An Eleven-microRNA Signature Related to Tumor-Associated Macrophages Predicts Prognosis of Breast Cancer
Abstract
1. Introduction
2. Results
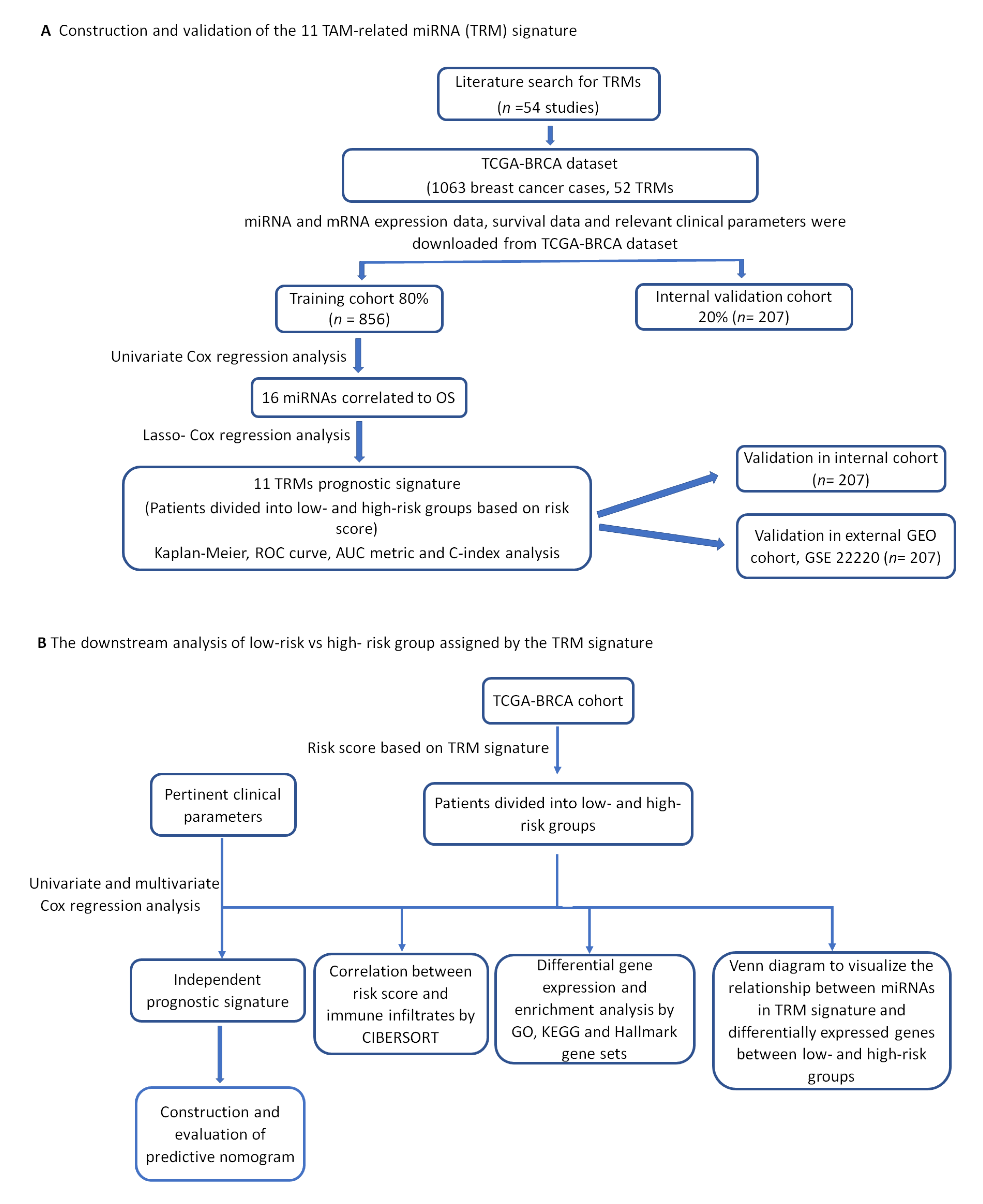
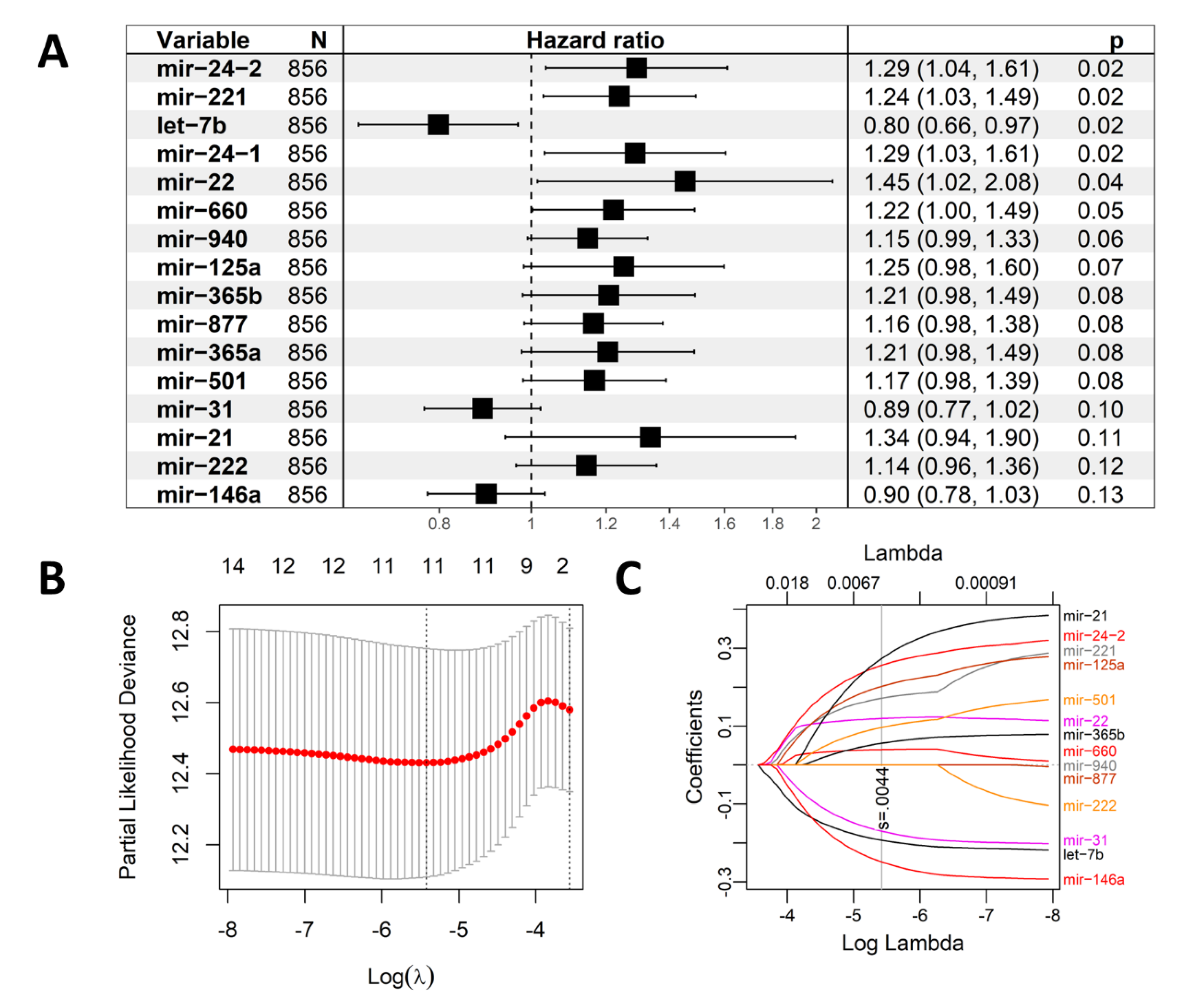
2.1. Construction of the Eleven-TAM-Related miRNA Signature
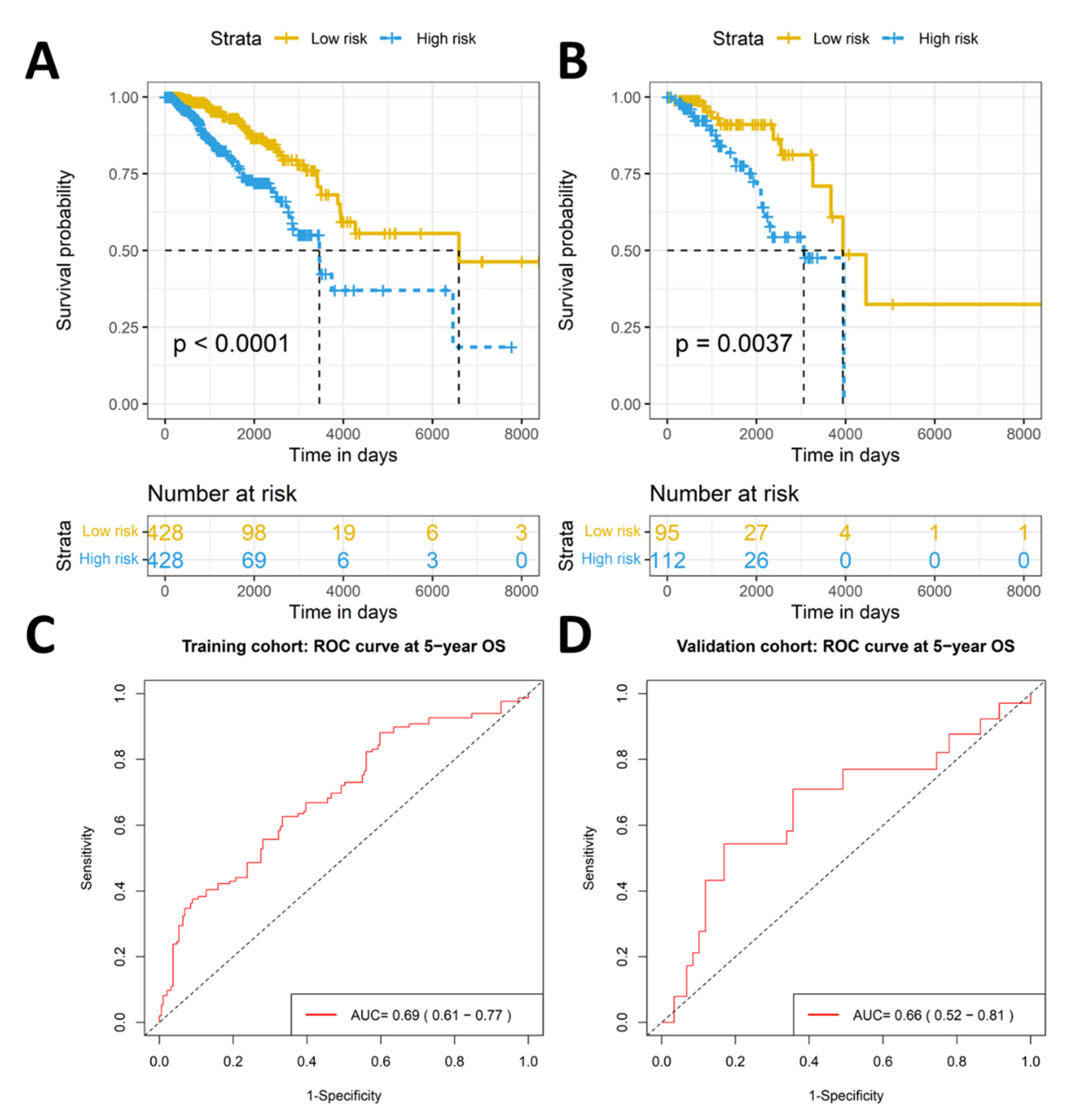
2.2. The Eleven-TAM-Related miRNA Signature Significantly Differentiate the High-Risk Group from the Low-Risk Group
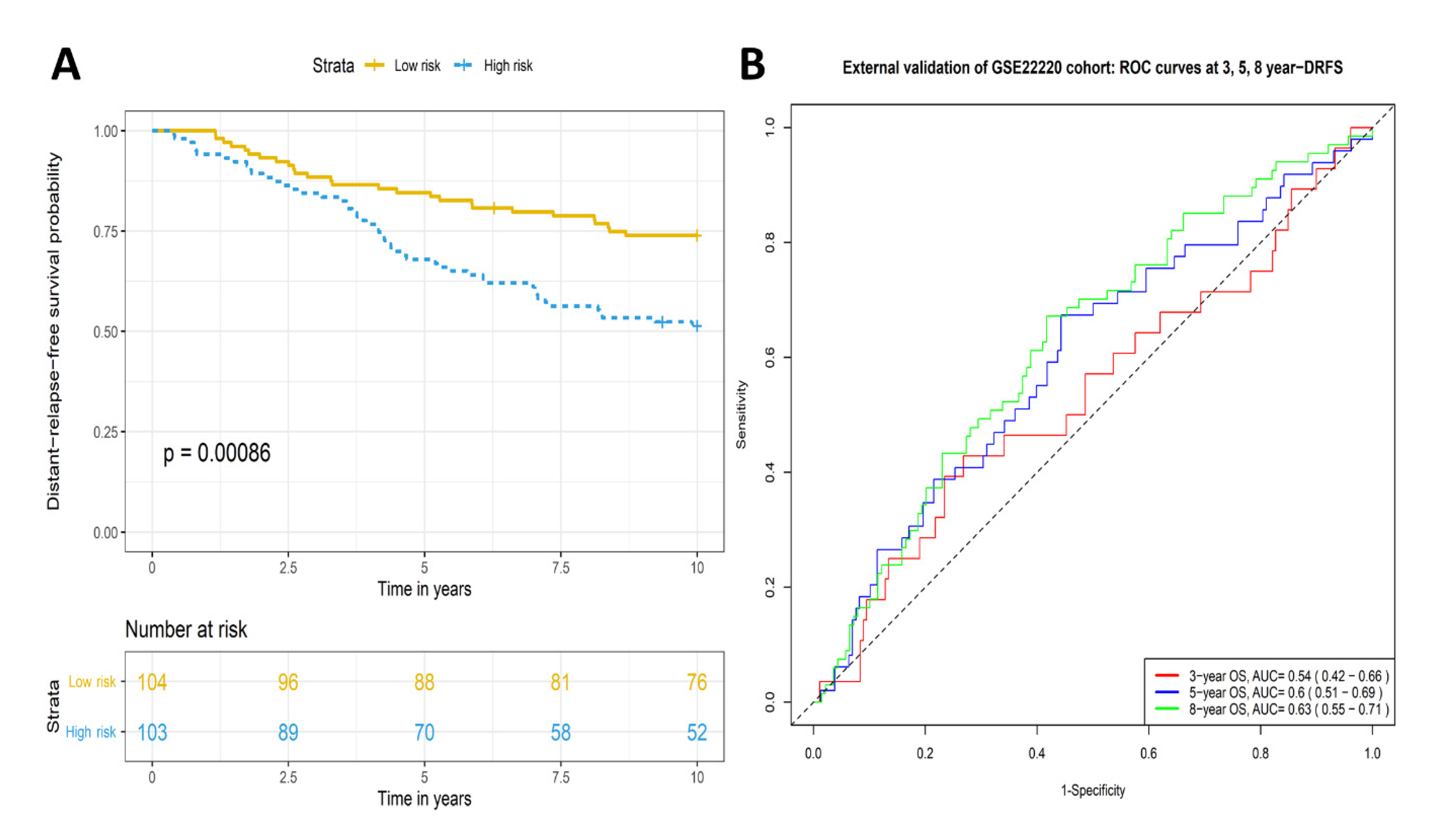
2.3. Validation of Prognostic Significance of the Eleven-TAM-Related miRNA Signature in the GEO Dataset
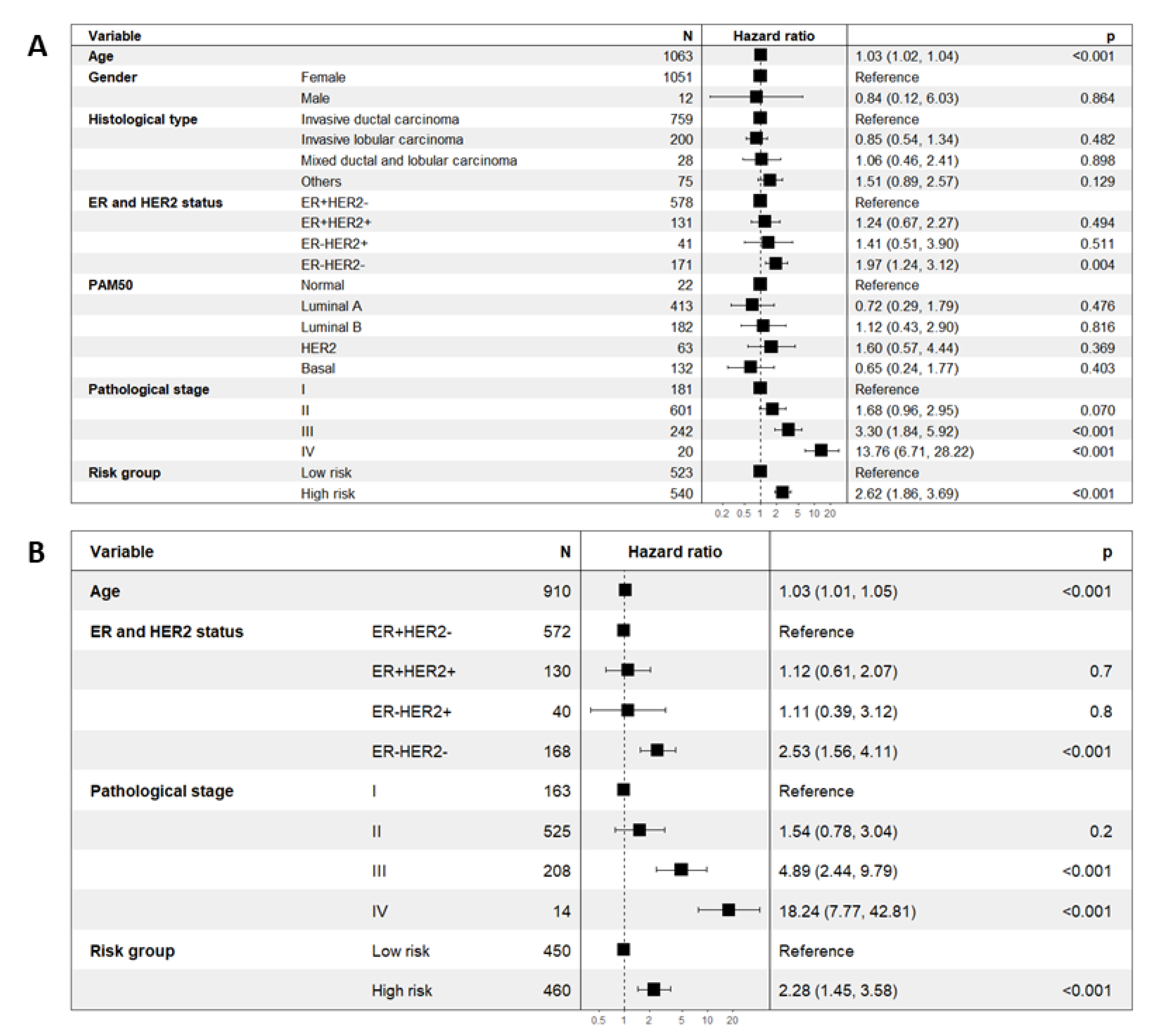
2.4. Prognostic Significance of the Eleven-TAM-Related miRNA Signature Is Independent of Clinicopathological Parameters
2.5. Among the Different Immune Infiltrate Populations, TAMs Had the Highest Correlation with the Risk Score Responsible for Poor Prognosis
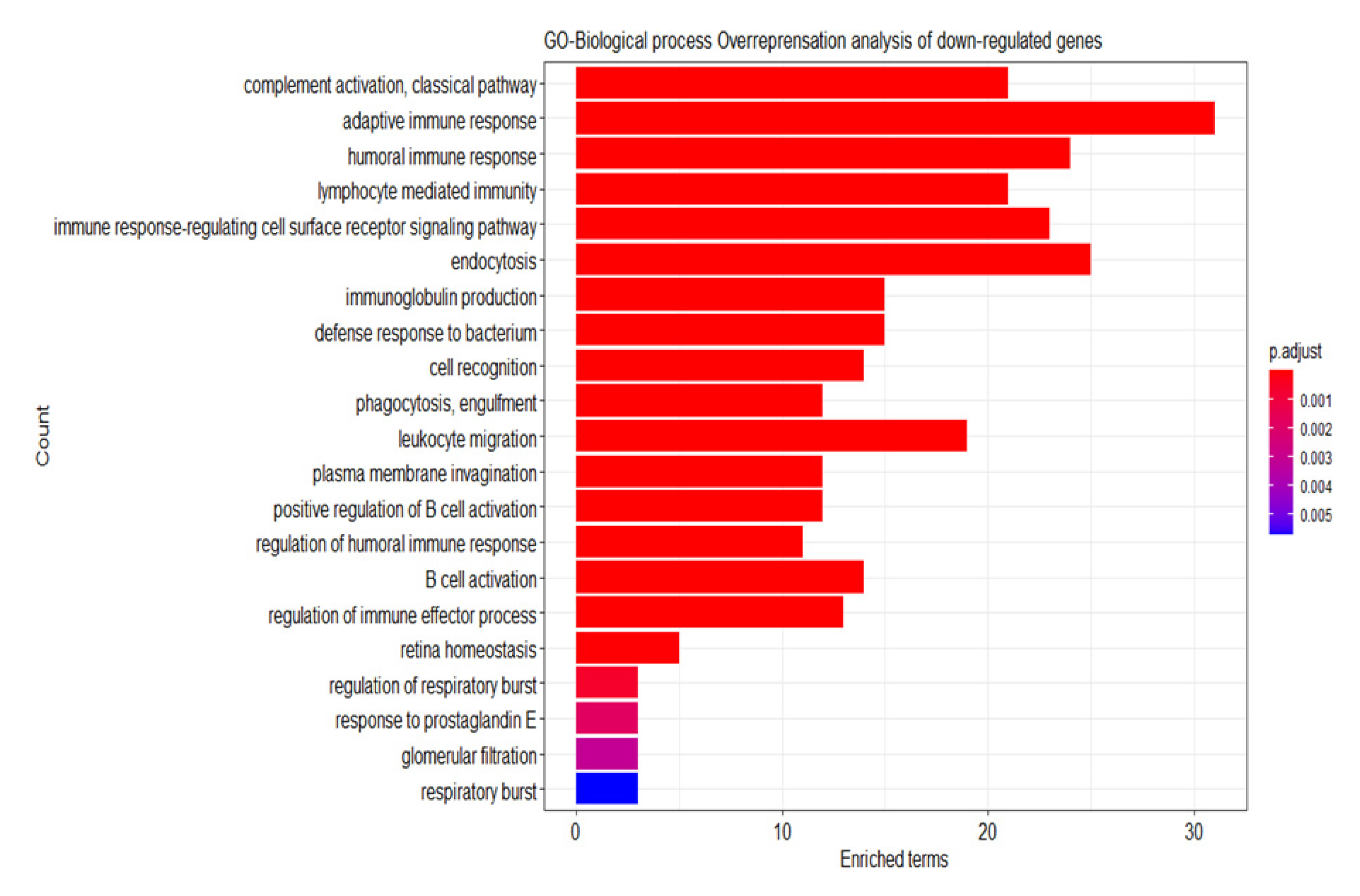
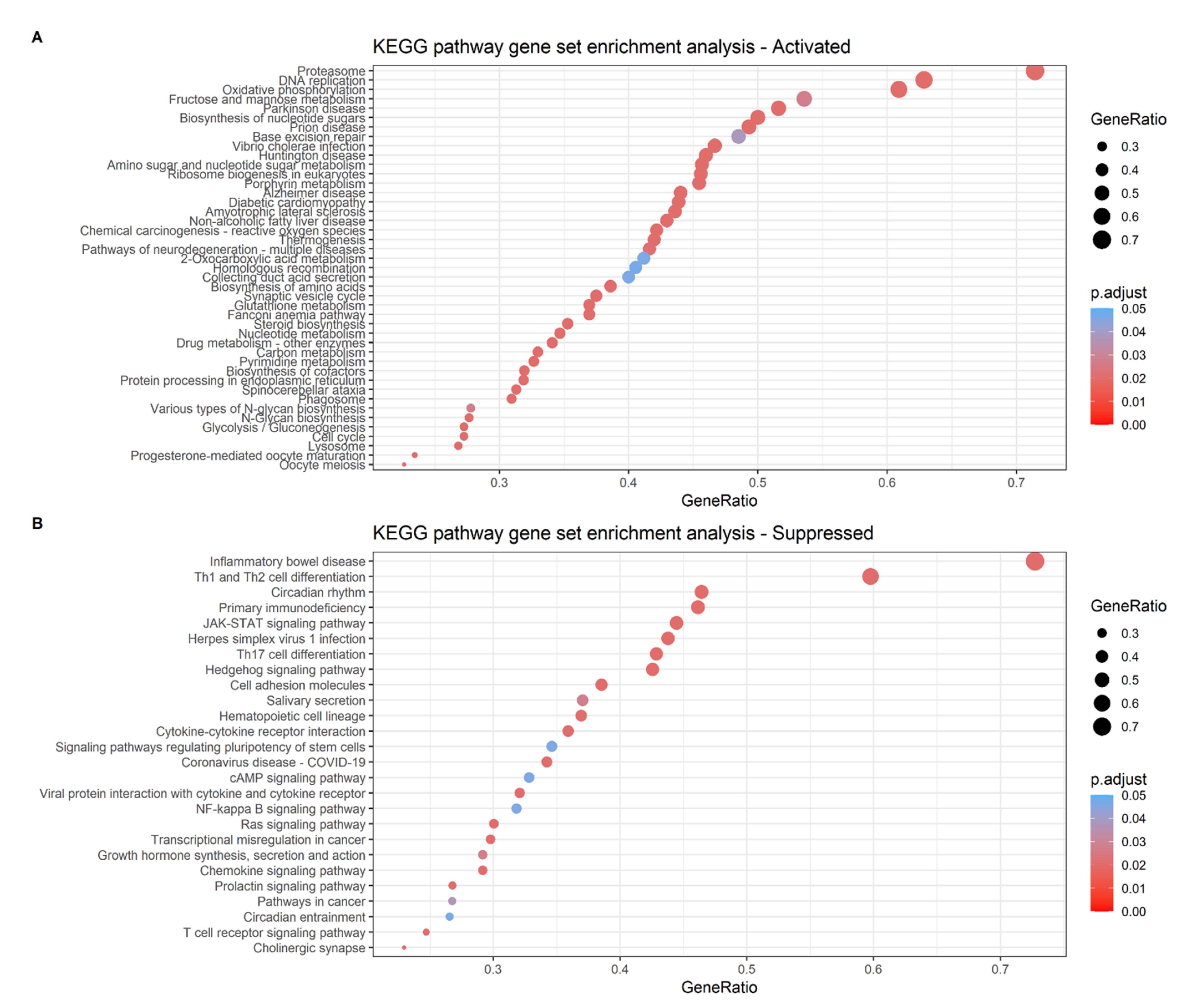
2.6. Analysis of the Differential Gene Expression and Gene Set Enrichment
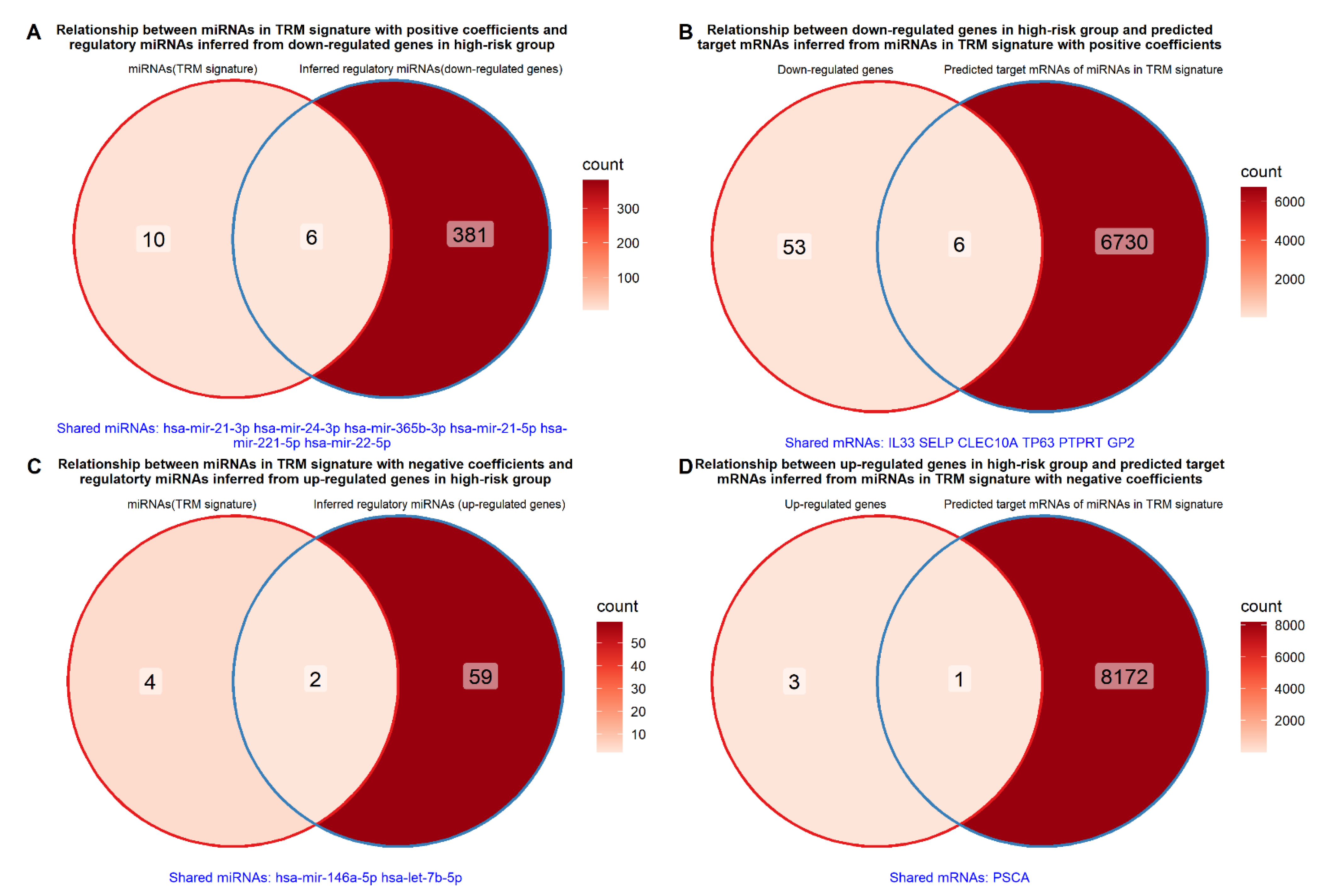
2.7. Relationship between miRNAs in TRM Signature and Differentially Expressed Genes between High- and Low-Risk Groups
3. Discussion
4. Methods
4.1. Literature Search for TAM-Related miRNAs
4.2. Data Mining of miRNAs, mRNAs and Clinicopathological Data
4.3. Prognostic TAM-Related miRNA Signature Construction
4.4. External Validation of the Prognostic Significance of TAM-Related miRNA Signature with the Gene Expression Omnibus (GEO) Dataset
4.5. Independent Prognostic Significance of the Risk Score Based on TAM-Related miRNAs Signature
4.6. Correlation between Prognostic Risk Score and Immune Infiltrate Populations
4.7. Differential Gene Expression and Enrichment Analysis
4.8. Relationship between miRNAs in the TRM Signature and the Differentially Expressed Genes of High- and Low-Risk Groups
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Jayasingam, S.D.; Citartan, M.; Thang, T.H.; Mat Zin, A.A.; Ang, K.C.; Ch’ng, E.S. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: Technicalities and challenges in routine clinical practice. Front. Oncol. 2020, 9, 1512. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, J.-m.; Luo, Y.-p. MicroRNAs in tumor immunity: Functional regulation in tumor-associated macrophages. J. Zhejiang Univ. Sci. B 2020, 21, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Meehan, J.; Gray, M.; Martínez-Pérez, C.; Kay, C.; Pang, L.Y.; Fraser, J.A.; Poole, A.V.; Kunkler, I.H.; Langdon, S.P.; Argyle, D. Precision medicine and the role of biomarkers of radiotherapy response in breast cancer. Front. Oncol. 2020, 10, 628. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Raue, R.; Frank, A.-C.; Syed, S.N.; Brüne, B. Therapeutic targeting of MicroRNAs in the tumor microenvironment. Int. J. Mol. Sci. 2021, 22, 2210. [Google Scholar] [CrossRef]
- Iacona, J.R.; Lutz, C.S. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip. Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef]
- Liu, H.-T.; Wang, Y.-W.; Xing, A.-Y.; Shi, D.-B.; Zhang, H.; Guo, X.-Y.; Xu, J.; Gao, P. Prognostic value of microRNA signature in patients with gastric cancers. Sci. Rep. 2017, 7, 42806. [Google Scholar] [CrossRef]
- Qi, Y.; Lai, Y.-L.; Shen, P.-C.; Chen, F.-H.; Lin, L.-J.; Wu, H.-H.; Peng, P.-H.; Hsu, K.-W.; Cheng, W.-C. Identification and validation of a miRNA-based prognostic signature for cervical cancer through an integrated bioinformatics approach. Sci. Rep. 2020, 10, 22270. [Google Scholar] [CrossRef]
- Tian, B.; Hou, M.; Zhou, K.; Qiu, X.; Du, Y.; Gu, Y.; Yin, X.; Wang, J. A Novel TCGA-Validated, MiRNA-Based Signature for Prediction of Breast Cancer Prognosis and Survival. Front. Cell Dev. Biol. 2021, 9, 717462. [Google Scholar] [CrossRef]
