Abstract
The peptidase M24 (Metallopeptidase 24, M24) superfamily is essential for plant growth, stress response, and pathogen defense. At present, there are few systematic reports on the identification and classification of members of the peptidase M24 proteins superfamily in wheat. In this work, we identified 53 putative candidate TaM24 genes. According to the protein sequences characteristics, these members can be roughly divided into three subfamilies: I, II, III. Most TaM24 genes are complex with multiple exons, and the motifs are relatively conserved in each sub-group. Through chromosome mapping analysis, we found that the 53 genes were unevenly distributed on 19 wheat chromosomes (except 3A and 3D), of which 68% were in triads. Analysis of gene duplication events showed that 62% of TaM24 genes in wheat came from fragment duplication events, and there were no tandem duplication events to amplify genes. Analysis of the promoter sequences of TaM24 genes revealed that cis-acting elements were rich in response elements to drought, osmotic stress, ABA, and MeJA. We also studied the expression of TaM24 in wheat tissues at developmental stages and abiotic stress. Then we selected TaM24-9 as the target for further analysis. The results showed that TaM24-9 genes strengthened the drought and salt tolerance of plants. Overall, our analysis showed that members of the peptidase M24 genes may participate in the abiotic stress response and provided potential gene resources for improving wheat resistance.
1. Introduction
Wheat (Triticum aestivum) is one of the most widely cultivated crops in the world, providing an important source of calories for the growing world population [1,2]. Wheat is an indelible part of food culture, and its output is directly related to people’s daily life and national food security. However, extreme climate change, the limitation of available cultivated land area, and reduced soil quality have in recent years posed great challenges to the safe production of wheat. Abiotic stress factors, mainly including drought, salt, extreme temperature, and poor land nutrition, have led to decreased agricultural productivity [2,3,4]. Drought stress can reduce wheat germination rate, seedling rate, yield, and quality. Under drought stress, the relative water content in plants decreases, and the transport of soluble nutrients decreases due to reduced water absorption by roots [5,6]. In addition, the photosynthetic system of plants is damaged, reducing photosynthesis [4,7]. On average, 20-30% of wheat production is lost due to drought every year, which may have serious consequences for the global food supply in the future [8,9].
High salt accumulation caused by farmland irrigation or natural accumulation is another major factor in decreased wheat yield [10]. Studies have shown that 45 million hectares (about 19.5% of available arable land) are threatened by salt globally [11,12]. In Asia, China occupies the largest area of land affected by salt [2]. For example, NaCl stress enhanced the autophagy activity of roots and leaves of wheat seedlings, causing the tissue death of wheat seedlings [13]. Therefore, in increasingly harsh environmental conditions, it is necessary to explore genes potentially related to stress and identify their functions for cultivating wheat varieties with desirable traits. Studies have shown that members of the peptidase M24 protein superfamily play an important role in plant stress resistance [14,15,16,17]. Therefore, it is of great significance to explore the peptidase M24 genes in wheat.
The peptidase M24 superfamily is a complex group composed of many different types of proteins. Peptidase M24 proteins' superfamily members are grouped together mainly because they have a conserved ‘pita bread’ fold that is made of both alpha helices and an anti-parallel beta sheet within two structurally similar domains. This peptidase M24 proteins superfamily includes methionine aminopeptidase (EC 3.4.11.18), aminopeptidase P (EC 3.4.11.9), prolidase (EC 3.4.13.9), intermediate cleavage peptidase (EC 3.4.11.26), creatinase (EC 3.5.3.3), ectoine hydrolase (EC 3.5.4.44), and non-peptidase homologues (such as Spt16 of the FACT complex, Proliferation-associated 2G4 like protein, ErbB-3 binding protein, CDC68-like protein, etc.) [18,19,20,21]. The research on the function of peptidase M24 proteins in plants is relatively late compared to animals and microorganisms. In recent years, peptidase M24 proteins in plants including methionine aminopeptidase (MAPs), aminopeptidase P (APPs), SPT16, ICP55, and ErbB-3 binding protein (EBP1) have been reported [22,23,24,25,26,27,28]. Many peptidase M24 proteins have aminopeptidase activity and can hydrolyze specific peptide bonds to ensure the normal synthesis and metabolism of proteins in plants. For example, multiple types of MAPs are found in Arabidopsis (A. thalianna), which are involved in the excision of methionine [29]. Previous studies in maize (Zea mays) and Arabidopsis have found that assumed MAPs are essential to maintain the normal structure of chloroplasts and participate in stable biological processes of chloroplast proteins [14,29]. Moreover, upon deletion of an assumed peptidase M24 gene, thylakoid arrangement is loose, chlorophyll formation and photosynthesis are destroyed, and cell death is eventually caused by the reactive oxygen species (ROS) pathway [14].These non-peptidase homologues directly bind to DNA or RNA, or combine with other proteins to act on nucleic acids. For example, Arabidopsis EBP1 (AtEBP1) can be phosphorylated by kinases to bind the CALMODULIN-like protein 38 (CLM38) gene promoter region and then participate in signal transmission in plants [17].
