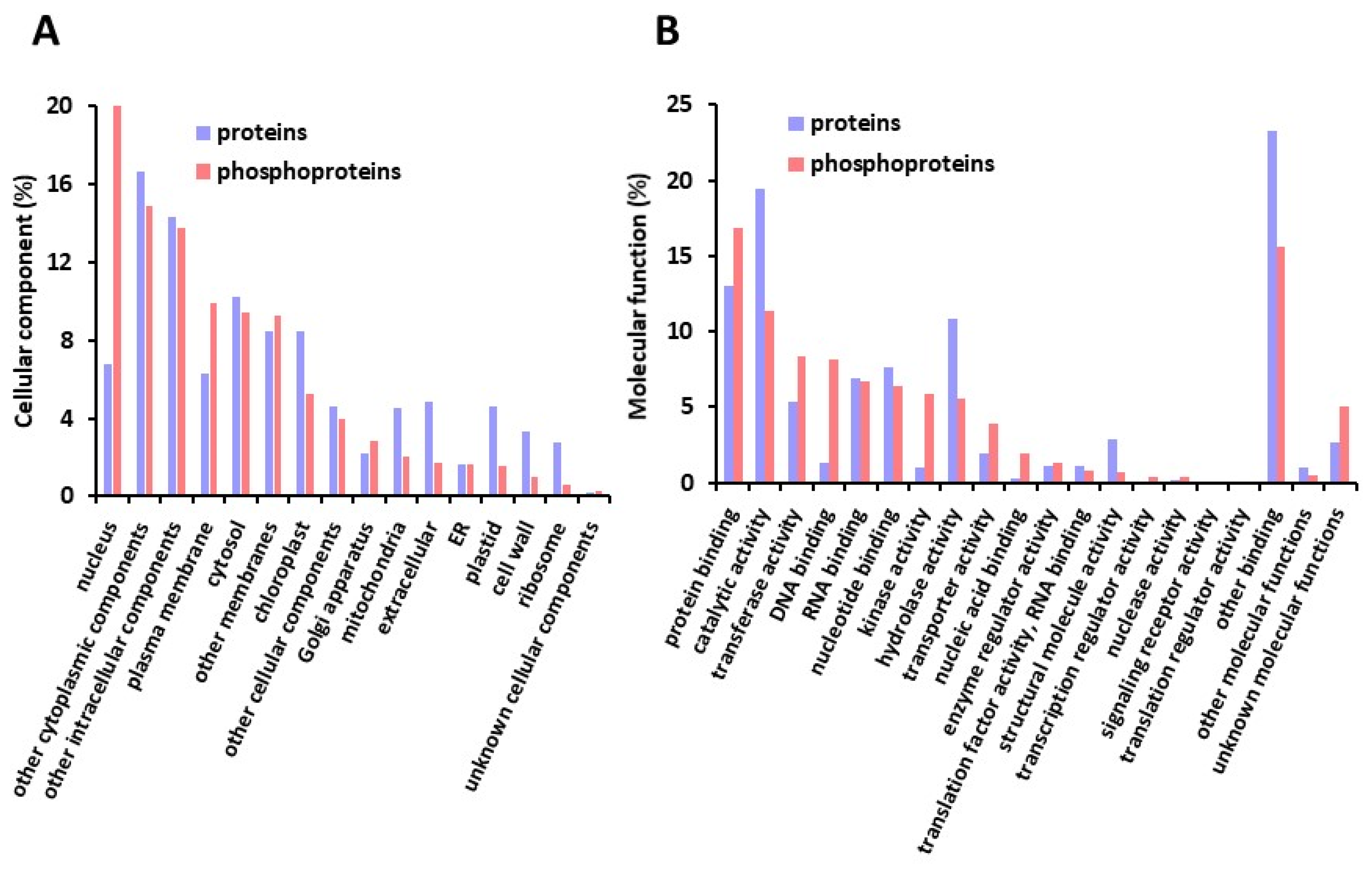
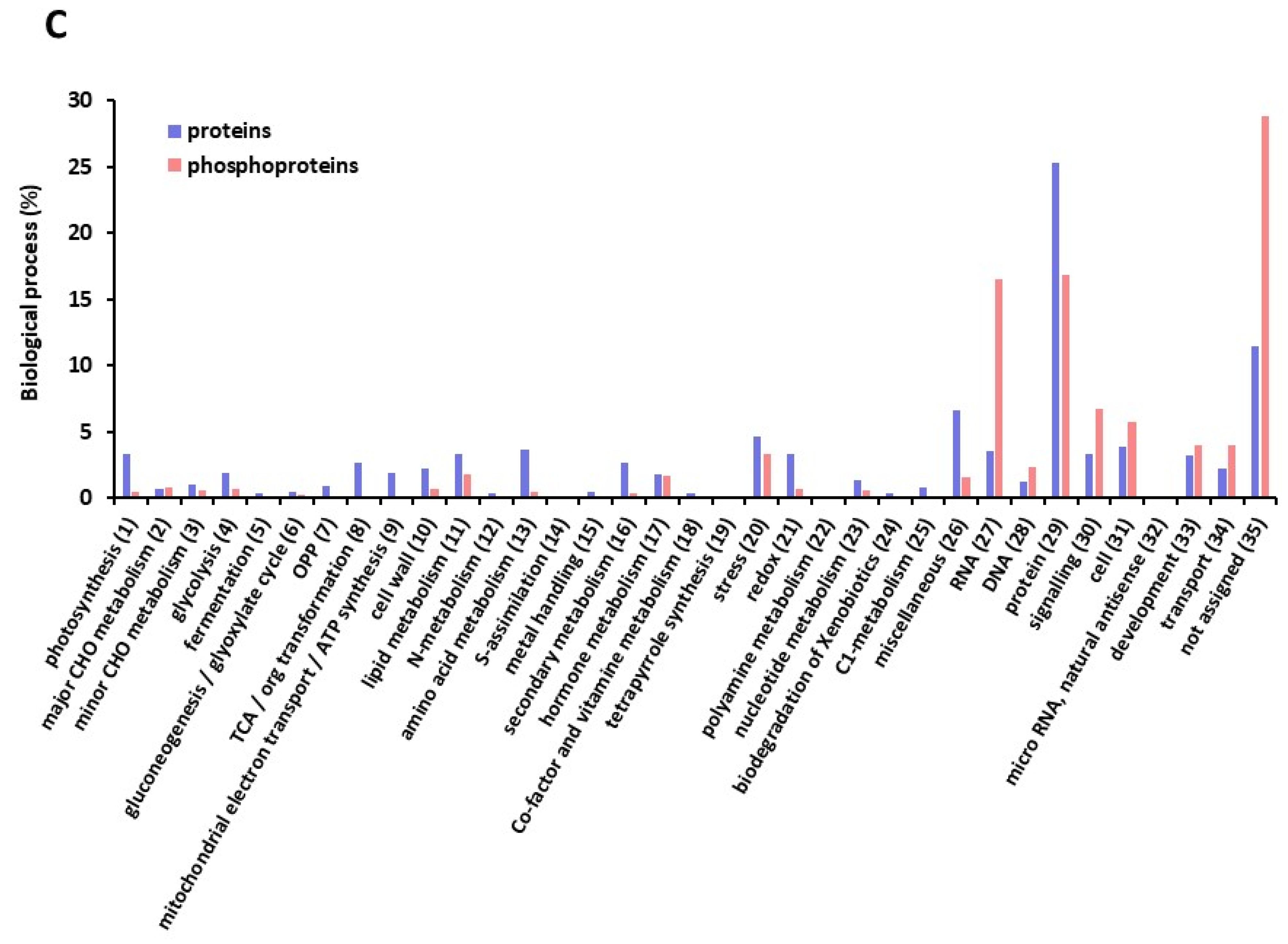
2.1. A Large Repertoire of Phosphoproteins Identified in Dry and Imbibed Arabidopsis Seeds
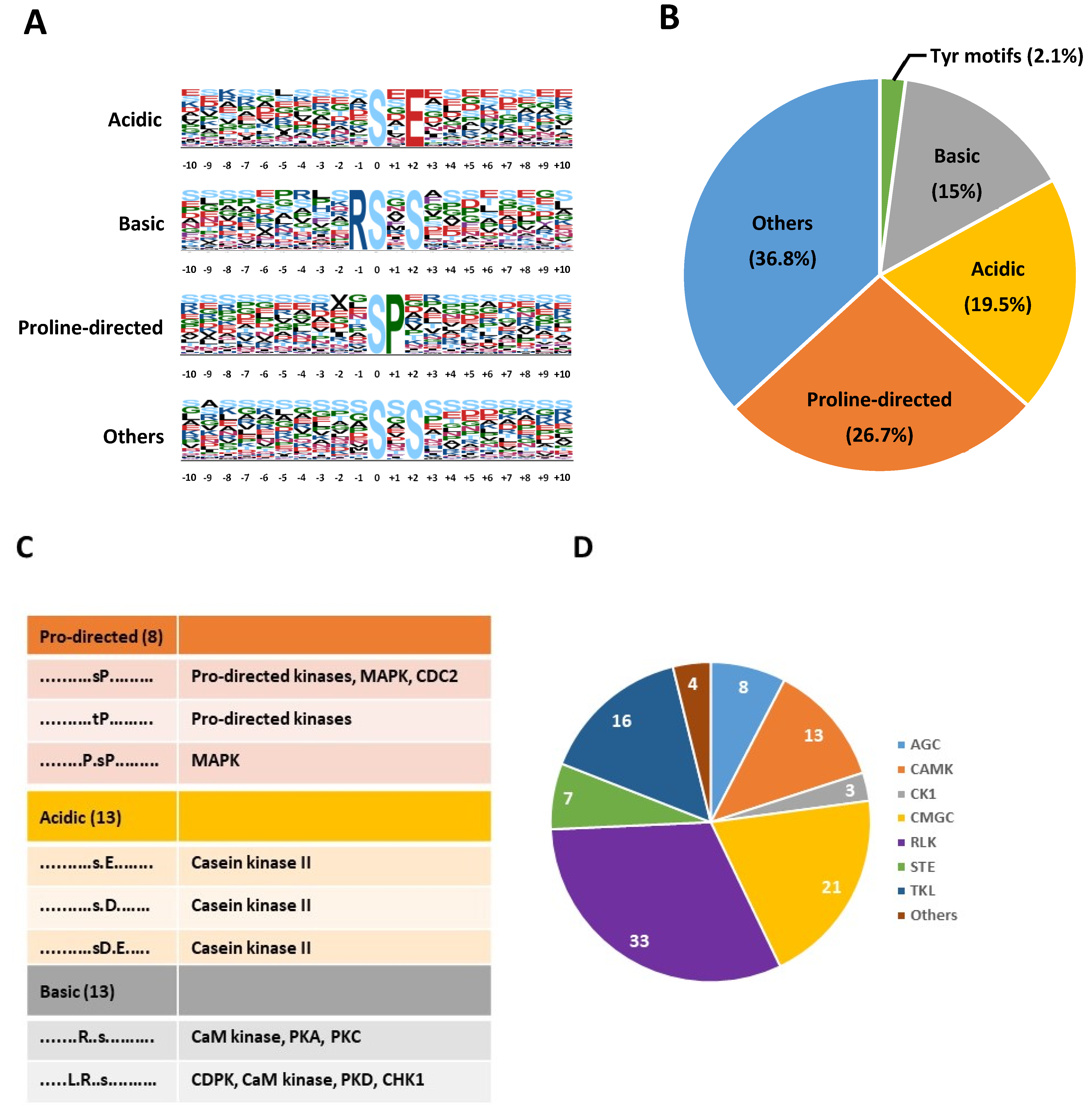
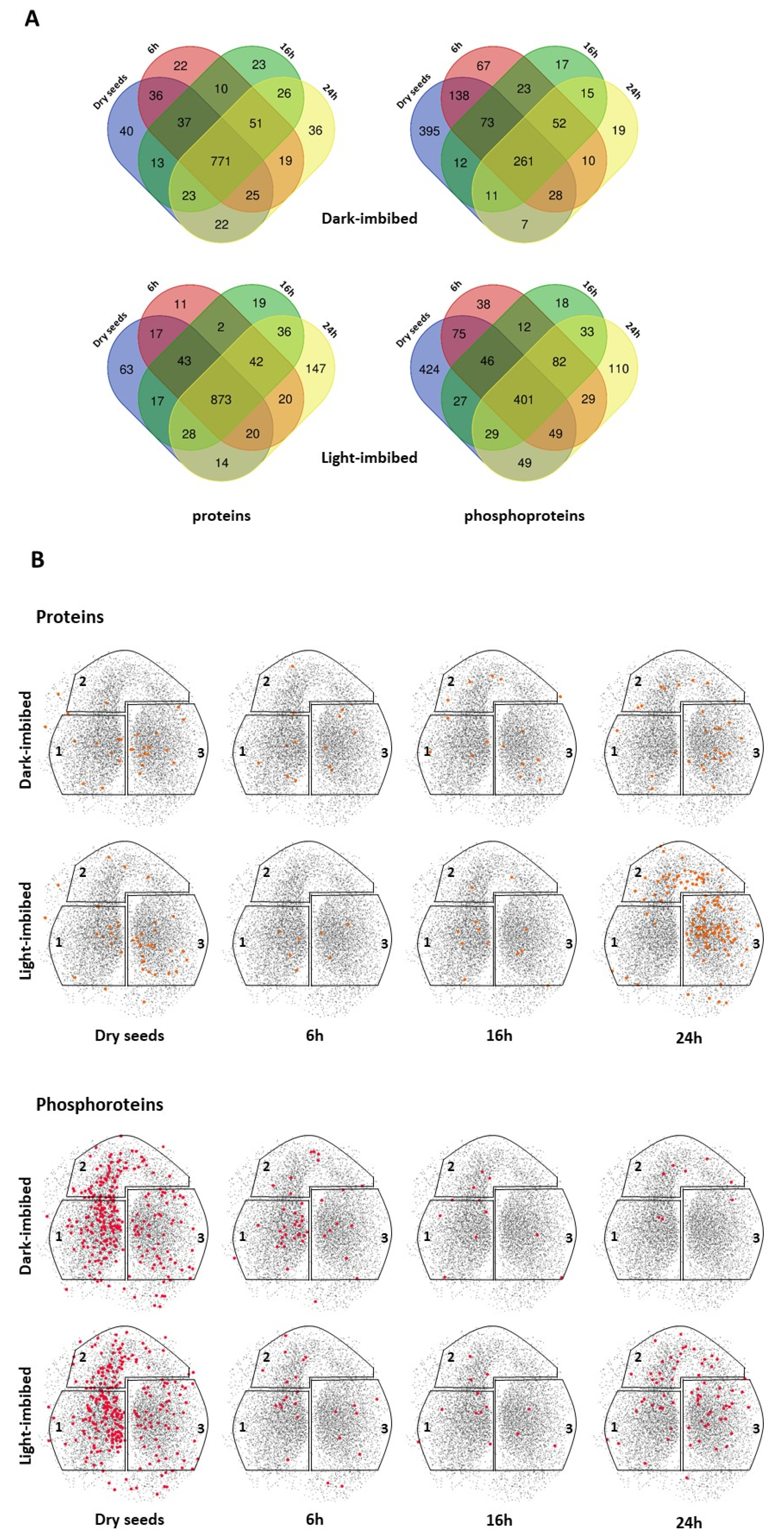
2.2. Modification of the Composition of Seed Phosphoproteome in Dark-Imbibed and Light-Imbibed Seeds
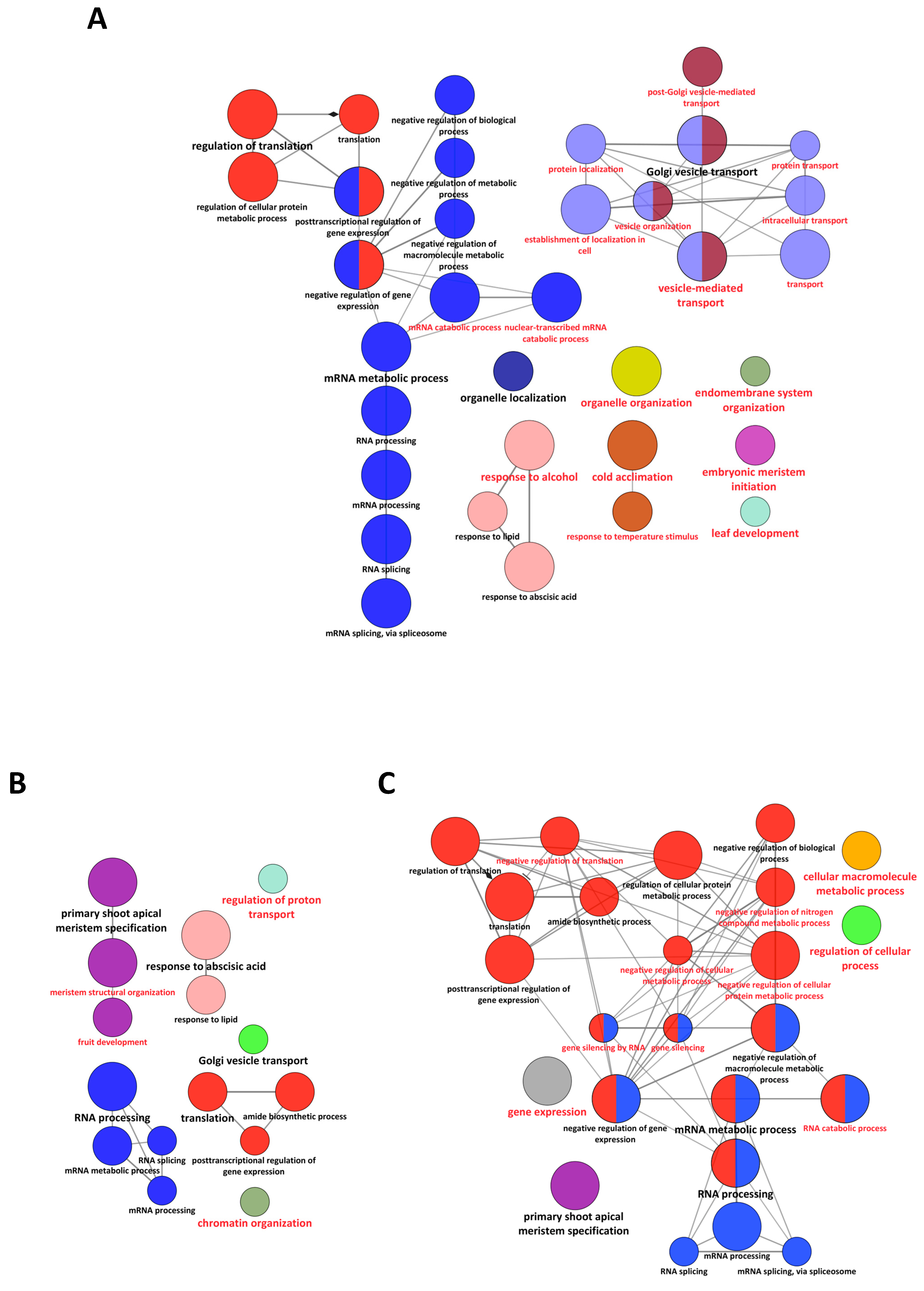
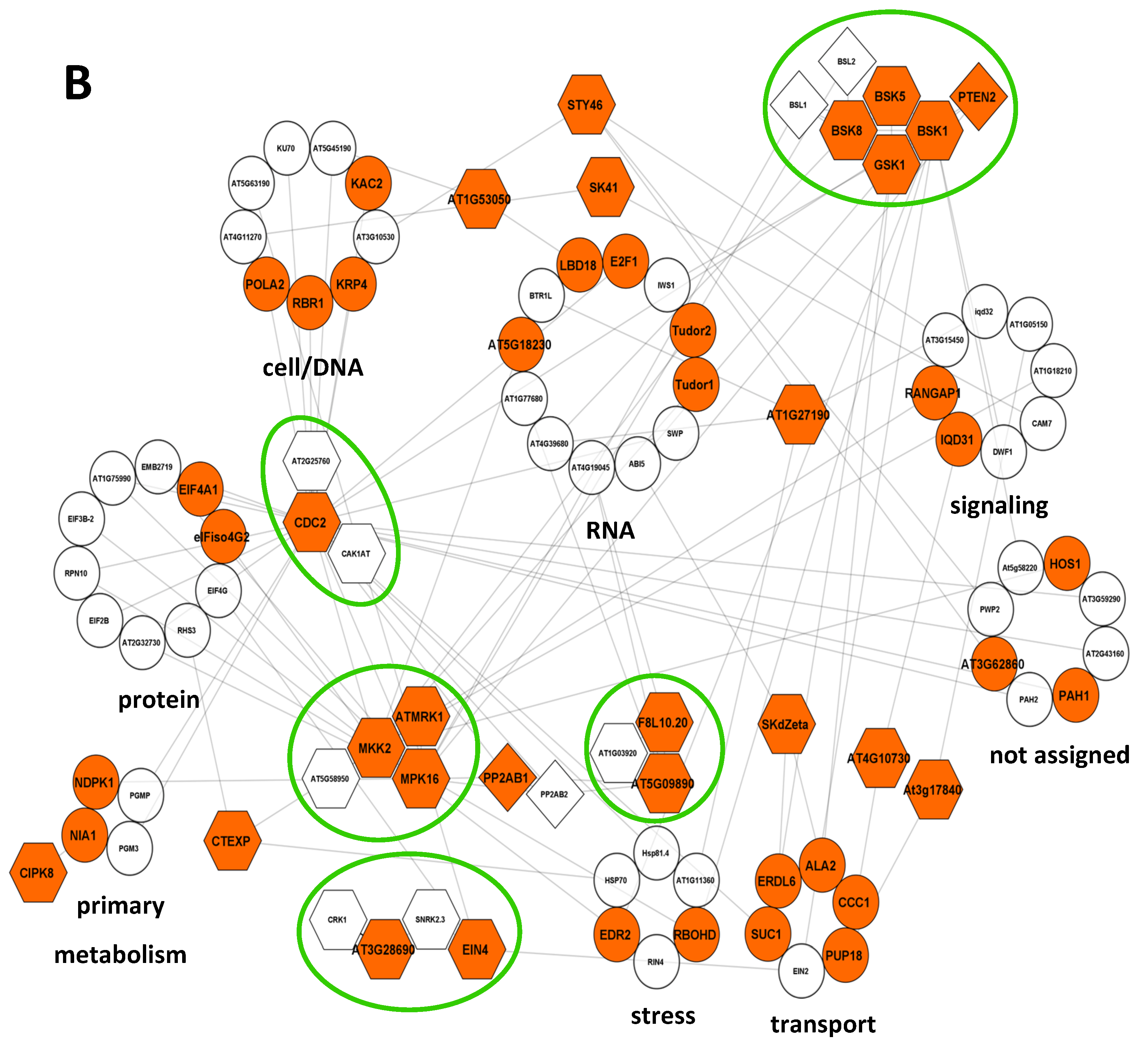
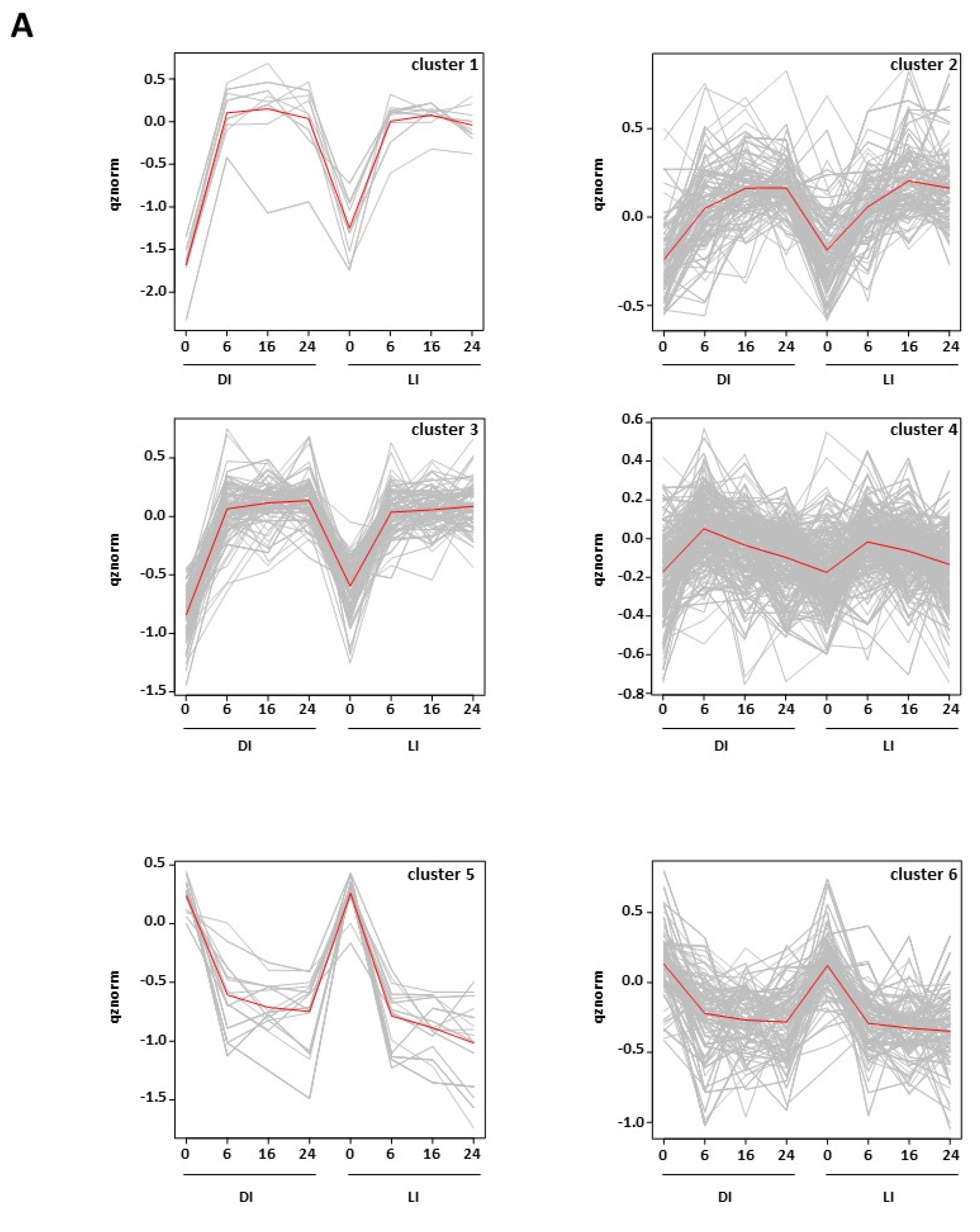
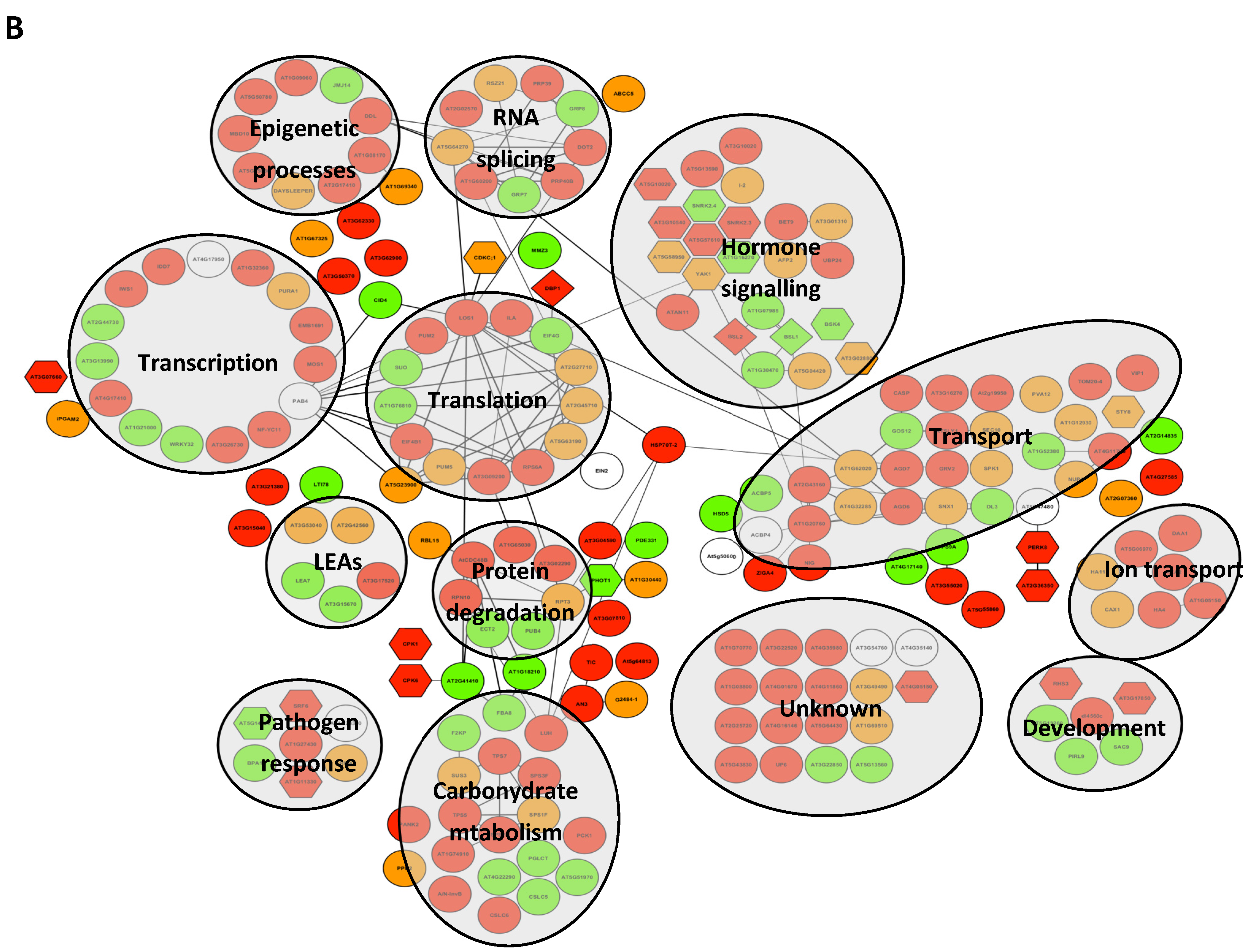
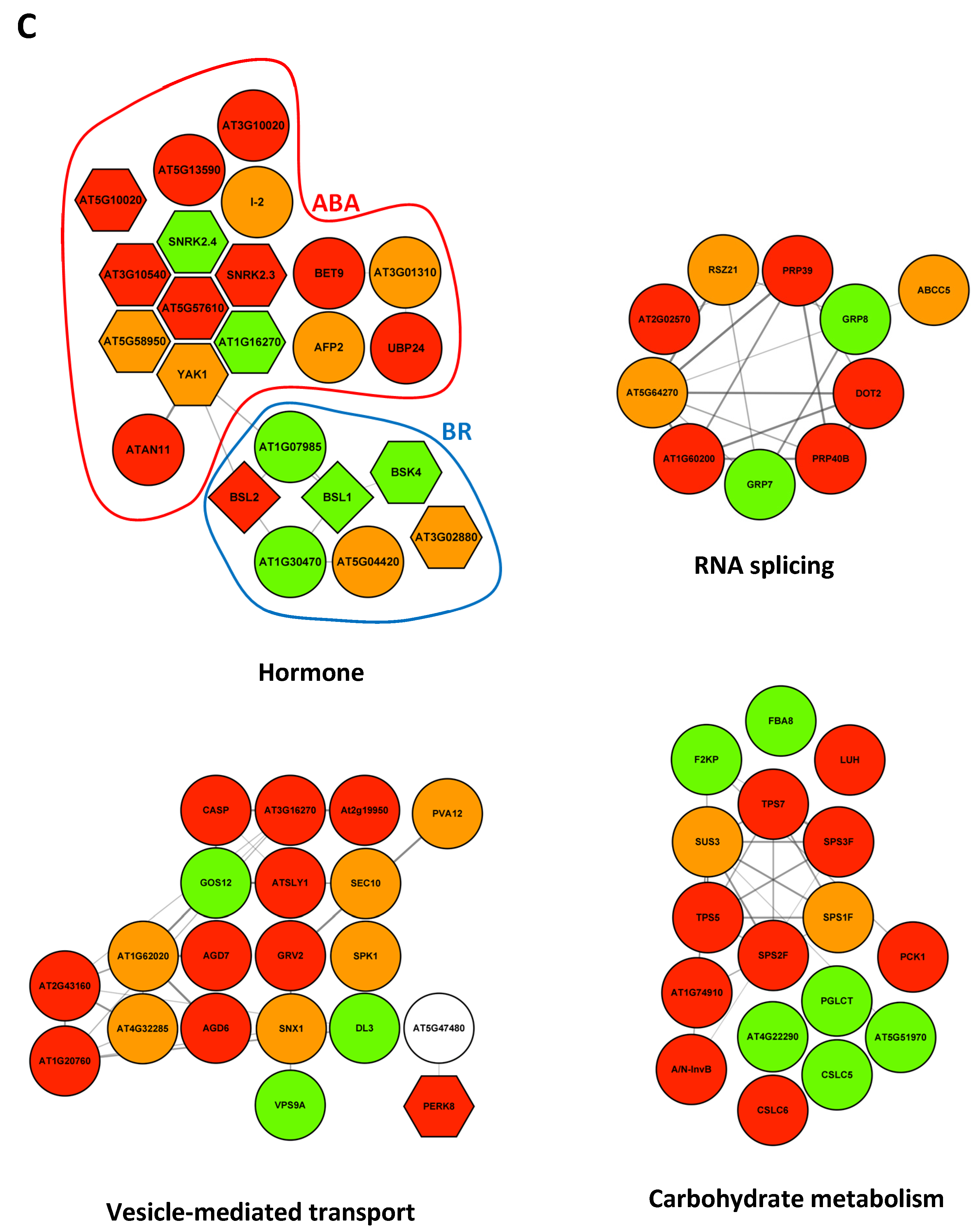
2.3. Modifications of the Abundance of Constitutive Seed Phosphoproteins in Dark-Imbibed and Light-Imbibed Seeds
3. Discussion
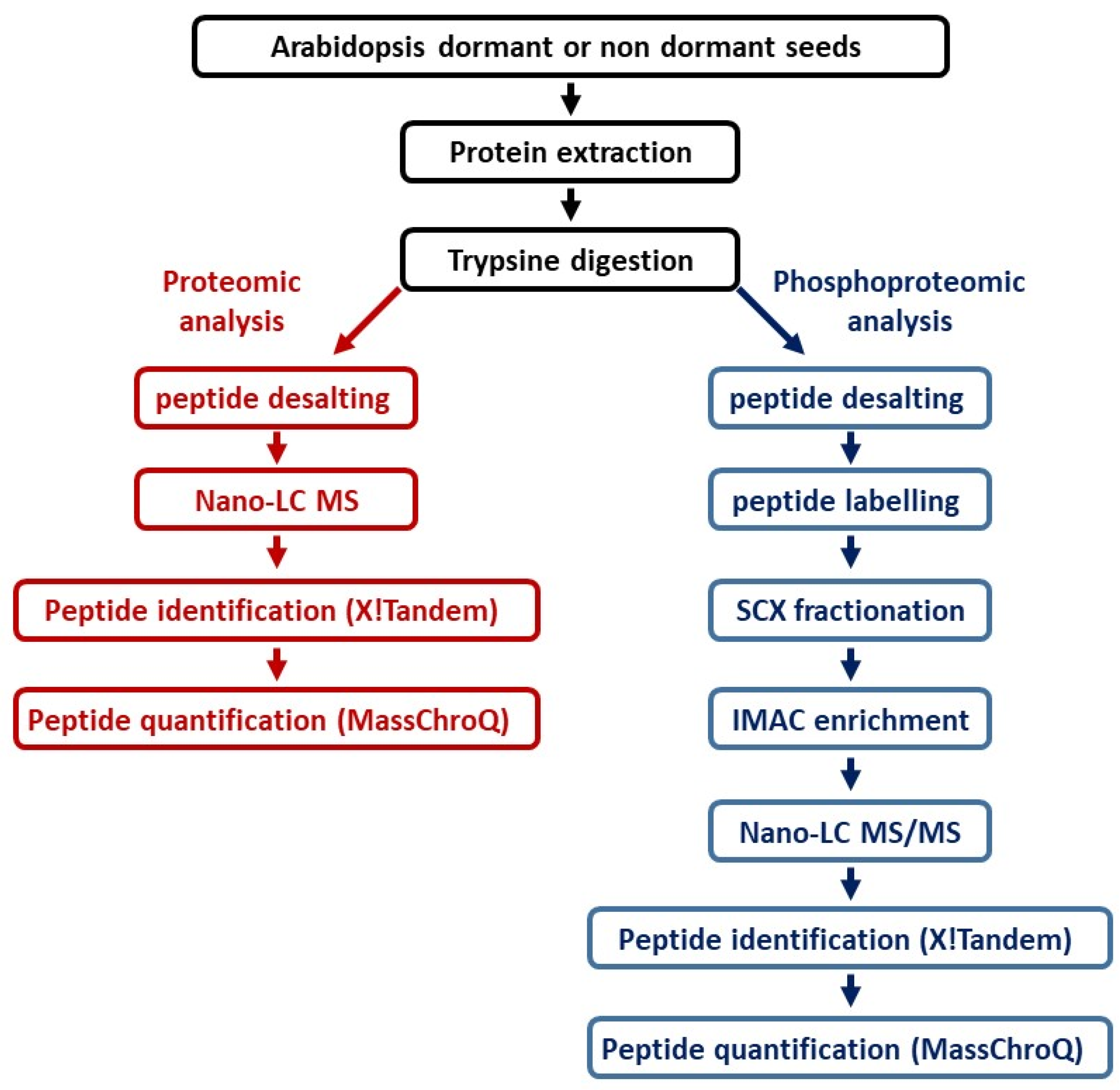
4. Materials and Methods
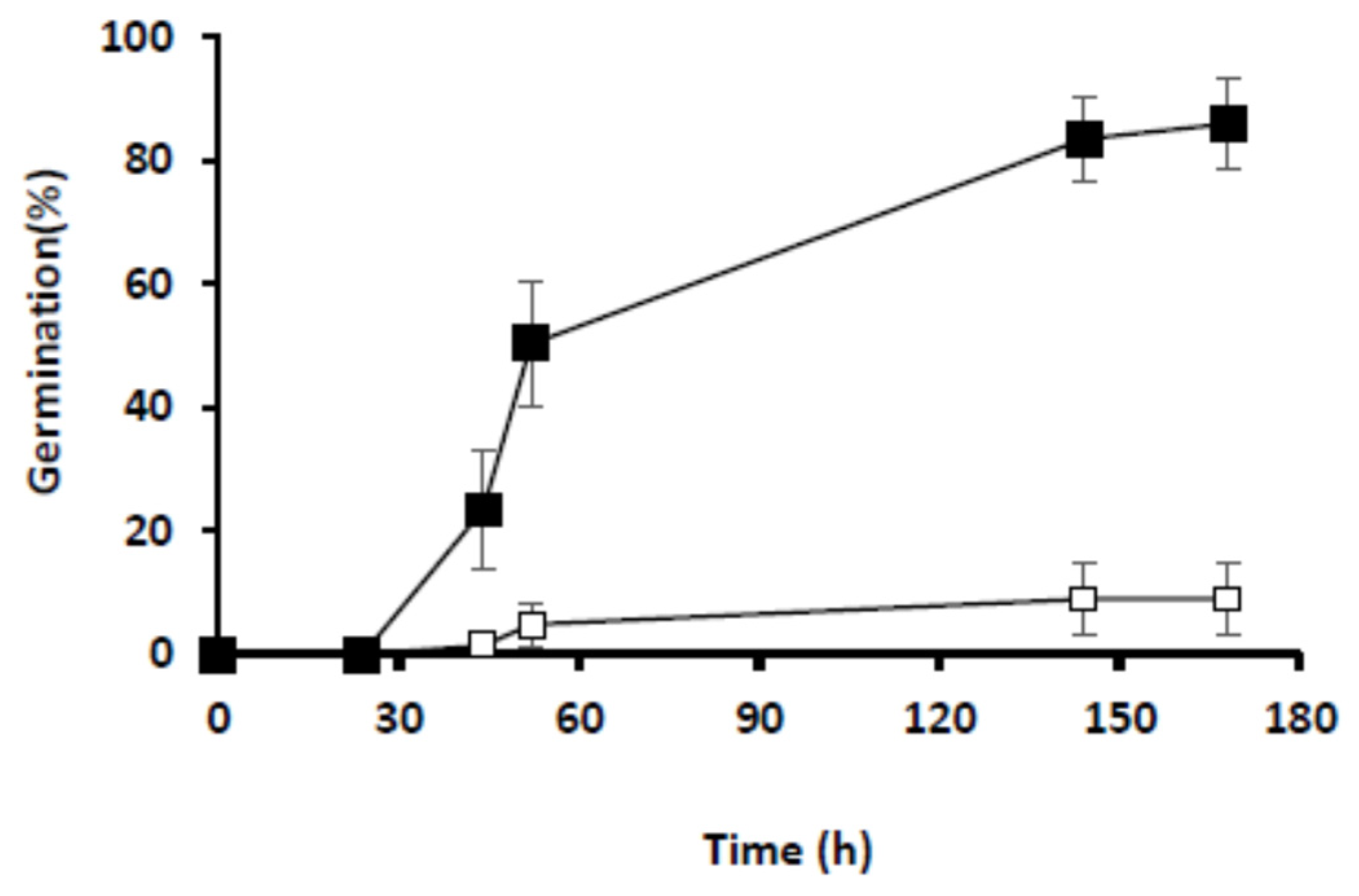
4.1. Seed Material
4.2. Germination Assays
4.3. Protein Extraction and In-Solution Digestion
4.4. Stable Isotope Dimethyl Labeling for Phosphoproteomic Approach
4.5. Peptide Fractionation Using Strong Cation Exchange Chromatography (SCX)
4.6. Enrichment of Phosphopeptides Using Immobilized Metal Ion Affinity Chromatography (IMAC)
4.7. nanoLC-MS/MS Analysis
4.8. Identification of Proteins and Phosphopeptides
4.9. Quantification of Peptides and Phosphopeptides
4.10. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Penfield, S. Seed Dormancy and Germination. Curr. Biol. CB 2017, 27, R874–R878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graeber, K.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J.J. Molecular Mechanisms of Seed Dormancy. Plant Cell Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular Aspects of Seed Dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed Germination and Vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-Q.; Liu, S.-J.; Song, S.-Q.; Møller, I.M. Proteomics of Seed Development, Desiccation Tolerance, Germination and Vigor. Plant Physiol. Biochem. 2015, 86, 1–15. [Google Scholar] [CrossRef]
- Galland, M.; Huguet, R.; Arc, E.; Cueff, G.; Job, D.; Rajjou, L. Dynamic Proteomics Emphasizes the Importance of Selective MRNA Translation and Protein Turnover during Arabidopsis Seed Germination. Mol. Cell. Proteomics MCP 2014, 13, 252–268. [Google Scholar] [CrossRef] [Green Version]
- Galland, M.; Rajjou, L. Regulation of MRNA Translation Controls Seed Germination and Is Critical for Seedling Vigor. Front. Plant Sci. 2015, 6, 284. [Google Scholar] [CrossRef] [Green Version]