- Wei, J.; Yin, Y.; Deng, Q.; Zhou, J.; Wang, Y.; Yin, G.; Yang, J.; Tang, Y. Integrative analysis of MicroRNA and gene interactions for revealing candidate signatures in prostate cancer. Front. Genet. 2020, 11, 176. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qu, A.; Wu, Q.; Zhang, X.; Wang, L.; Li, C.; Dong, Z.; Du, L.; Wang, C. Prognostic value of a hypoxia-related microRNA signature in patients with colorectal cancer. Aging 2020, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Yu, S.; Yuan, T.; Yao, S.; Zhang, L. Exogenous let-7a-5p induces A549 lung cancer cell death through BCL2L1-mediated PI3Kγ signaling pathway. Front. Oncol. 2019, 9, 808. [Google Scholar] [CrossRef]
- Huang, Z.; Gan, J.; Long, Z.; Guo, G.; Shi, X.; Wang, C.; Zang, Y.; Ding, Z.; Chen, J.; Zhang, J. Targeted delivery of let-7b to reprogramme tumor-associated macrophages and tumor infiltrating dendritic cells for tumor rejection. Biomaterials 2016, 90, 72–84. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, L.; Hu, Y.; Huang, Y.; Zhang, Y.; Zheng, X.; Wang, S.; Wang, Y.; Yu, Y.; Zhang, M. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci. Rep. 2016, 6, 25602. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, D.; Wu, A.; Qiu, X.; Di, W.; Huang, L.; Qiu, L. TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. 2017, 393, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Tong, F.; Mao, X.; Zhang, S.; Xie, H.; Yan, B.; Wang, B.; Sun, J.; Wei, L. HPV + HNSCC-derived exosomal miR-9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett. 2020, 478, 34–44. [Google Scholar] [CrossRef]
- Li, J.; Xue, J.; Ling, M.; Sun, J.; Xiao, T.; Dai, X.; Sun, Q.; Cheng, C.; Xia, H.; Wei, Y. MicroRNA-15b in extracellular vesicles from arsenite-treated macrophages promotes the progression of hepatocellular carcinomas by blocking the LATS1-mediated Hippo pathway. Cancer Lett. 2021, 497, 137–153. [Google Scholar] [CrossRef]
- Li, Z.; Suo, B.; Long, G.; Gao, Y.; Song, J.; Zhang, M.; Feng, B.; Shang, C.; Wang, D. Exosomal miRNA-16-5p derived from M1 macrophages enhances T cell-dependent immune response by regulating PD-L1 in gastric cancer. Front. Cell Dev. Biol. 2020, 8, 1362. [Google Scholar] [CrossRef]
- Peng, Y.; Li, X.; Liu, H.; Deng, X.; She, C.; Liu, C.; Wang, X.; Liu, A. microRNA-18a from M2 Macrophages Inhibits TGFBR3 to Promote Nasopharyngeal Carcinoma Progression and Tumor Growth via TGF-β Signaling Pathway. Nanoscale Res. Lett. 2020, 15, 196. [Google Scholar] [CrossRef]
- Teng, Y.; Mu, J.; Hu, X.; Samykutty, A.; Zhuang, X.; Deng, Z.; Zhang, L.; Cao, P.; Yan, J.; Miller, D. Grapefruit-derived nanovectors deliver miR-18a for treatment of liver metastasis of colon cancer by induction of M1 macrophages. Oncotarget 2016, 7, 25683. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.; Li, N.; Gomez-Cabrero, A.; Reisfeld, R.; Xiang, R. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Yin, H.B.; Li, X.Y.; Zhu, G.M.; He, W.Y.; Gou, X. Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int. J. Oncol. 2020, 56, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Qin, J.F.; Han, X.; Jin, F.J.; Zhang, J.H.; Lan, L.; Wang, Y. miR-21a negatively modulates tumor suppressor genes PTEN and miR-200c and further promotes the transformation of M2 macrophages. Immunol. Cell Biol. 2018, 96, 68–80. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, J.; Chen, Z.; Wang, H.; Xue, H.; Yang, C.; Guo, Q.; Qi, Y.; Guo, X.; Qian, M. Transfer of microRNA via macrophage-derived extracellular vesicles promotes proneural-to-mesenchymal transition in glioma stem cells. Cancer Immunol. Res. 2020, 8, 966–981. [Google Scholar] [CrossRef]
- Ma, S.; Liu, M.; Xu, Z.; Li, Y.; Guo, H.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget 2016, 7, 13502. [Google Scholar] [CrossRef]