Members of the peptidase M24 proteins superfamily function in plant growth and development and the abiotic stress response. The protein encoded by the peptidase M24 gene found in barley (Hordeum vulgare) has methionine aminopeptidase activity. After cold stress, protein localization shifted from the nucleus to the cytoplasm, and the transgenic plant lines also showed frost resistance [30]. The EBP1 of peptidase M24 protein superfamily was identified in Ammopiptanthus mongolicus [31]. In addition, the expression of the cold tolerance gene KIN1 in transgenic Arabidopsis was higher without stress treatment, and improved survival rates of transgenic E. coli and Arabidopsis under low temperatures [31]. The members of peptidase M24 genes can not only improve the cold tolerance of plants but also respond to the drought stress and salt stress of plants. The Hevea Brasiliensis EBP1 gene (HbEBP1) provided stronger drought resistance for the developed root system in Arabidopsis [15]. At the same time, transgenic Arabidopsis that overexpresses the Atriplex canescens EBP1 gene (ACEBP1) also has greater drought resistance by reducing water consumption, promoting peroxidase (POD) activity, and reducing the content of ROS [16]. In addition, the gene expression of the Ammopiptanthus mongolicus EBP1 gene (AmEBP1) increases continuously under salinity stress over 48 h. AtAPP1 can also interact with superoxide dismutase (SOD), POD and E2-ubiquitin-conjugating enzymes in response to hormone signalling [32,33]. In addition, another study showed that the initial immune response to biotic stress was related to the same core proteases from C1, C48, C65, M24, M41, S10, S9, S8, and A1 families in wheat [34].
Peptidase M24 proteins' superfamily members have been studied in a variety of plants, but they have not been systematically classified or identified. The goal of this study was to supplement the lack of identification of peptidase M24 superfamily members in wheat. We identified 53 members of the peptidase M24 genes superfamily in wheat. In addition, we performed phylogenetic analysis of the peptidase M24 genes' superfamily, showed positional relationships of these genes on wheat chromosomes, conducted motif analysis, and also determined expression patterns in wheat. The results contribute to an understanding of the evolutionary pattern of the peptidase M24 proteins superfamily and they participate in the growth, development and regulation of abiotic stress response in wheat.
2. Results
2.1. Identification and Characterization of TaM24 Genes
A total of 53 assumed members of the peptidase M24 superfamily were identified in the wheat genome (Figure 1). The basic information of these genes is in Table S1. Based on the position of these genes on the wheat chromosome and referring to previous articles about new gene family naming rules [35,36], we renamed these genes TaM24-1 to TaM24-53. The common feature of peptidase M24 proteins members in wheat is that they all have the same complete conserved domain, PF00557.
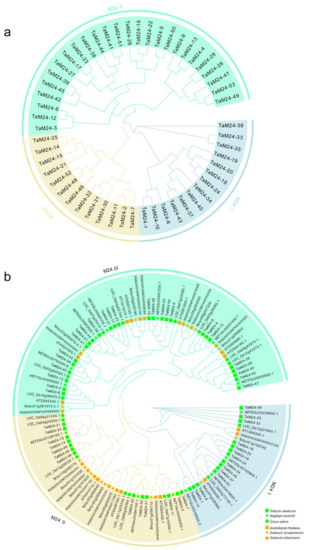
Figure 1.