- Layat, E.; Leymarie, J.; El-Maarouf-Bouteau, H.; Caius, J.; Langlade, N.; Bailly, C. Translatome Profiling in Dormant and Nondormant Sunflower (Helianthus Annuus) Seeds Highlights Post-Transcriptional Regulation of Germination. New Phytol. 2014, 204, 864–872. [Google Scholar] [CrossRef]
- Basbouss-Serhal, I.; Soubigou-Taconnat, L.; Bailly, C.; Leymarie, J. Germination Potential of Dormant and Nondormant Arabidopsis Seeds Is Driven by Distinct Recruitment of Messenger RNAs to Polysomes. Plant Physiol. 2015, 168, 1049–1065. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A Postgermination Developmental Arrest Checkpoint Is Mediated by Abscisic Acid and Requires the ABI5 Transcription Factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [Green Version]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.-P. Della Proteins and Gibberellin-Regulated Seed Germination and Floral Development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arc, E.; Galland, M.; Cueff, G.; Godin, B.; Lounifi, I.; Job, D.; Rajjou, L. Reboot the System Thanks to Protein Post-Translational Modifications and Proteome Diversity: How Quiescent Seeds Restart Their Metabolism to Prepare Seedling Establishment. Proteomics 2011, 11, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Xue, Q.; Mccray, T.; Margavage, K.; Chen, F.; Lee, J.-H.; Nezames, C.D.; Guo, L.; Terzaghi, W.; Wan, J.; et al. The PP6 Phosphatase Regulates ABI5 Phosphorylation and Abscisic Acid Signaling in Arabidopsis. Plant Cell 2013, 25, 517–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.-H. ABI5 Acts Downstream of ABI3 to Execute an ABA-Dependent Growth Arrest during Germination. Plant J. Cell Mol. Biol. 2002, 32, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Cao, D.; Cheng, H.; Wen, Z.; Peng, J. Identification of the Conserved Serine/Threonine Residues Important for Gibberellin-Sensitivity of Arabidopsis RGL2 Protein. Plant J. Cell Mol. Biol. 2005, 44, 88–99. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H.-W. Rice Early Flowering1, a CKI, Phosphorylates DELLA Protein SLR1 to Negatively Regulate Gibberellin Signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Zhang, W.; Wang, X. Post-Translational Control of ABA Signalling: The Roles of Protein Phosphorylation and Ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, Involved in ABA Signaling Are Essential for the Control of Seed Development and Dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 and AtMPK6 Are Involved in Abscisic Acid and Sugar Signaling in Arabidopsis Seed Germination. Plant Mol. Biol. 2009, 70, 725–736. [Google Scholar] [CrossRef]