- Mi, X.; Xu, R.; Hong, S.; Xu, T.; Zhang, W.; Liu, M. M2 macrophage-derived exosomal lncRNA AFAP1-AS1 and microRNA-26a affect cell migration and metastasis in esophageal cancer. Mol. Ther.-Nucleic Acids 2020, 22, 779–790. [Google Scholar] [CrossRef]
- Cai, J.; Qiao, B.; Gao, N.; Lin, N.; He, W. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am. J. Physiol.—Cell Physiol. 2019, 316, C731–C740. [Google Scholar] [CrossRef]
- Lu, L.; Ling, W.; Ruan, Z. Tumor associated macrophages-derived extracellular vesicles containing microRNA-29a-3p explains the deterioration of ovarian cancer. Mol. Ther. Nucleic Acids 2021, 25, 468–482. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, Z.; Chen, M.; Jing, Y.; Shu, W.; Xie, Z.; Li, Z.; Xu, J.; He, F.; Jiao, P. Macrophage-Derived Exosomal miR-31-5p Promotes Oral Squamous Cell Carcinoma Tumourigenesis Through the Large Tumor Suppressor 2-Mediated Hippo Signalling Pathway. J. Biomed. Nanotechnol. 2021, 17, 822–837. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.; Möller, C.; Jirström, K.; Lee, A.; Busch, S.; Lamb, R.; Landberg, G. Downregulation of miR-92a is associated with aggressive breast cancer features and increased tumour macrophage infiltration. PLoS ONE 2012, 7, e36051. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Ouyang, X.; Sun, Y.; Xiao, Y.; You, B.; Gao, Y.; Yeh, S.; Li, Y.; Chang, C. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020, 27, 3258–3272. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Peng, R.; Fang, F.; Mao, L.; Chen, Z.; Yang, S.; Dai, C.; Wu, H.; Wang, C.; Feng, N. Tumor-associated macrophages promote prostate cancer progression via exosome-mediated miR-95 transfer. J. Cell. Physiol. 2020, 235, 9729–9742. [Google Scholar] [CrossRef]
- Yin, Z.; Zhou, Y.; Ma, T.; Chen, S.; Shi, N.; Zou, Y.; Hou, B.; Zhang, C. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J. Cell. Mol. Med. 2020, 24, 5028–5038. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Xiao, S.; Li, Y.; Chen, Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J. Cell. Biochem. 2019, 120, 3046–3055. [Google Scholar] [CrossRef]
- Lin, L.; Lin, H.; Wang, L.; Wang, B.; Hao, X.; Shi, Y. miR-130a regulates macrophage polarization and is associated with non-small cell lung cancer. Oncol. Rep. 2015, 34, 3088–3096. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, W.; Yue, P.; Li, X. M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J. Exp. Clin. Cancer Res. 2020, 39, 134. [Google Scholar] [CrossRef]
- Zhang, J.; Shan, W.-f.; Jin, T.-t.; Wu, G.-q.; Xiong, X.-x.; Jin, H.-y.; Zhu, S.-m. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J. Transl. Med. 2014, 12, 279. [Google Scholar] [CrossRef]
- Xu, S.; Wei, J.; Wang, F.; Kong, L.-Y.; Ling, X.-Y.; Nduom, E.; Gabrusiewicz, K.; Doucette, T.; Yang, Y.; Yaghi, N.K. Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy against murine glioblastoma. JNCI J. Natl. Cancer Inst. 2014, 106, dju162. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Shi, B.; Ma, S.; Xu, Z.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. Functions of miR-146a and miR-222 in tumor-associated macrophages in breast cancer. Sci. Rep. 2015, 5, 18648. [Google Scholar] [CrossRef]
- Zhou, Y.-x.; Zhao, W.; Mao, L.-w.; Wang, Y.-l.; Xia, L.-q.; Cao, M.; Shen, J.; Chen, J. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int. J. Biochem. Cell Biol. 2018, 104, 25–33. [Google Scholar] [CrossRef]
