Identification and Characterization of Abiotic Stress–Responsive NF-YB Family Genes in Medicago
Abstract
1. Introduction
2. Results
2.1. Identification of NF-YB Family Genes in Medicago
2.2. Multiple Sequence Alignment of NF-YB Genes in Medicago
2.3. Phylogenetic Analysis of NF-YB Proteins
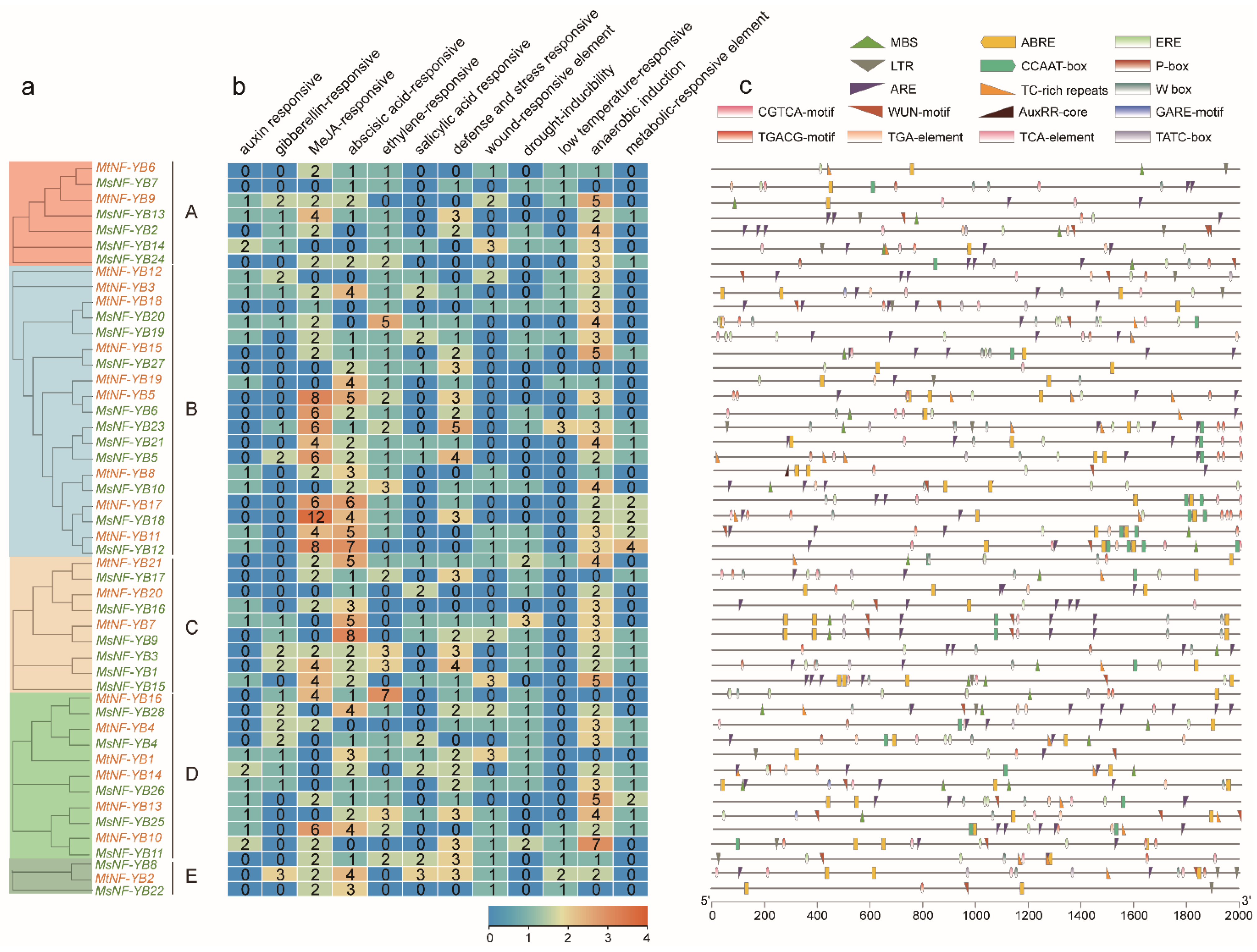
2.4. Analysis of Gene Structure and Conserved Motifs of NF-YB Genes in Medicago
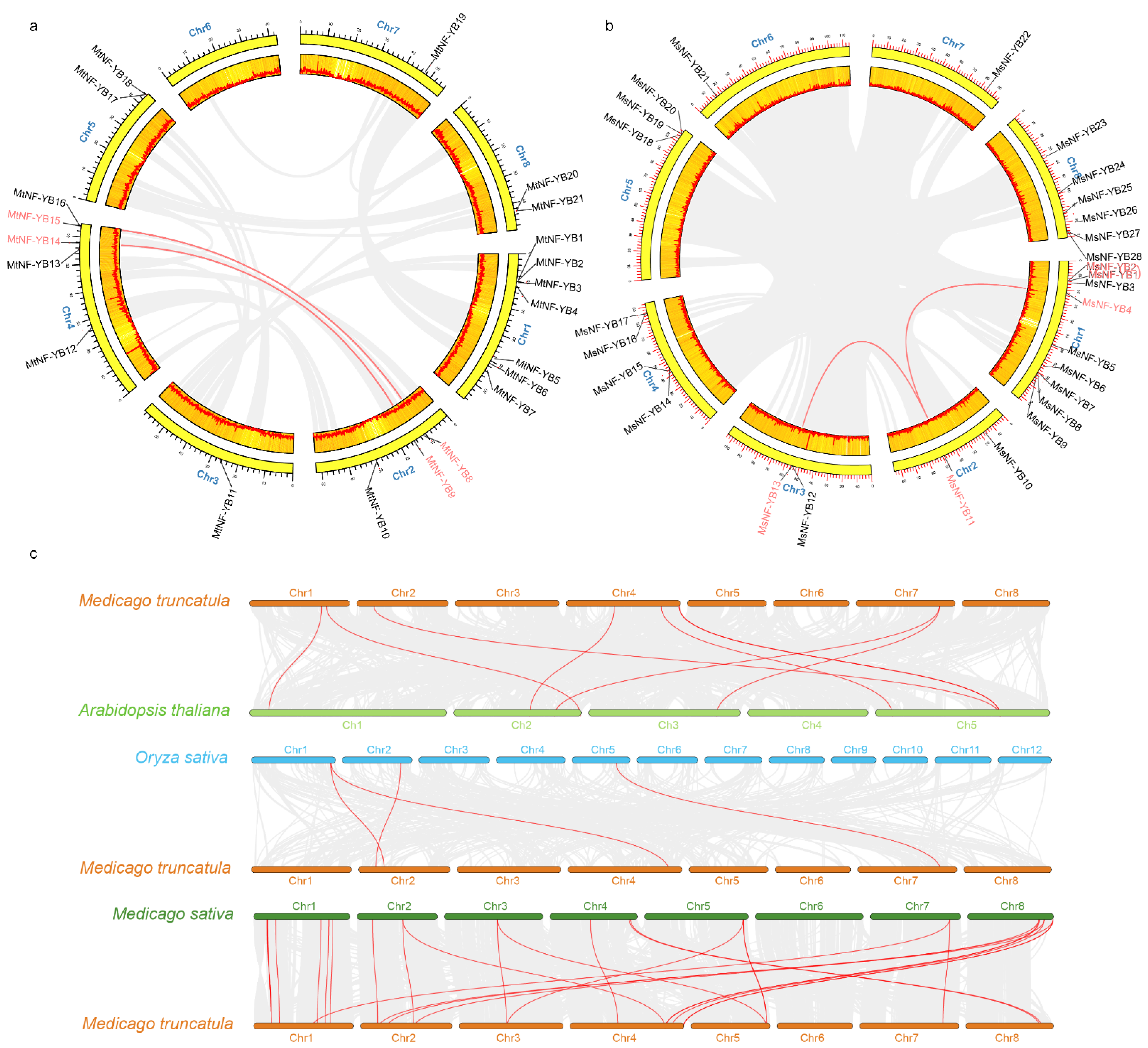
2.5. Analyses of Chromosomal Distribution and Synteny of NF-YB Genes in Medicago
2.6. Analysis of Cis-Elements in the Promoter Sequences of NF-YB Genes in Medicago
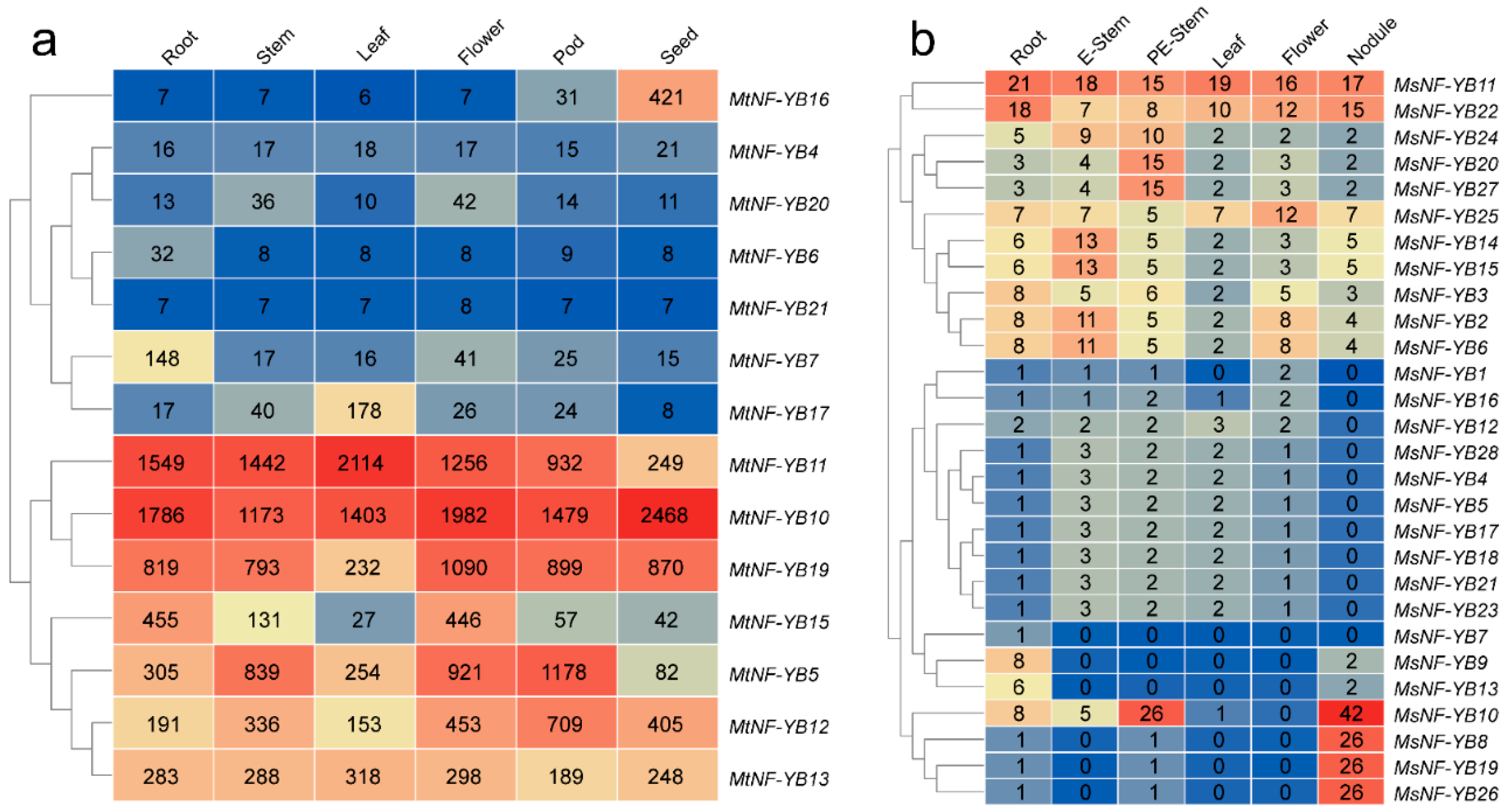
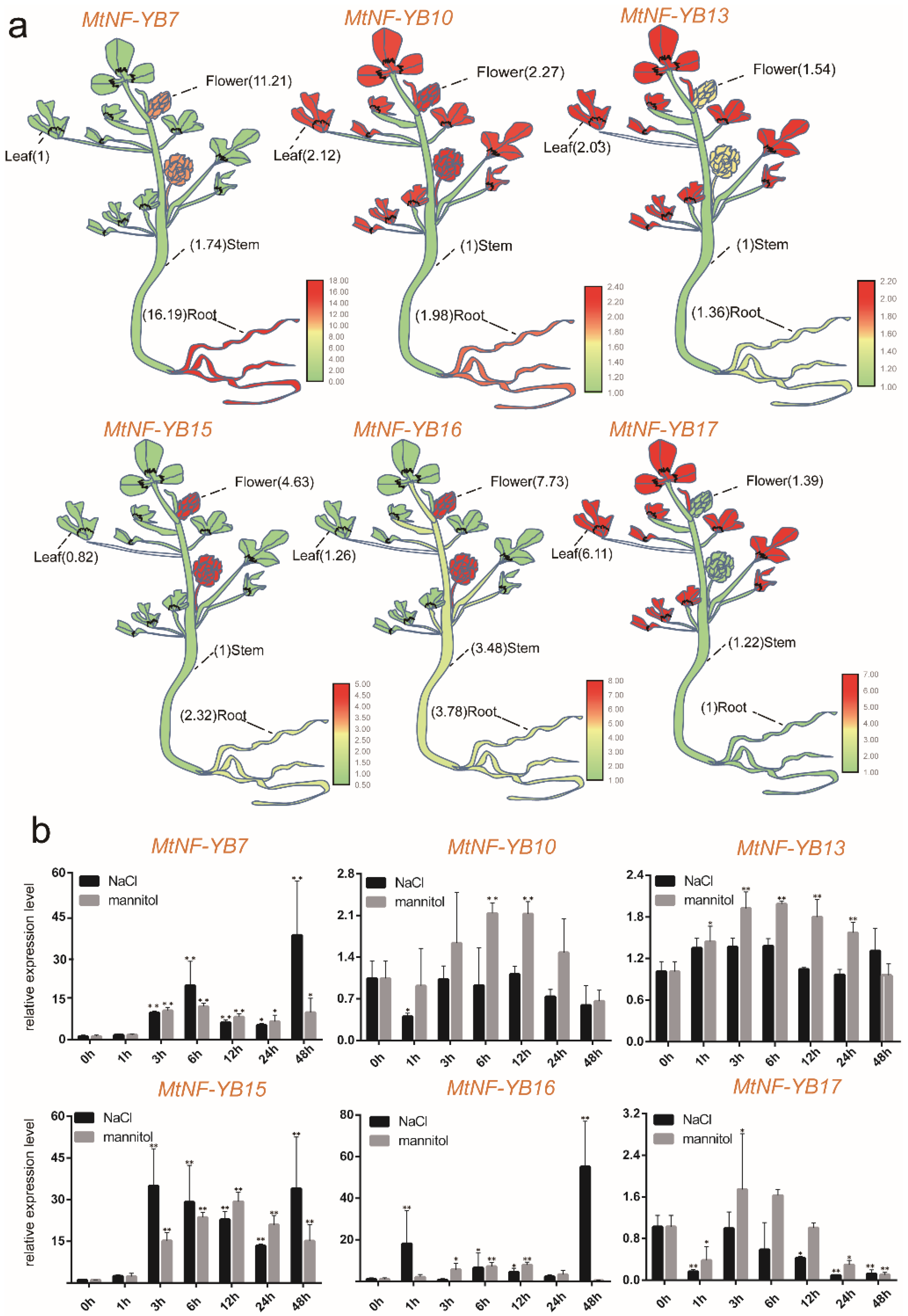
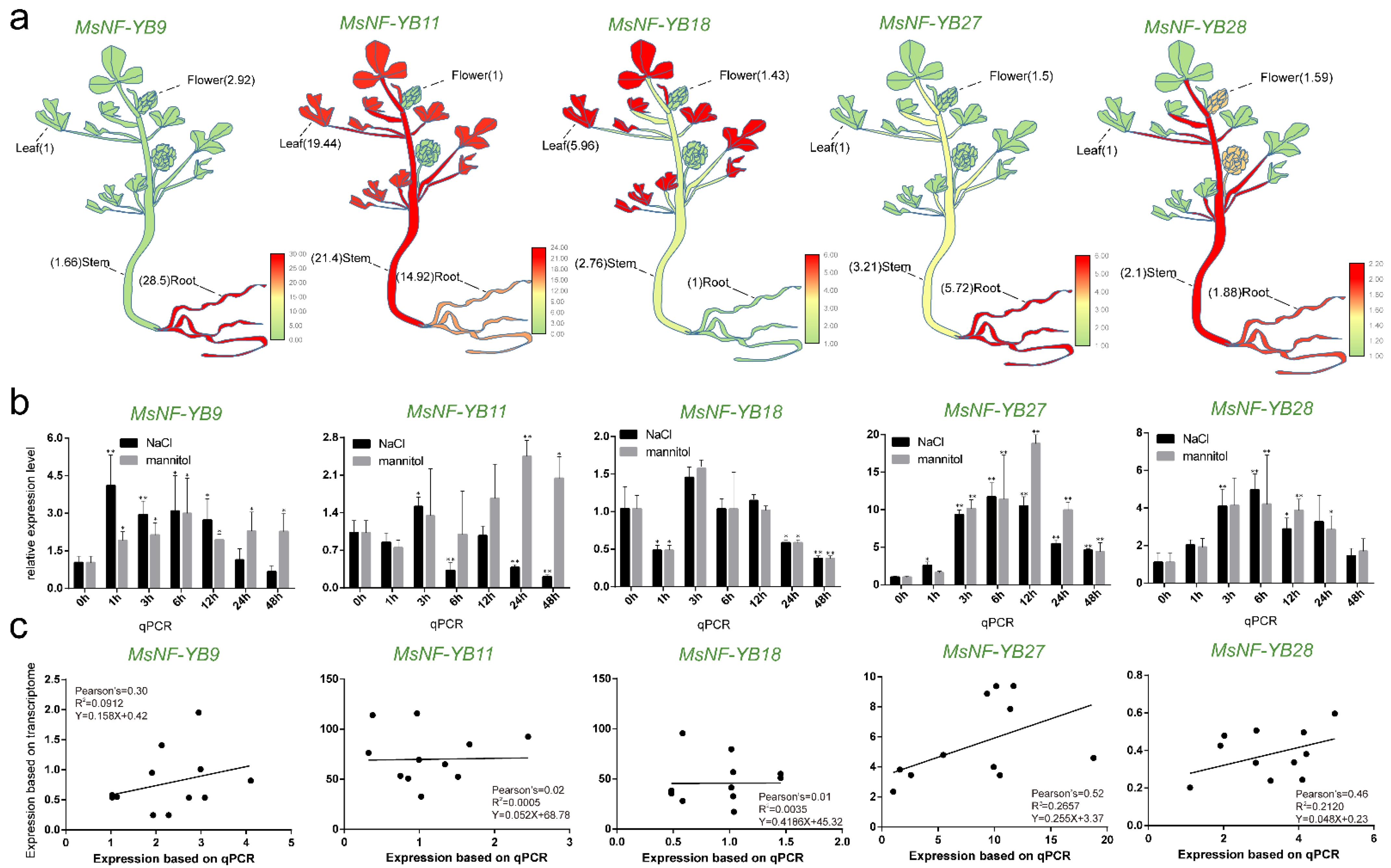
2.7. Expression Patterns of NF-YB Genes in Different Tissues
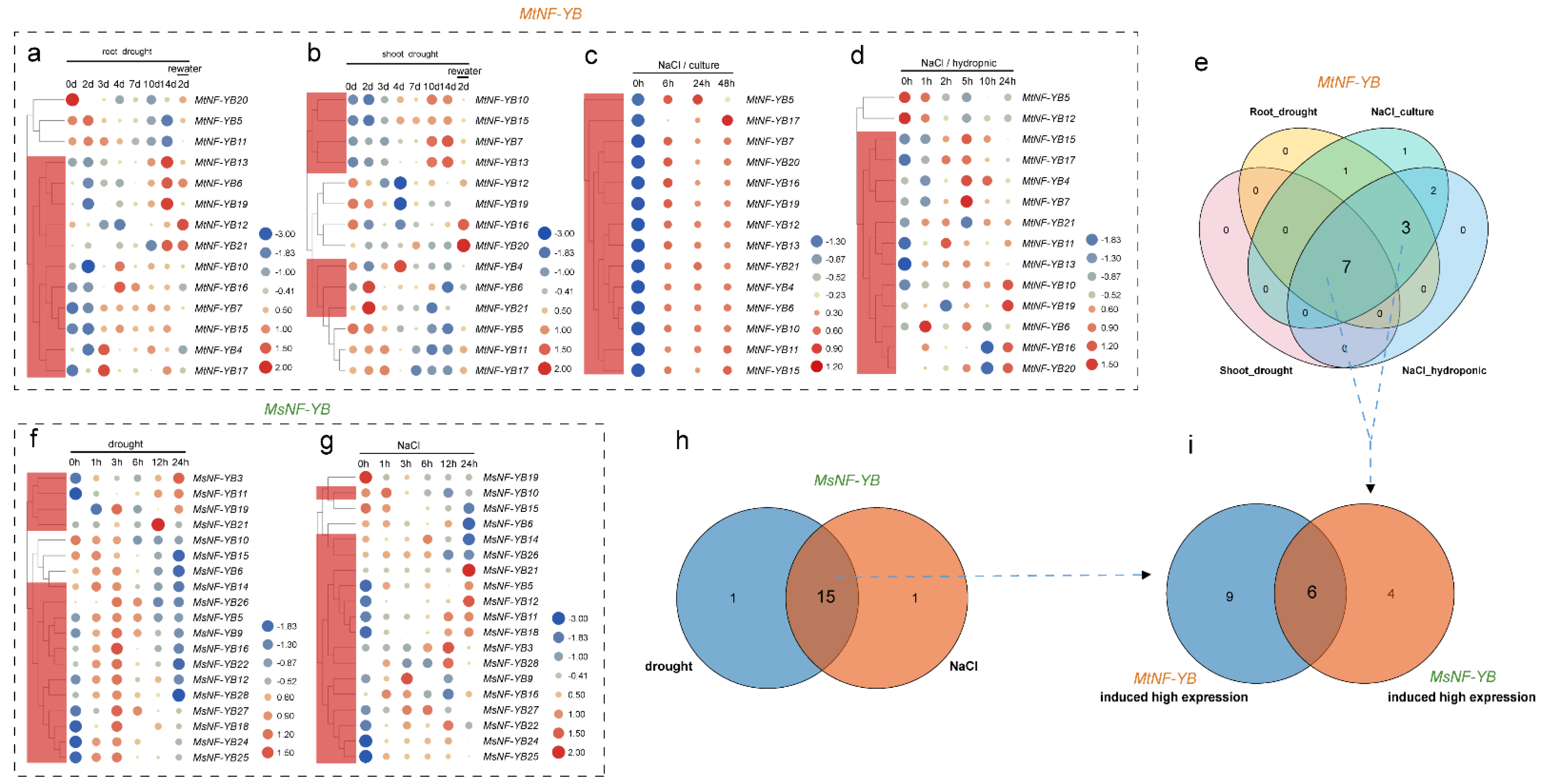
2.8. Expression Levels of NF-YB Genes in Medicago under Stress
2.9. Validation of the Expression Profile of Stress-Responsive NF-YB Genes by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Identification of NF-YB Family Members in Medicago
4.2. Analyses of Sequence, Conserved Motif, and Structural Characterization
4.3. Analyses of Phylogenetic Relationship
4.4. Analysis of Chromosome Locations and Collinearity
4.5. Identification of Stress-Response Cis-Elements of the NF-YB Promoter
4.6. Analysis of Gene Expression Profile
4.7. Plant Materials and Treatments
4.8. Gene Expression Analyses by RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edwards, D.D.; Murray, J.A.; Smith, A.G. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 1998, 117, 1015–1022. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Myers, Z.A.; Holt, B.F. NUCLEAR FACTOR-Y: Still complex after all these years? Curr. Opin. Plant Biol. 2018, 45, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Chaves-Sanjuan, A.; Nardini, M. Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 571–580. [Google Scholar] [CrossRef]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Mei, X.P.; Liu, C.X.; Yu, T.T.; Liu, X.L.; Xu, D.; Wang, J.Q.; Wang, G.Q.; Cai, Y.L. Identification and characterization of paternal-preferentially expressed gene NF-YC8 in maize endosperm. Mol. Genet. Genom. 2015, 290, 1819–1831. [Google Scholar] [CrossRef]
- Junker, A.; Monke, G.; Rutten, T.; Keilwagen, J.; Seifert, M.; Thi, T.M.; Renou, J.P.; Balzergue, S.; Viehover, P.; Hahnel, U.; et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 2012, 71, 427–442. [Google Scholar] [CrossRef]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef]
- Combier, J.P.; Frugier, F.; de Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernie, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef]
- Cai, X.; Ballif, J.; Endo, S.; Davis, E.; Liang, M.; Chen, D.; DeWald, D.; Kreps, J.; Zhu, T.; Wu, Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007, 145, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [PubMed]
- Warpeha, K.M.; Upadhyay, S.; Yeh, J.; Adamiak, J.; Hawkins, S.I.; Lapik, Y.R.; Anderson, M.B.; Kaufman, L.S. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007, 143, 1590–1600. [Google Scholar] [CrossRef]
- Ballif, J.; Endo, S.; Kotani, M.; MacAdam, J.; Wu, Y. Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol. Biochem. 2011, 49, 579–583. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef]
- Han, X.; Tang, S.; An, Y.; Zheng, D.C.; Xia, X.L.; Yin, W.L. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 2013, 64, 4589–4601. [Google Scholar] [CrossRef]