- Zhao, R.; Sun, H.-L.; Mei, C.; Wang, X.-J.; Yan, L.; Liu, R.; Zhang, X.-F.; Wang, X.-F.; Zhang, D.-P. The Arabidopsis Ca(2+) -Dependent Protein Kinase CPK12 Negatively Regulates Abscisic Acid Signaling in Seed Germination and Post-Germination Growth. New Phytol. 2011, 192, 61–73. [Google Scholar] [CrossRef]
- Zhang, W.; Cochet, F.; Ponnaiah, M.; Lebreton, S.; Matheron, L.; Pionneau, C.; Boudsocq, M.; Resentini, F.; Huguet, S.; Blázquez, M.Á.; et al. The MPK8-TCP14 Pathway Promotes Seed Germination in Arabidopsis. New Phytol. 2019, 100, 677–692. [Google Scholar]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination3 Encodes a Protein Phosphatase 2C (AtPP2CA) That Strongly Regulates Abscisic Acid Signaling during Germination among Arabidopsis Protein Phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C Protein Phosphatases Directly Regulate Abscisic Acid-Activated Protein Kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.; Yang, P.; Sakata, K.; Komatsu, S. Quantitative Proteomics Reveals the Role of Protein Phosphorylation in Rice Embryos during Early Stages of Germination. J. Proteome Res. 2014, 13, 1766–1782. [Google Scholar] [CrossRef]
- Li, M.; Yin, X.; Sakata, K.; Yang, P.; Komatsu, S. Proteomic Analysis of Phosphoproteins in the Rice Nucleus During the Early Stage of Seed Germination. J. Proteome Res. 2015, 14, 2884–2896. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Barrero, J.; Takahashi, F.; Peck, S.; Gubler, F.; Shinozaki, K.; Umezawa, T. Comparative Phosphoproteomic Analysis of Barley Embryos with Different Dormancy during Imbibition. Int. J. Mol. Sci. 2019, 20, 451. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, S.; Barrero, J.M.; Takahashi, F.; Nakagami, H.; Peck, S.C.; Gubler, F.; Shinozaki, K.; Umezawa, T. Comparative Phosphoproteomic Analysis Reveals a Decay of ABA Signaling in Barley Embryos during After-Ripening. Plant Cell Physiol. 2019, 60, 2758–2768. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, B.; Née, G.; Kramer, K.; Finkemeier, I.; Soppe, W.J.J. Sequence Polymorphisms at the REDUCED DORMANCY5 Pseudophosphatase Underlie Natural Variation in Arabidopsis Dormancy. Plant Physiol. 2016, 171, 2659–2670. [Google Scholar] [CrossRef] [Green Version]
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary Seed Dormancy: A Temporally Multilayered Riddle Waiting to Be Unlocked. J. Exp. Bot. 2017, 68, 857–869. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Cadman, C.S.C.; Toorop, P.E.; Lynn, J.R.; Hilhorst, H.W.M. Seed Dormancy Release in Arabidopsis Cvi by Dry After-Ripening, Low Temperature, Nitrate and Light Shows Common Quantitative Patterns of Gene Expression Directed by Environmentally Specific Sensing. Plant J. 2007, 51, 60–78. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. ITAK: A Program for Genome-Wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassel, G.W.; Lan, H.; Glaab, E.; Gibbs, D.J.; Gerjets, T.; Krasnogor, N.; Bonner, A.J.; Holdsworth, M.J.; Provart, N.J. Genome-Wide Network Model Capturing Seed Germination Reveals Coordinated Regulation of Plant Cellular Phase Transitions. Proc. Natl. Acad. Sci. USA 2011, 108, 9709–9714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinforma. Oxf. Engl. 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein–Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Meyer, L.J.; Gao, J.; Xu, D.; Thelen, J.J. Phosphoproteomic Analysis of Seed Maturation in Arabidopsis, Rapeseed, and Soybean. Plant Physiol. 2012, 159, 517–528. [Google Scholar] [CrossRef] [Green Version]
- van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-Analysis of Arabidopsis Thaliana Phospho-Proteomics Data Reveals Compartmentalization of Phosphorylation Motifs. Plant Cell 2014, 26, 2367–2389. [Google Scholar] [CrossRef] [Green Version]
- Heazlewood, J.L.; Durek, P.; Hummel, J.; Selbig, J.; Weckwerth, W.; Walther, D.; Schulze, W.X. PhosPhAt: A Database of Phosphorylation Sites in Arabidopsis Thaliana and a Plant-Specific Phosphorylation Site Predictor. Nucleic Acids Res. 2008, 36, D1015–D1021. [Google Scholar] [CrossRef]
- Yao, Q.; Bollinger, C.; Gao, J.; Xu, D.; Thelen, J.J. P(3)DB: An Integrated Database for Plant Protein Phosphorylation. Front. Plant Sci. 2012, 3, 206. [Google Scholar] [CrossRef] [Green Version]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-Spectrometry-Based Draft of the Arabidopsis Proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; del Carmen Rodríguez-Gacio, M.; Matilla, A.J. Delay of Germination-1 (DOG1): A Key to Understanding Seed Dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-J.; Lee, M.H.; Kim, J.-I.; Kim, S.Y. Arabidopsis Putative MAP Kinase Kinase Kinases Raf10 and Raf11 Are Positive Regulators of Seed Dormancy and ABA Response. Plant Cell Physiol. 2015, 56, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.-Y.; Yu, X.-C.; Wang, X.-J.; Zhao, R.; Li, Y.; Fan, R.-C.; Shang, Y.; Du, S.-Y.; Wang, X.-F.; Wu, F.-Q.; et al. Two Calcium-Dependent Protein Kinases, CPK4 and CPK11, Regulate Abscisic Acid Signal Transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quettier, A.