- Yin, C.; Han, Q.; Xu, D.; Zheng, B.; Zhao, X.; Zhang, J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology 2019, 8, e1601479. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, X.; Ying, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wu, Y.; Wang, X. Suppression of endothelial cell migration by tumor associated macrophage-derived exosomes is reversed by epithelial ovarian cancer exosomal lncRNA. Cancer Cell Int. 2017, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Tong, L.; LiuAnwei Liu, F.; Liu, A.; Zeng, S.; Xiong, Q.; Yang, Z.; He, X.; Sun, Y.; Xu, C. Tumor-infiltrating M2 macrophages driven by specific genomic alterations are associated with prognosis in bladder cancer. Oncol. Rep. 2019, 42, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, L.-J.; Qin, J.-J.; Zhang, L.; Zhuang, G.-H. MicroRNA-155 inversely correlates with esophageal cancer progression through regulating tumor-associated macrophage FGF2 expression. Biochem. Biophys. Res. Commun. 2018, 503, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Z.; Ni, Y.; Bian, C.; Huang, J.; Chen, L.; Xie, X.; Wang, J. Tumor-associated macrophages secret exosomal miR-155 and miR-196a-5p to promote metastasis of non-small-cell lung cancer. Transl. Lung Cancer Res. 2021, 10, 1338. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef]
- Li, X.; Tang, M. Exosomes released from M2 macrophages transfer miR-221-3p contributed to EOC progression through targeting CDKN1B. Cancer Med. 2020, 9, 5976–5988. [Google Scholar] [CrossRef]
- Liu, W.; Long, Q.; Zhang, W.; Zeng, D.; Hu, B.; Liu, S.; Chen, L. miRNA-221-3p derived from M2-polarized tumor-associated macrophage exosomes aggravates the growth and metastasis of osteosarcoma through SOCS3/JAK2/STAT3 axis. Aging (Albany NY) 2021, 13, 19760. [Google Scholar] [CrossRef]
- Chen, W.-X.; Wang, D.-D.; Zhu, B.; Zhu, Y.-Z.; Zheng, L.; Feng, Z.-Q.; Qin, X.-H. Exosomal miR-222 from adriamycin-resistant MCF-7 breast cancer cells promote macrophages M2 polarization via PTEN/Akt to induce tumor progression. Aging 2021, 13, 10415. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 81. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.-D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ma, J.; Cheng, Y.; Zheng, P. Exosomal transfer of macrophage-derived miR-223 confers doxorubicin resistance in gastric Cancer. OncoTargets Ther. 2020, 13, 12169. [Google Scholar] [CrossRef]
- Bai, Z.-z.; Li, H.-y.; Li, C.-h.; Sheng, C.-l.; Zhao, X.-n. M1 Macrophage-Derived Exosomal MicroRNA-326 Suppresses Hepatocellular Carcinoma Cell Progression Via Mediating NF-κB Signaling Pathway. Nanoscale Res. Lett. 2020, 15, 221. [Google Scholar] [CrossRef]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; David, G.B.; Shlomi, T.; Gil, Z. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef]
- Li, X.; Xu, H.; Yi, J.; Dong, C.; Zhang, H.; Wang, Z.; Miao, L.; Zhou, W. miR-365 secreted from M2 Macrophage-derived extracellular vesicles promotes pancreatic ductal adenocarcinoma progression through the BTG2/FAK/AKT axis. J. Cell. Mol. Med. 2021, 25, 4671–4683. [Google Scholar] [CrossRef]
- Yang, X.; Cai, S.; Shu, Y.; Deng, X.; Zhang, Y.; He, N.; Wan, L.; Chen, X.; Qu, Y.; Yu, S. Exosomal miR-487a derived from m2 macrophage promotes the progression of gastric cancer. Cell Cycle 2021, 20, 434–444. [Google Scholar] [CrossRef]
- Yin, Z.; Ma, T.; Huang, B.; Lin, L.; Zhou, Y.; Yan, J.; Zou, Y.; Chen, S. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 1–20. [Google Scholar] [CrossRef]
- Lei, J.; Chen, P.; Zhang, F.; Zhang, N.; Zhu, J.; Wang, X.; Jiang, T. M2 macrophages-derived exosomal microRNA-501-3p promotes the progression of lung cancer via targeting WD repeat domain 82. Cancer Cell Int. 2021, 21, 91. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Fan, P.; Zhang, C.; Xie, J.; Gu, X.; Lei, S.; Chen, Z.; Huang, Z. Exosomal microRNA-503-3p derived from macrophages represses glycolysis and promotes mitochondrial oxidative phosphorylation in breast cancer cells by elevating DACT2. Cell Death Discov. 2021, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, X.; Wu, X.; Zhang, T.; Zhu, Q.; Wang, X.; Wang, H.; Wang, K.; Lin, Y.; Wang, X. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol. Res. 2018, 6, 1578–1592. [Google Scholar] [CrossRef]