- Feng, Z.J.; He, G.H.; Zheng, W.J.; Lu, P.P.; Chen, M.; Gong, Y.M.; Ma, Y.Z.; Xu, Z.S. Foxtail millet NF-Y families: Genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 2015, 6, 1142. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.; Ran, Q.; Li, P.; Peng, Z.; Zhang, J. ZmNF-YB16 overexpression improves drought resistance and yield by enhancing photosynthesis and the antioxidant capacity of maize plants. Front. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research. Mol. Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.P.; Shan, C.J.; Zhao, H.; Li, J.C.; Jia, G.L.; Jiang, H.; Wu, S.Q.; Wang, Q. The difference in antioxidant capacity of four alfalfa cultivars in response to Zn. Ecotoxicol. Environ. Saf. 2015, 114, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Beiyuan, J.; Hu, W.; Zhang, Z.; Duan, C.; Cui, Q.; Zhu, X.; He, H.; Huang, X.; Fang, L. Phytoremediation of potentially toxic elements (PTEs) contaminated soils using alfalfa (Medicago sativa L.): A comprehensive review. Chemosphere 2022, 293, 133577. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yang, J.; Ma, L.; Yan, S.; Pang, Y. Genome-Wide Identification and Analyses of Drought/Salt-Responsive Cytochrome P450 Genes in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 9957. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, Z.; Li, R.; Xu, Y.; Kong, Y.; Zhou, G.; Meng, C.; Hu, R. Genome-wide identification and expression profiling of HD-ZIP gene family in Medicago truncatula. Genomics 2020, 112, 3624–3635. [Google Scholar] [CrossRef]
- Yang, Y.; Qi, L.; Nian, L.; Zhu, X.; Yi, X.; Jiyu, Z.; Qiu, J. Genome-Wide Identification and Expression Analysis of the SRS Gene Family in Medicago sativa. DNA Cell Biol. 2021, 40, 1539–1553. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, J.; Sun, Z.; Tang, Y.; Wu, Y. Genome-Wide Identification and Analysis of the Polycomb Group Family in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 7537. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, X.; Nie, X.; Li, H.; Yuan, S.; Dai, B.; Zhan, J.; Wen, Z.; Jiang, J.; Chen, C.; et al. Nuclear miR-665 aggravates heart failure via suppressing phosphatase and tensin homolog transcription. Sci. China Life Sci. 2020, 63, 724–736. [Google Scholar] [CrossRef]