-L.; Bertrand, C.; Habricot, Y.; Miginiac, E.; Agnes, C.; Jeannette, E.; Maldiney, R. The Phs1-3 Mutation in a Putative Dual-Specificity Protein Tyrosine Phosphatase Gene Provokes Hypersensitive Responses to Abscisic Acid in Arabidopsis Thaliana. Plant J. 2006, 47, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.M.; Putti, F.F.; White, P.J.; dos Reis, A.R. Phytic Acid Accumulation in Plants: Biosynthesis Pathway Regulation and Role in Human Diet. Plant Physiol. Biochem. 2021, 164, 132–146. [Google Scholar] [CrossRef]
- Quiroga, I.; Regente, M.; Pagnussat, L.; Maldonado, A.; Jorrín, J.; de la Canal, L. Phosphorylated 11S Globulins in Sunflower Seeds. Seed Sci. Res. 2013, 23, 199–204. [Google Scholar] [CrossRef]
- Wan, L.; Ross, A.R.S.; Yang, J.; Hegedus, D.D.; Kermode, A.R. Phosphorylation of the 12 S Globulin Cruciferin in Wild-Type and Abi1-1 Mutant Arabidopsis Thaliana (Thale Cress) Seeds. Biochem. J. 2007, 404, 247–256. [Google Scholar] [CrossRef] [Green Version]
- López-Pedrouso, M.; Alonso, J.; Zapata, C. Evidence for Phosphorylation of the Major Seed Storage Protein of the Common Bean and Its Phosphorylation-Dependent Degradation during Germination. Plant Mol. Biol. 2014, 84, 415–428. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; Kinoshita, N.; Chua, N.-H. AFP Is a Novel Negative Regulator of ABA Signaling That Promotes ABI5 Protein Degradation. Genes Dev. 2003, 17, 410–418. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhu, Y.; Zhai, H.; Cai, H.; Ji, W.; Luo, X.; Li, J.; Bai, X. AtPP2CG1, a Protein Phosphatase 2C, Positively Regulates Salt Tolerance of Arabidopsis in Abscisic Acid-Dependent Manner. Biochem. Biophys. Res. Commun. 2012, 422, 710–715. [Google Scholar] [CrossRef]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.-J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.-K. Quantitative Phosphoproteomics Identifies SnRK2 Protein Kinase Substrates and Reveals the Effectors of Abscisic Acid Action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanno, T.; Venhuizen, P.; Wen, T.-N.; Lin, W.-D.; Chiou, P.; Kalyna, M.; Matzke, A.J.M.; Matzke, M. PRP4KA, a Putative Spliceosomal Protein Kinase, Is Important for Alternative Splicing and Development in Arabidopsis Thaliana. Genetics 2018, 210, 1267–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawa, D.; Meyer, A.J.; Dekker, H.L.; Abd-El-Haliem, A.M.; Gevaert, K.; Van De Slijke, E.; Maszkowska, J.; Bucholc, M.; Dobrowolska, G.; De Jaeger, G.; et al. SnRK2 Protein Kinases and MRNA Decapping Machinery Control Root Development and Response to Salt. Plant Physiol. 2020, 182, 361–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tognacca, R.S.; Botto, J.F. Post-Transcriptional Regulation of Seed Dormancy and Germination: Current Understanding and Future Directions. Plant Commun. 2021, 2, 100169. [Google Scholar] [CrossRef]