Phylogenetic analysis of peptidase M24 proteins. (a) In total, 53 peptidase M24 protein sequences in wheat. (b) A phylogenetic tree of peptidase M24 protein in monocotyledons (Triticum aestivum, Oryza sativa, Aegilops tauschii) and dicotyledons (Arabidopsis thaliana, Solanum tuberosum, Solanum lycopersicum). Cluster Omega was used to compare the amino acid sequences based on a hidden Markov model. The phylogenetic tree was inferred using the maximum-likelihood method of PhyML (Shimodaira Hasegawa test, 1000 times), which divided M24 proteins into three subfamilies. These subfamilies are represented by different colors: M24I (azure), M24II (peach), M24III (cyan). The monocotyledons and dicotyledons are represented by orange and grass green, respectively. The different colored shapes represent different species.
We found that the transcripts of all 53 TaM24 genes (including untranslated regions (UTRs) and coding sequence (CDS)) had an average length of 1922 bp, ranging from 701 bp (TaM24-42) to 4708 bp (TaM24-48). The amino acid number of peptide chains encoded by the TaM24 genes ranged from 133 (TaM24-5; TaM24-50) to 1086 (TaM24-21), with an average of 517. The molecular weights of TaM24 proteins ranged from 766.16 KDa (TaM24-14) to 14.68 KDa (TaM24-5), and the average molecular weight was 70.34 KDa, with isoelectric point (pI) values ranging from 5 (TaM24-45) to 8.74 (TaM24-17). Moreover, 70% (37/53) of the predicted TaM24 proteins were stable (Instability Index < 40). Subcellular location prediction showed that TaM24 proteins were mostly distributed in the nucleus, cytoplasm, peroxisome, mitochondria, chloroplast, extracellular, etc. Therefore, we speculate that the peptidase M24 is of great significance for the plant’s multiple biological activities.
2.2. Phylogenetic Analysis and Classification of TaM24 Proteins
In order to identify the phylogenetic relationships of peptidase M24 members, we constructed a phylogenetic tree using protein sequences of monocotyledon (Triticum aestivum, Oryza sativa, Aegilops tauschii) and dicotyledon (Arabidopsis thaliana, Solanum tuberosum, Solanum lycopersicum) peptidase M24 superfamily members (Figure 1). In the MEROPS database, peptidase M24 is divided into two categories: M24 A and M24 B [37,38]. Based on this classification, and according to the protein sequences and structural characteristics of peptidase M24 proteins, we divided M24 proteins in plants into three subfamilies: peptidase M24 I (M24I), peptidase M24 II (M24II), and peptidase M24 III (M24III). The phylogenetic tree shows that the three subfamilies of peptidase M24 are distributed in all six species, indicating that the peptidase M24 superfamily was conserved over evolutionary time. In addition, compared with Arabidopsis, the peptidase M24 superfamily of Aegilops and rice (Oryza sativa), which are monocotyledonous plants, have a closer phylogenetic relationship with those from wheat. We found that 26.4%, 24.5%, and 49.1% of the peptidase M24 proteins were located in the M24I, M24II, and M24III subfamilies, respectively (Figure S1 and Table S2).
2.3. Chromosome Mapping and Homologous Gene Identification of TaM24 Genes
In this work, TaM24 genes were mapped to 19 chromosomes of wheat and showed an uneven distribution in the wheat genome (Figure 2). Table S1 lists the precise location information of TaM24 genes on the chromosomes. Wheat chromosomes 1 to 7 have 13, 14, two, three, four, nine, and eight genes, respectively. The number of TaM24 genes distributed on chromosome 2B is the largest with six, followed by 1B with five, while there were no TaM24 genes in 3A and 3D. The uneven distribution of TaM24 genes indicates that there is an independent genetic variation mechanism of TaM24 genes in the process of chromosome evolution.