- Zhong, Y.; Yi, C. MicroRNA-720 suppresses M2 macrophage polarization by targeting GATA3. Biosci. Rep. 2016, 36, e00363. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Ye, X.; Zhou, T.; Gan, D.; Qian, H.; Fang, W.; Yao, M.; Zhang, D.; Shi, H.; Chen, T. PGRN−/− TAMs-derived exosomes inhibit breast cancer cell invasion and migration and its mechanism exploration. Life Sci. 2021, 264, 118687. [Google Scholar] [CrossRef]
- An, M.; Zang, X.; Wang, J.; Kang, J.; Tan, X.; Fu, B. Comprehensive analysis of differentially expressed long noncoding RNAs, miRNAs and mRNAs in breast cancer brain metastasis. Epigenomics 2021, 13, 14. [Google Scholar] [CrossRef]
- Liang, Y.-K.; Lin, H.-Y.; Dou, X.-W.; Chen, M.; Wei, X.-L.; Zhang, Y.-Q.; Wu, Y.; Chen, C.-F.; Bai, J.-W.; Xiao, Y.-S. MiR-221/222 promote epithelial-mesenchymal transition by targeting Notch3 in breast cancer cell lines. NPJ Breast Cancer 2018, 4, 20. [Google Scholar] [CrossRef]
- Lv, C.; Li, F.; Li, X.; Tian, Y.; Zhang, Y.; Sheng, X.; Song, Y.; Meng, Q.; Yuan, S.; Luan, L. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 2017, 8, 1036. [Google Scholar] [CrossRef]
- Shen, Y.; Ye, Y.; Ruan, L.; Bao, L.; Wu, M.; Zhou, Y. Inhibition of miR-660-5p expression suppresses tumor development and metastasis in human breast cancer. Genet. Mol. Res. 2017, 16, gmr16019479. [Google Scholar] [CrossRef]
- Yan, L.; Yu, M.C.; Gao, G.L.; Liang, H.W.; Zhou, X.Y.; Zhu, Z.T.; Zhang, C.Y.; Wang, Y.B.; Chen, X. MiR-125a-5p functions as a tumour suppressor in breast cancer by downregulating BAP1. J. Cell. Biochem. 2018, 119, 8773–8783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Wang, D.; Wei, X. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol. Res. 2017, 50, 27. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Sun, H.-F.; Wang, W.-J.; Li, Q.; Ding, J.; Di, W. miRNA-365b promotes hepatocellular carcinoma cell migration and invasion by downregulating SGTB. Future Oncol. 2019, 15, 2019–2028. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Chen, Y.T.; Chen, C.F.; Tang, Y.S.; Huang, T.T.; Lin, Y.C.; Yeh, T.S.; Huang, K.H.; Lee, H.C.; Hsu, M.T. MicroRNA-23a/27a/24-2 cluster promotes gastric cancer cell proliferation synergistically. Oncol. Lett. 2018, 16, 2319–2325. [Google Scholar] [CrossRef] [PubMed]
- Maghsudlu, M.; Yazd, E.F.; Amiriani, T. Increased expression of MiR-27a and MiR-24-2 in esophageal squamous cell carcinoma. J. Gastrointest. Cancer 2020, 51, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, B.; Hendrayani, S.-F.; Silva, G.; Aboussekhra, A. Let-7b inhibits cancer-promoting effects of breast cancer-associated fibroblasts through IL-8 repression. Oncotarget 2018, 9, 17825. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.-J.; Han, Z.-Q.; Zhang, H.-B.; Wang, Z.-A. Let-7b attenuates cisplatin resistance and tumor growth in gastric cancer by targeting AURKB. Cancer Gene Ther. 2018, 25, 300–308. [Google Scholar] [CrossRef]
- Kuang, Y.; Xu, H.; Lu, F.; Meng, J.; Yi, Y.; Yang, H.; Hou, H.; Wei, H.; Su, S. Inhibition of microRNA let-7b expression by KDM2B promotes cancer progression by targeting EZH2 in ovarian cancer. Cancer Sci. 2021, 112, 231. [Google Scholar] [CrossRef]
- Yu, T.; Ma, P.; Wu, D.; Shu, Y.; Gao, W. Functions and mechanisms of microRNA-31 in human cancers. Biomed. Pharmacother. 2018, 108, 1162–1169. [Google Scholar] [CrossRef]
- Ge, F.; Wang, C.; Wang, W.; Liu, W.; Wu, B. MicroRNA-31 inhibits tumor invasion and metastasis by targeting RhoA in human gastric cancer. Oncol. Rep. 2017, 38, 1133–1139. [Google Scholar] [CrossRef]
- Li, X.; Truong, B.; Xu, T.; Liu, L.; Li, J.; Le, T.D. Uncovering the roles of microRNAs/lncRNAs in characterising breast cancer subtypes and prognosis. BMC Bioinform. 2021, 22, 300. [Google Scholar] [CrossRef] [PubMed]