- Bai, B.; van der Horst, S.; Cordewener, J.H.G.; America, T.A.H.P.; Hanson, J.; Bentsink, L. Seed-Stored MRNAs That Are Specifically Associated to Monosomes Are Translationally Regulated during Germination. Plant Physiol. 2020, 182, 378–392. [Google Scholar] [CrossRef] [Green Version]
- Basbouss-Serhal, I.; Pateyron, S.; Cochet, F.; Leymarie, J.; Bailly, C. 5’ to 3’ MRNA Decay Contributes to the Regulation of Arabidopsis Seed Germination by Dormancy. Plant Physiol. 2017, 173, 1709–1723. [Google Scholar] [CrossRef] [Green Version]
- Tognacca, R.S.; Servi, L.; Hernando, C.E.; Saura-Sanchez, M.; Yanovsky, M.J.; Petrillo, E.; Botto, J.F. Alternative Splicing Regulation During Light-Induced Germination of Arabidopsis Thaliana Seeds. Front. Plant Sci. 2019, 10, 1076. [Google Scholar] [CrossRef]
- de la Fuente van Bentem, S.; Anrather, D.; Roitinger, E.; Djamei, A.; Hufnagl, T.; Barta, A.; Csaszar, E.; Dohnal, I.; Lecourieux, D.; Hirt, H. Phosphoproteomics Reveals Extensive In Vivo Phosphorylation of Arabidopsis Proteins Involved in RNA Metabolism. Nucleic Acids Res. 2006, 34, 3267–3278. [Google Scholar] [CrossRef]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y.V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive Transcriptomic and Epigenomic Remodelling Occurs during Arabidopsis Thaliana Germination. Genome Biol. 2017, 18, 172. [Google Scholar] [CrossRef]
- Sajeev, N.; Baral, A.; America, A.H.P.; Willems, L.A.J.; Merret, R.; Bentsink, L. The MRNA-Binding Proteome of a Critical Phase Transition during Arabidopsis Seed Germination. New Phytol. 2022, 233, 251–264. [Google Scholar] [CrossRef]
- Le, H.; Browning, K.S.; Gallie, D.R. The Phosphorylation State of Poly(A)-Binding Protein Specifies Its Binding to Poly(A) RNA and Its Interaction with Eukaryotic Initiation Factor (EIF) 4F, EIFiso4F, and EIF4B. J. Biol. Chem. 2000, 275, 17452–17462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Yan, Z.; Wang, Y.; Yan, X.; Han, Y. Tudor-SN, a Component of Stress Granules, Regulates Growth under Salt Stress by Modulating GA20ox3 MRNA Levels in Arabidopsis. J. Exp. Bot. 2014, 65, 5933–5944. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jia, J.; Gao, Y.; Zhang, B.; Han, Y. The AtTudor2, a Protein with SN-Tudor Domains, Is Involved in Control of Seed Germination in Arabidopsis. Planta 2010, 232, 197–207. [Google Scholar] [CrossRef]