- Wu, P.H.; Isaji, M.; Carthew, R.W. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol. Cell 2013, 52, 113–123. [Google Scholar] [CrossRef]
- Maity, S.N.; Crombrugghe, B. Biochemical analysis of the B subunit of the heteromeric CCAAT-binding factor. J. Biol. Chem. 1992, 267, 8286–8292. [Google Scholar] [CrossRef]
- Liang, M.; Yin, X.; Lin, Z.; Zheng, Q.; Liu, G.; Zhao, G. Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 2014, 239, 107–126. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Bertini, L.; Palazzi, L.; Proietti, S.; Pollastri, S.; Arrigoni, G.; Polverino de Laureto, P.; Caruso, C. Proteomic analysis of meJa-induced defense responses in rice against wounding. Int. J. Mol. Sci. 2019, 20, 2525. [Google Scholar] [CrossRef]
- Lee, S.C.; Kim, S.Y.; Kim, S.R. Drought inducible OsDhn1 promoter is activated by OsDREB1A and OsDREB1D. J. Plant Biol. 2013, 56, 115–121. [Google Scholar] [CrossRef]
- Zanetti, M.E.; Ripodas, C.; Niebel, A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 645–654. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z.; Xiong, Y.; Yao, J. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J. 2017, 5, 21–31. [Google Scholar] [CrossRef]
- Xie, Z.; Li, X.; Glover, B.J.; Bai, S.; Rao, G.Y.; Luo, J.; Yang, J. Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 2008, 25, 1581–1592. [Google Scholar] [CrossRef]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 1994, 256, 119–124. [Google Scholar]
- Van de Peer, Y.; Fawcett, J.A.; Proost, S.; Sterck, L.; Vandepoele, K. The flowering world: A tale of duplications. Trends Plant Sci. 2009, 14, 680–688. [Google Scholar] [CrossRef]