- Blanche, P.; Kattan, M.W.; Gerds, T.A. The c-index is not proper for the evaluation of-year predicted risks. Biostatistics 2019, 20, 347–357. [Google Scholar] [CrossRef]
- Fan, Y.; He, S. The Characteristics of Tumor Microenvironment in Triple Negative Breast Cancer. Cancer Manag. Res. 2022, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Shimoda, M.; Sota, Y.; Miyake, T.; Tanei, T.; Kagara, N.; Naoi, Y.; Kim, S.J.; Noguchi, S.; Shimazu, K. Enhanced humoral immunity in breast cancer patients with high serum concentration of anti-HER2 autoantibody. Cancer Med. 2021, 10, 1418–1430. [Google Scholar] [CrossRef] [PubMed]
- Pandya, P.H.; Murray, M.E.; Pollok, K.E.; Renbarger, J.L. The immune system in cancer pathogenesis: Potential therapeutic approaches. J. Immunol. Res. 2016, 2016, 4273943. [Google Scholar] [CrossRef] [PubMed]
- Zagirova, D.; Autenried, R.; Nelson, M.E.; Rezvani, K. Proteasome Complexes and Their Heterogeneity in Colorectal, Breast and Pancreatic Cancers. J. Cancer 2021, 12, 2472. [Google Scholar] [CrossRef] [PubMed]
- Zacksenhaus, E.; Shrestha, M.; Liu, J.C.; Vorobieva, I.; Chung, P.E.; Ju, Y.; Nir, U.; Jiang, Z. Mitochondrial OXPHOS induced by RB1 deficiency in breast cancer: Implications for anabolic metabolism, stemness, and metastasis. Trends Cancer 2017, 3, 768–779. [Google Scholar] [CrossRef]
- Raninga, P.V.; Lee, A.; Sinha, D.; Dong, L.-f.; Datta, K.K.; Lu, X.; Kalita-de Croft, P.; Dutt, M.; Hill, M.; Pouliot, N. Marizomib suppresses triple-negative breast cancer via proteasome and oxidative phosphorylation inhibition. Theranostics 2020, 10, 5259. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, J.; Ge, S.; Chen, C.; Li, S.; Wu, X.; Feng, X.; Wang, Y.; Cai, D. Saikosaponin A inhibits breast cancer by regulating Th1/Th2 balance. Front. Pharmacol. 2019, 10, 624. [Google Scholar] [CrossRef]
- Eftekhari, R.; Esmaeili, R.; Mirzaei, R.; Bidad, K.; de Lima, S.; Ajami, M.; Shirzad, H.; Hadjati, J.; Majidzadeh-A., K. Study of the tumor microenvironment during breast cancer progression. Cancer Cell Int. 2017, 17, 123. [Google Scholar] [CrossRef][Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, F.; Xie, P.; Yuan, L.; Zhou, M. PTPRT Could Be a Treatment Predictive and Prognostic Biomarker for Breast Cancer. BioMed Res. Int. 2021, 2021, 3301402. [Google Scholar] [CrossRef] [PubMed]
- Kurze, A.K.; Buhs, S.; Eggert, D.; Oliveira-Ferrer, L.; Müller, V.; Niendorf, A.; Wagener, C.; Nollau, P. Immature O-glycans recognized by the macrophage glycoreceptor CLEC10A (MGL) are induced by 4-hydroxy-tamoxifen, oxidative stress and DNA-damage in breast cancer cells. Cell Commun. Signal. 2019, 17, 1–18. [Google Scholar] [CrossRef]
- Neophytou, C.; Boutsikos, P.; Papageorgis, P. Molecular mechanisms and emerging therapeutic targets of triple-negative breast cancer metastasis. Front. Oncol. 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Link, T.; Kuithan, F.; Ehninger, A.; Kuhlmann, J.D.; Kramer, M.; Werner, A.; Gatzweiler, A.; Richter, B.; Ehninger, G.; Baretton, G. Exploratory investigation of PSCA-protein expression in primary breast cancer patients reveals a link to HER2/neu overexpression. Oncotarget 2017, 8, 54592. [Google Scholar] [CrossRef]
- Vietri, M.T.; D’elia, G.; Benincasa, G.; Ferraro, G.; Caliendo, G.; Nicoletti, G.F.; Napoli, C. DNA methylation and breast cancer: A way forward. Int. J. Oncol. 2021, 59, 98. [Google Scholar] [CrossRef]
- Li, Z.; Wang, D.; Wang, W.; Chen, X.; Tang, A.; Hou, P.; Li, M.; Zheng, J.; Bai, J. Macrophages-stimulated PRMT1-mediated EZH2 methylation promotes breast cancer metastasis. Biochem. Biophys. Res. Commun. 2020, 533, 679–684. [Google Scholar] [CrossRef]
- Lai, J.; Wang, H.; Pan, Z.; Su, F. A novel six-microRNA-based model to improve prognosis prediction of breast cancer. Aging (Albany NY) 2019, 11, 649. [Google Scholar] [CrossRef]