- Su, C.; Gao, X.; Yang, W.; Zhao, Y.; Fu, X.; Cui, X.; Zhang, C.; Xin, L.; Ren, Y.; Li, L.; et al. Phosphorylation of Tudor-SN, a Novel Substrate of JNK, Is Involved in the Efficient Recruitment of Tudor-SN into Stress Granules. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2017, 1864, 562–571. [Google Scholar] [CrossRef]
- Frick, E.M.; Strader, L.C. Kinase MPK17 and the Peroxisome Division Factor PMD1 Influence Salt-Induced Peroxisome Proliferation. Plant Physiol. 2018, 176, 340–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreeramulu, S.; Mostizky, Y.; Sunitha, S.; Shani, E.; Nahum, H.; Salomon, D.; Hayun, L.B.; Gruetter, C.; Rauh, D.; Ori, N.; et al. BSKs Are Partially Redundant Positive Regulators of Brassinosteroid Signaling in Arabidopsis. Plant J. 2013, 74, 905–919. [Google Scholar] [CrossRef]
- Ruan, J.; Chen, H.; Zhu, T.; Yu, Y.; Lei, Y.; Yuan, L.; Liu, J.; Wang, Z.-Y.; Kuang, J.-F.; Lu, W.-J.; et al. Brassinosteroids Repress the Seed Maturation Program during the Seed-to-Seedling Transition. Plant Physiol. 2021, 186, 534–548. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Xu, Z.-S.; He, G.-Y.; Yang, G.-X.; Chen, M.; Li, L.-C.; Ma, Y.-Z. A Mutation in Arabidopsis BSK5 Encoding a Brassinosteroid-Signaling Kinase Protein Affects Responses to Salinity and Abscisic Acid. Biochem. Biophys. Res. Commun. 2012, 426, 522–527. [Google Scholar] [CrossRef]
- Leymarie, J.; Vitkauskaité, G.; Hoang, H.H.; Gendreau, E.; Chazoule, V.; Meimoun, P.; Corbineau, F.; El-Maarouf-Bouteau, H.; Bailly, C. Role of Reactive Oxygen Species in the Regulation of Arabidopsis Seed Dormancy. Plant Cell Physiol. 2012, 53, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Liu, S.; Lin, R. The Role of Light in Regulating Seed Dormancy and Germination. J. Integr. Plant Biol. 2020, 62, 1310–1326. [Google Scholar] [CrossRef]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-Type Zinc Finger Protein in Arabidopsis, Negatively Regulates Light-Dependent Seed Germination Downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.; Park, J.; Kim, K.; Choi, G. The Transcriptional Coregulator LEUNIG_HOMOLOG Inhibits Light-Dependent Seed Germination in Arabidopsis. Plant Cell 2015, 27, 2301–2313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xavier, L.R.; Almeida, F.A.; Pinto, V.B.; Passamani, L.Z.; Santa-Catarina, C.; de Souza Filho, G.A.; Mooney, B.P.; Thelen, J.J.; Silveira, V. Integrative Proteomics and Phosphoproteomics Reveals Phosphorylation Networks Involved in the Maintenance and Expression of Embryogenic Competence in Sugarcane Callus. J. Plant Physiol. 