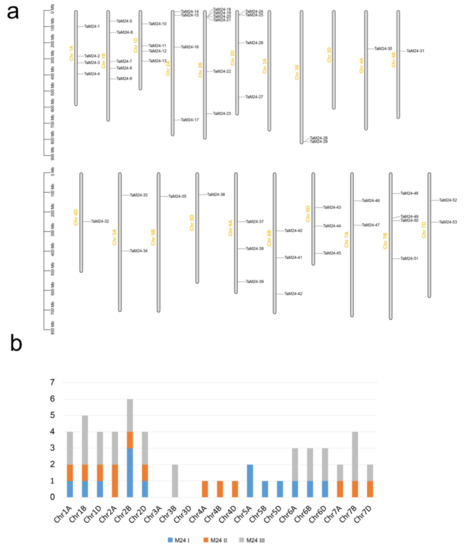
Figure 2.
The distribution of 53 TaM24 genes on wheat chromosomes. (a) The physical location of 53 TaM24 genes on wheat chromosomes. (b) The number of TaM24 genes per chromosome.
The homoeologous groups of TaM24 were also analyzed, and the results are shown in Table 1 and Table S3. Approximately 68% (36/53) of TaM24 genes have a homoeolog in the A, B, and D sub-genomes. However, in the whole genome of wheat (IWGSC, 2018), only 35.8% of genes were present in triads (homoeologous groups of 3). The proportion of orphans/singletons is also relatively low at 6%. In addition, we identified peptidase M24 genes in other plants (A. thaliana, S. tuberosum, S. lycopersicum, Hordeum vulgare, O. sativa, Zea mays, ect.), but found that wheat still has many members of the peptidase M24 genes superfamily (Table S4). Wheat, an allohexaploid, has a genome size that reaches 17Gb, and has a high retention rate of homoeologous TaM24 genes, which can explain why the number of TaM24 genes is higher in wheat than in other species.

Table 1.
Homoeologous TaM24 genes in wheat.
2.4. Analyzing Gene Duplication Events and Natural Selection
Gene duplication includes tandem duplication and segmental duplication, which are particularly important in the expansion of polyploid plant genes and the functional differentiation of genes. In order to clarify the evolutionary mechanism of TaM24 expansion, we analyzed tandem duplication and whole genome/segmental duplication events in the wheat genome (Figure 3). There were 33 TaM24 genes located in synonymous regions of wheat chromosomes, and 27 duplicate gene pairs were formed (Table S5). Among them, 58% (19/33) of TaM24 genes clustered on chromosomes 1 and 2, exactly corresponding to a large number of genes located on these chromosomes. Results showed that 62% (33/53) of TaM24 amplification in wheat came from the WGD/segmental duplication event, and there was no tandem duplication to amplify the gene. Next, we calculated the Ka/Ks values of all repeated gene pairs to explore the evolutionary selection pressure on TaM24 (Figure S2, Table S5). The results showed that the Ka/Ks values of all TaM24 repeated gene pairs were lower than 0.5, indicating that the replicated genes were under the pressure of purification and selection. This evidence suggests that the expansion (or contraction) of TaM24 may enable it to adapt to environmental changes, thus contributing to its wider distribution.
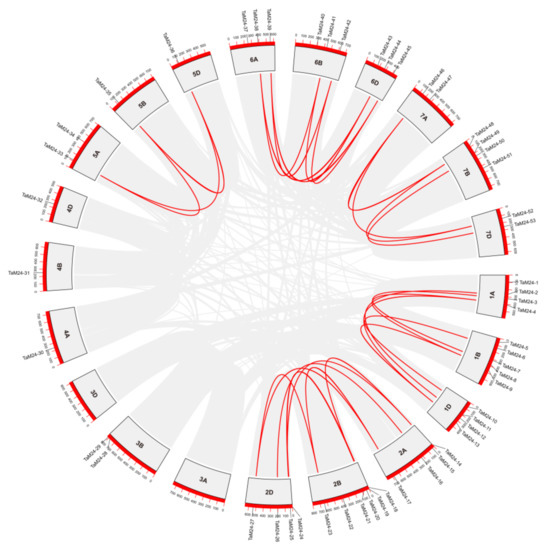
Figure 3.
The result of collinearity analysis of TaM24 genes in the wheat genome. TaM24 genes are mapped to their respective positions in the wheat genome chromosomes. The whole genome/segmental and tandem duplications of the wheat genome are represented in the gray area in a circular diagram using Circos. The red line is a WGD/segmental duplicate TaM24 gene pair connected by fragment replication.