- Shannon, M.C.; Rhoades, J.D.; Draper, J.H.; Scardaci, S.C.; Spyres, M.D. Assessment of Salt Tolerance in Rice Cultivars in Response to Salinity Problems in California. Crop Ecol. Prod. Manag. 1998, 38, 394–398. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana Nuclear Factor Y Transcription Factors. Front. Plant Sci. 2016, 7, 2045. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Kumimoto, R.W.; Gnesutta, N.; Calogero, A.M.; Mantovani, R.; Holt, B.F. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 2014, 26, 1009–1017. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Lee, H.; Fischer, R.; Goldberg, R.; Harada, J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT biinding transcription factor. Proc. Natl. Acad. Sci. USA 2003, 18, 2152–2156. [Google Scholar] [CrossRef]
- Huang, M.; Hu, Y.; Liu, X.; Li, Y.; Hou, X. Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front. Plant Sci. 2015, 6, 955. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kakar, K.U.; Nawaz, Z.; Kakar, K.; Ali, E.; Almoneafy, A.A.; Ullah, R.; Ren, X.L.; Shu, Q.Y. Comprehensive genomic analysis of the CNGC gene family in Brassica oleracea: Novel insights into synteny, structures, and transcript profiles. BMC Genom. 2017, 18, 869. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gong, Q.; Qin, W.; Yang, Z.; Cheng, Y.; Lu, L.; Ge, X.; Zhang, C.; Wu, Z.; Li, F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.F.; Ma, L.; Jiang, W.B.; Yao, Y.; Tang, Y.H.; Pang, Y.Z. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021, 158, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van, P.Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Luo, D.; Zhou, Q.; Wu, Y.; Chai, X.; Liu, W.; Wang, Y.; Yang, Q.; Wang, Z.; Liu, Z. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Seo, H.D.; Hennighausen, L.; Lee, D.; Kang, K. Octopus-toolkit: A workflow to automate mining of public epigenomic and transcriptomic next-generation sequencing data. Nucleic Acids Res. 2018, 46, e53. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene Name | TIGR Locus | Homologous Gene | PI | MW (kDa) | Protein Length | Subcellular Localization | Plant TFDB |