2022, 268, 153587. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Hall, A. A Role for Multiple Circadian Clock Genes in the Response to Signals That Break Seed Dormancy in Arabidopsis. Plant Cell 2009, 21, 1722–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zha, P.; Liu, S.; Li, Y.; Ma, T.; Yang, L.; Jing, Y.; Lin, R. The Evening Complex and the Chromatin-Remodeling Factor PICKLE Coordinately Control Seed Dormancy by Directly Repressing DOG1 in Arabidopsis. Plant Commun. 2020, 1, 100011. [Google Scholar] [CrossRef]
- Boersema, P.J.; Raijmakers, R.; Lemeer, S.; Mohammed, S.; Heck, A.J.R. Multiplex Peptide Stable Isotope Dimethyl Labeling for Quantitative Proteomics. Nat. Protoc. 2009, 4, 484–494. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open Source Software for Rapid Proteomics Tools Development. Bioinforma. Oxf. Engl. 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Langella, O.; Valot, B.; Balliau, T.; Blein-Nicolas, M.; Bonhomme, L.; Zivy, M. X!TandemPipeline: A Tool to Manage Sequence Redundancy for Protein Inference and Phosphosite Identification. J. Proteome Res. 2017, 16, 494–503. [Google Scholar] [CrossRef]
- Valot, B.; Langella, O.; Nano, E.; Zivy, M. MassChroQ: A Versatile Tool for Mass Spectrometry Quantification. Proteomics 2011, 11, 3572–3577. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. Cell Mol. Biol. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baudouin, E.; Puyaubert, J.; Meimoun, P.; Blein-Nicolas, M.; Davanture, M.; Zivy, M.; Bailly, C. Dynamics of Protein Phosphorylation during Arabidopsis Seed Germination. Int. J. Mol. Sci. 2022, 23, 7059. https://doi.org/10.3390/ijms23137059
Baudouin E, Puyaubert J, Meimoun P, Blein-Nicolas M, Davanture M, Zivy M, Bailly C. Dynamics of Protein Phosphorylation during Arabidopsis Seed Germination. International Journal of Molecular Sciences. 2022; 23(13):7059. https://doi.org/10.3390/ijms23137059
Chicago/Turabian StyleBaudouin, Emmanuel, Juliette Puyaubert, Patrice Meimoun, Mélisande Blein-Nicolas, Marlène Davanture, Michel Zivy, and Christophe Bailly. 2022. "Dynamics of Protein Phosphorylation during Arabidopsis Seed Germination" International Journal of Molecular Sciences 23, no. 13: 7059. https://doi.org/10.3390/ijms23137059
APA StyleBaudouin, E., Puyaubert, J., Meimoun, P., Blein-Nicolas, M., Davanture, M., Zivy, M., & Bailly, C. (2022). Dynamics of Protein Phosphorylation during Arabidopsis Seed Germination. International Journal of Molecular Sciences, 23(13), 7059. https://doi.org/10.3390/ijms23137059