- Tang, J.; Ma, W.; Zeng, Q.; Tan, J.; Cao, K.; Luo, L. Identification of miRNA-based signature as a novel potential prognostic biomarker in patients with breast cancer. Dis. Markers 2019, 2019, 3815952. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Lu, L.; Wu, Y.; Feng, M.; Xue, X.; Fan, Y. A novel seven-miRNA prognostic model to predict overall survival in head and neck squamous cell carcinoma patients. Mol. Med. Rep. 2019, 20, 4340–4348. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.; Robertson, G.; Brooks, D.; Mungall, A.J.; Birol, I.; Coope, R.; Ma, Y.; Jones, S.; Marra, M.A. Large-scale profiling of microRNAs for the cancer genome atlas. Nucleic Acids Res. 2016, 44, e3. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]



| No | miRNA | Cancer Type | Function | Ref | Precursor miRNAs |
|---|---|---|---|---|---|
| 1 | let-7a | Lung | transferred from TAMs to lung cancer to inhibit cell proliferation, migration, and invasion | [13] | let-7a-1 let-7a-2 let-7a-3 |
| 2 | let-7b | Breast Prostate | repolarizes M2 TAMs to M1 in tumor cells modulates macrophage polarization to promote angiogenesis and mobility | [14] [15] | |
| 3 | miR-7 | Ovarian | released by TAMs to inhibit cell metastasis | [16] | mir-7-1 mir-7-2 mir-7-3 |
| 4 | miR-9 | HNSCC | induces M1 TAM polarization and increases tumor radiosensitivity | [17] | mir-9-1 mir-9-2 mir-9-3 |
| 5 | miR-15b | HCC | derived from M2 TAMs to promote cancer progression | [18] | |
| 6 | miR-16 | Gastric | transferred from M1 TAMs to cancer cells to inhibit tumor formation | [19] | mir-16-1 mir-16-2 |
| 7 | miR-18a | Nasopharynx Liver | derived from M2 TAMs to promote cancer progression and tumor growth induces M1 TAMs to inhibit tumor metastasis | [20] [21] | |
| 8 | miR-19a | Breast Gastric | downregulates M2 TAMs to inhibit cancer progression and metastasis derived from M2 TAMs to reduce chemosensitivity and tumor cell apoptosis | [22] [23] | |
| 9 | miR-21 | Bladder Breast | promotes cancer progression by polarizing TAMs to M2 phenotype promotes M2 TAM transformation to induce metastasis | [24] [25] | |
| 10 | miR-22 | Glioma | derived from TAMs to promote mesenchymal phenotype and induce radiotherapy resistance | [26] | |
| 11 | miR-23a | Breast | regulates TAM polarization | [27] | |
| 12 | miR-24-2 | Breast | regulates M1 and M2 TAM polarization | [27] | mir-24-1 mir-24-2 |
| 13 | miR-26a | Esophageal | M2 TAMs downregulate miR-26a to promote invasion and metastasis of cancer | [28] | mir-26a-1 mir-26a-2 |
| 14 | miR-27a | Glioma | derived from TAMs to promote mesenchymal phenotype and induce radiotherapy resistance | [26] | |
| 15 | miR-29a | OSCC Ovarian | promotes M2 TAMs polarization to enhance proliferation and invasion of cancer cells derived by TAM to facilitate cancer cell proliferation and immune escape | [29] [30] | |
| 16 | miR-31 | OSCC | derived by M2 TAMs to facilitate cancer progression | [31] | |
| 17 | miR-92a | Breast Liver | suppresses the infiltration of TAMs in tumor cells derived from TAMs to increase liver cancer cells invasion | [32] [33] | mir-92a-1 mir-92a-2 |
| 18 | miR-95 | Prostate | derived by M2 TAMs to promote cancer progression | [34] | |
| 19 | miR-122 | Pancreatic | M2 TAMs increases miR-122-5p expression to inhibit PC progression | [35] | |
| 20 | miR-125a | HCC | inhibits TAMs mediated in cancer stem cells | [36] | |
| 21 | miR-125b | HCC | inhibits TAMs mediated in cancer stem cells | [36] | mir-125b-1 mir-125b-2 |
| 22 | miR-130a | Lung | suppresses the polarization of M2 TAMs and enhances M1 polarization | [37] | |
| 23 | miR-130b | Gastric | transferred from M2 TAM to promote survival, migration, invasion, and angiogenesis | [38] | |