|---|---|---|---|---|---|---|---|
| MtNF-YB1 | MtrunA17Chr1g0158951 | MsNF-YB4 | 4.64 | 12.44 | 112 | Nuclear | Medtr1g028480.1 |
| MtNF-YB2 | MtrunA17Chr1g0159271 | MsNF-YB1 | 5.28 | 11.57 | 108 | Nuclear | Medtr1g029070.1 |
| MtNF-YB3 | MtrunA17Chr1g0159291 | MsNF-YB2 | 4.66 | 10.36 | 91 | Nuclear | Medtr1g029100.1 |
| MtNF-YB4 | MtrunA17Chr1g0165041 | MsNF-YB4 | 5.59 | 21.65 | 191 | Nuclear | Medtr1g039040.1 |
| MtNF-YB5 | MtrunA17Chr1g0185911 | MsNF-YB6 | 5.75 | 19.25 | 175 | Nuclear | Medtr1g072790.2 |
| MtNF-YB6 | MtrunA17Chr1g0191981 | MsNF-YB7 | 6.90 | 14.46 | 128 | Nuclear | Medtr1g083070.1 |
| MtNF-YB7 | MtrunA17Chr1g0195851 | MsNF-YB9 | 5.72 | 22.24 | 195 | Nuclear | Medtr1g088860.1 |
| MtNF-YB8 | MtrunA17Chr2g0290491 | MsNF-YB10 | 5.78 | 17.99 | 167 | Nuclear | Medtr2g026710.1 |
| MtNF-YB9 | MtrunA17Chr2g0296321 | MsNF-YB26 | 8.88 | 28.96 | 257 | Nuclear | Medtr0392s0020.1 |
| MtNF-YB10 | MtrunA17Chr2g0307551 | MsNF-YB11 | 4.63 | 17.43 | 157 | Nuclear | Medtr2g056000.1 |
| MtNF-YB11 | MtrunA17Chr3g0102351 | MsNF-YB12 | 6.21 | 20.11 | 191 | Nuclear | Medtr3g058980.1 |
| MtNF-YB12 | MtrunA17Chr4g0026311 | MsNF-YB14 | 5.25 | 15.54 | 141 | Nuclear | Medtr4g052950.1 |
| MtNF-YB13 | MtrunA17Chr4g0062271 | MsNF-YB11 | 4.80 | 17.48 | 157 | Nuclear | Medtr4g112380.1 |
| MtNF-YB14 | MtrunA17Chr4g0067091 | MsNF-YB26 | 6.31 | 24.94 | 218 | Nuclear | Medtr4g119500.1 |
| MtNF-YB15 | MtrunA17Chr4g0076321 | MsNF-YB27 | 5.36 | 15.33 | 137 | Nuclear | Medtr4g133938.1 |
| MtNF-YB16 | MtrunA17Chr4g0076381 | MsNF-YB28 | 6.26 | 26.78 | 241 | Nuclear | Medtr4g133952.1 |
| MtNF-YB17 | MtrunA17Chr5g0446491 | MsNF-YB18 | 5.96 | 19.98 | 185 | Nuclear | Medtr5g095740.1 |
| MtNF-YB18 | MtrunA17Chr5g0446601 | MsNF-YB20 | 4.57 | 21.16 | 192 | Extracellular | Medtr5g095900.1 |
| MtNF-YB19 | MtrunA17Chr7g0263601 | MsNF-YB6 | 6.10 | 18.77 | 172 | Nuclear | Medtr7g100650.1 |
| MtNF-YB20 | MtrunA17Chr8g0382931 | MsNF-YB16 | 5.80 | 22.55 | 204 | Nuclear | Medtr8g091720.1 |
| MtNF-YB21 | MtrunA17Chr8g0384451 | MsNF-YB17 | 6.04 | 24.96 | 215 | Nuclear | Medtr8g093920.1 |
| MsNF-YB1 | MsG0180001010.01.T01 | MtNF-YB2 | 5.25 | 18.87 | 173 | Nuclear | — |
| MsNF-YB2 | MsG0180001011.01.T01 | MtNF-YB3 | 5.17 | 22.86 | 204 | Extracellular | — |
| MsNF-YB3 | MsG0180001031.01.T01 | MtNF-YB2 | 5.09 | 20.95 | 192 | Nuclear | — |
| MsNF-YB4 | MsG0180001571.01.T01 | MtNF-YB4 | 5.32 | 21.27 | 188 | Nuclear | — |
| MsNF-YB5 | MsG0180003209.01.T01 | MtNF-YB17 | 4.47 | 10.76 | 99 | Nuclear | — |
| MsNF-YB6 | MsG0180004009.01.T01 | MtNF-YB5 | 5.75 | 19.26 | 175 | Nuclear | — |
| MsNF-YB7 | MsG0180004588.01.T01 | MtNF-YB6 | 6.55 | 10.8 | 97 | Nuclear | — |
| MsNF-YB8 | MsG0180004726.01.T01 | MtNF-YB14 | 6.42 | 21.34 | 182 | Nuclear | — |
| MsNF-YB9 | MsG0180004882.01.T01 | MtNF-YB7 | 5.51 | 21.18 | 186 | Nuclear | — |
| MsNF-YB10 | MsG0280007510.01.T01 | MtNF-YB8 | 5.78 | 17.67 | 163 | Nuclear | — |
| MsNF-YB11 | MsG0280009235.01.T01 | MtNF-YB10 | 6.02 | 26.5 | 235 | Nuclear | — |
| MsNF-YB12 | MsG0380014538.01.T01 | MtNF-YB11 | 6.21 | 20.18 | 191 | Nuclear | — |
| MsNF-YB13 | MsG0380014546.01.T02 | MtNF-YB17 | 6.16 | 22.05 | 194 | Extracellular | — |
| MsNF-YB14 | MsG0480020531.01.T01 | MtNF-YB12 | 5.40 | 14.64 | 130 | Extracellular | — |
| MsNF-YB15 | MsG0480020532.01.T01 | MtNF-YB12 | 7.67 | 15.19 | 135 | Extracellular | — |
| MsNF-YB16 | MsG0480023372.01.T01 | MtNF-YB20 | 5.80 | 21.81 | 197 | Nuclear | — |
| MsNF-YB17 | MsG0480023477.01.T02 | MtNF-YB21 | 6.14 | 26.18 | 226 | Nuclear | — |
| MsNF-YB18 | MsG0580029966.01.T01 | MtNF-YB17 | 4.92 | 15.87 | 144 | Nuclear | — |