| 24 | miR-142 | HCC Glioblastoma | transferred from TAM to cancer cells to inhibit proliferation, tumor growth and invasion inhibits glioma growth and induces apoptosis in M2 TAMs | [39] [40] | |
| 25 | miR-146a | Breast Endometrial HCC | promotes M2 TAM expression inhibits M2 TAM polarization promotes M2 polarization | [41] [42] [43] | |
| 26 | miR-146b | Ovarian Bladder | inhibits the migration of endothelial cells promotes M2 TAM infiltration | [44] [45] | |
| 27 | miR-155 | Esophageal Lung Colon | derived from TAMs to suppress cancer proliferation, migration, invasion and vasculature formation secreted by M2 TAMs to promote metastasis derived from M2 TAMs to promote cell migration and invasion | [46] [47] [48] | |
| 28 | miR-221 | Ovarian Osteosarcoma Glioma | released from M2 TAMs to promote cancer cell proliferation and progression derived from M2 TAMs to aggravate cancer growth and metastasis derived from TAMs to promote mesenchymal phenotype and induce radiotherapy resistance | [49] [50] [26] | |
| 29 | miR-222 | Breast Ovarian | delivered to TAMs to induce M2 polarization regulates polarization of M2 TAMs | [51] [52] | |
| 30 | miR-223 | Ovarian Breast Gastric | derived from TAM to enhance tumor malignancy and chemoresistance released by M2 TAMs to promote cancer cell invasion derived by M2 TAMs to promote drug resistance | [53] [54] [55] | |
| 31 | miR-326 | HCC | derived by M1 TAMs to inhibit cancer cell proliferation, colony formation, migration and invasion | [56] | |
| 32 | miR-365 | Pancreatic | secreted by M2 TAMs to induce drug resistance and promote cancer progression | [57] [58] | mir-365a mir-365b |
| 33 | miR-487a | Gastric | derived from M2 TAMs to promote cancer proliferation and tumorigenesis | [59] | |
| 34 | miR-501 | Pancreatic Lung | derived by M2 TAMs to inhibit tumor suppressor TGFBR3 gene and facilitate cancer development derived by M2 TAMs to promote cancer progression | [60] [61] | |
| 35 | miR-503 | Breast | derived from TAMs to suppress cancer progression | [62] | |
| 36 | miR-660 | Ovarian | upregulated in TAMs that promote cancer progression | [63] | |
| 37 | miR-720 | Breast | inhibits M2 TAM polarization | [64] | |
| 38 | miR-877 | Breast | increases expression in the late 4T1 tumor TAMs | [41] | |
| 39 | miR-940 | Ovarian | induces M2 TAMs polarization | [65] | |
| 40 | miR-4291 | Breast | downregulated in TAMs that promote cancer progression | [66] | |
| 41 | miR-5100 | Breast | inhibits invasion and migration of cancer | [66] | |
| 42 | miR-5196 | Breast | downregulated in TAMs that promote cancer progression | [66] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayasingam, S.D.; Citartan, M.; Mat Zin, A.A.; Rozhdestvensky, T.S.; Tang, T.-H.; Ch’ng, E.S. An Eleven-microRNA Signature Related to Tumor-Associated Macrophages Predicts Prognosis of Breast Cancer. Int. J. Mol. Sci. 2022, 23, 6994. https://doi.org/10.3390/ijms23136994
Jayasingam SD, Citartan M, Mat Zin AA, Rozhdestvensky TS, Tang T-H, Ch’ng ES. An Eleven-microRNA Signature Related to Tumor-Associated Macrophages Predicts Prognosis of Breast Cancer. International Journal of Molecular Sciences. 2022; 23(13):6994. https://doi.org/10.3390/ijms23136994
Chicago/Turabian StyleJayasingam, Sharmilla Devi, Marimuthu Citartan, Anani Aila Mat Zin, Timofey S. Rozhdestvensky, Thean-Hock Tang, and Ewe Seng Ch’ng. 2022. "An Eleven-microRNA Signature Related to Tumor-Associated Macrophages Predicts Prognosis of Breast Cancer" International Journal of Molecular Sciences 23, no. 13: 6994. https://doi.org/10.3390/ijms23136994
APA StyleJayasingam, S. D., Citartan, M., Mat Zin, A. A., Rozhdestvensky, T. S., Tang, T.-H., & Ch’ng, E. S. (2022). An Eleven-microRNA Signature Related to Tumor-Associated Macrophages Predicts Prognosis of Breast Cancer. International Journal of Molecular Sciences, 23(13), 6994. https://doi.org/10.3390/ijms23136994