| MsNF-YB19 | MsG0580029976.01.T01 | MtNF-YB18 | 4.51 | 20.54 | 188 | Nuclear | — |
| MsNF-YB20 | MsG0580029978.01.T01 | MtNF-YB18 | 4.64 | 20.68 | 187 | Nuclear | — |
| MsNF-YB21 | MsG0680031203.01.T01 | MtNF-YB17 | 8.66 | 88.39 | 80 | Nuclear | — |
| MsNF-YB22 | MsG0780040897.01.T01 | MtNF-YB19 | 8.67 | 90.71 | 289 | Extracellular | — |
| MsNF-YB23 | MsG0880043949.01.T01 | MtNF-YB17 | 9.34 | 8.52 | 78 | Nuclear | — |
| MsNF-YB24 | MsG0880046588.01.T01 | MtNF-YB13 | 6.92 | 31.34 | 277 | Extracellular | — |
| MsNF-YB25 | MsG0880046644.01.T01 | MtNF-YB13 | 4.70 | 13.72 | 124 | Nuclear | — |
| MsNF-YB26 | MsG0880047027.01.T01 | MtNF-YB14 | 5.27 | 14.69 | 130 | Nuclear | — |
| MsNF-YB27 | MsG0880047742.01.T01 | MtNF-YB15 | 5.35 | 15.28 | 137 | Nuclear | — |
| MsNF-YB28 | MsG0880047744.01.T01 | MtNF-YB16 | 6.25 | 23.97 | 214 | Nuclear | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, W.; Yang, J.; Li, Q.; He, C.; Pang, Y. Identification and Characterization of Abiotic Stress–Responsive NF-YB Family Genes in Medicago. Int. J. Mol. Sci. 2022, 23, 6906. https://doi.org/10.3390/ijms23136906
Du W, Yang J, Li Q, He C, Pang Y. Identification and Characterization of Abiotic Stress–Responsive NF-YB Family Genes in Medicago. International Journal of Molecular Sciences. 2022; 23(13):6906. https://doi.org/10.3390/ijms23136906
Chicago/Turabian StyleDu, Wenxuan, Junfeng Yang, Qian Li, Chunfeng He, and Yongzhen Pang. 2022. "Identification and Characterization of Abiotic Stress–Responsive NF-YB Family Genes in Medicago" International Journal of Molecular Sciences 23, no. 13: 6906. https://doi.org/10.3390/ijms23136906
APA StyleDu, W., Yang, J., Li, Q., He, C., & Pang, Y. (2022). Identification and Characterization of Abiotic Stress–Responsive NF-YB Family Genes in Medicago. International Journal of Molecular Sciences, 23(13), 6906. https://doi.org/10.3390/ijms23136906