In order to better understand the evolutionary relationships of the peptidase M24 superfamily, we carried out the analysis of gene duplication events between wheat and other plants (Figure 4, Table S6). T. aestivum had higher collinearity with monocotyledons A. tauschii and O. sativa, demonstrating that the number of peptidase M24 members expanded slowly and that this family was very conserved in the process of species evolution.
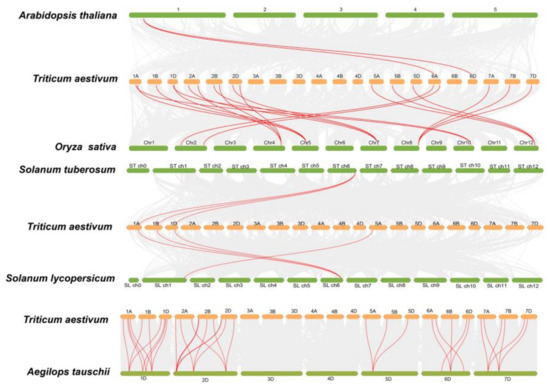
Figure 4.
Syntenic relationships of the peptidase M24 genes in wheat and other species. Gray lines in the background represent the synteny blocks of wheat and other plants, while the red lines highlight the M24 genes pairs.
2.5. Motif Analysis and Gene Structure Analysis
Using the full-length amino acid sequence of the predicted TaM24 proteins, we inferred a phylogenetic tree, and visualized their conserved domain, motif, and gene structure (Figure 5). A conserved domain analysis showed that TaM24 contained the common domain PF00557. We identified ten motifs, and motif length ranged between 15 and 50 amino acids (Table 2). The type and number of motifs varied in different protein sequences. Most proteins with a similar motif structure clustered on the same branch of the phylogenetic tree. For example, motif positions, number and types are mostly the same in distribution in M24I. Motif 2 and motif 5 are relatively conserved and are present in 53 members, indicating that these two motifs are important in the structure and function of TaM24. Interestingly, some motifs appear only in specific subfamilies, such as motif 9 and motif 1. It is speculated that these motifs may be related to specific protein functions. We also analyzed the structure of TaM24 genes and identified clear differences in the number of exons, which ranged from 1 to 19. The gene structure of the III sub-group is relatively conserved, the number of introns and exons is relatively large, and the gene structure is relatively complex.

Figure 5.
Phylogenetic analysis of M24 proteins in wheat (a), conserved motifs (b), conserved domain (c), and gene structure (d). (a) A phylogenetic tree was constructed based on the full-length sequences of TaM24 proteins in PhyML. (b) The ten hypothetical motifs are represented by rectangles of different colors. (c) The green rectangle indicates the peptidase_M24 domain (PF00557). (d) The orange rectangle represents CDS, the green rectangle represents UTR, and the introns are represented by black lines.

Table 2.
List of the identified motifs in TaM24 proteins.
2.6. Gene Ontology (GO) Analysis and Protein Interaction Network (PPI) Analysis
For the sake of better understanding the function of TaM24 genes, we submitted protein sequences to the AgriGO database. Specific details are in Table S7, and it shows that two genes were not annotated, TaM24-5 and TaM24-50, both of which belong to M24III. Since there is more GO term, we only show the most popular content in Figure S3. For cellular components, TaM24 proteins are important components of the cytoplasm, plant organelles, the nucleus, and chromosomes. In terms of molecular function, the main function of TaM24 proteins is in the activity of metalloaminopeptidase and catalyzing the hydrolysis of N-terminal amino acid residues of polypeptide chains. For the biological process, TaM24 protein members are mainly involved in modifying proteins by breaking peptide bonds. Based on the existing annotation information, we speculate that members of this family may participate in plant stress resistance by promoting the synthesis, translation, processing, and modification of target proteins.
In addition, to better understand the protein-protein interaction network involved in TaM24 genes, we compared the identified TaM24 orthologous proteins in Arabidopsis to predict relationships among proteins (Figure 6). We found that the members of the peptidase M24 superfamily are involved in growth and development (such as secondary cell wall synthesis-related genes NMD3, RSW10 [39,40]), organ development-related regulatory genes (such as APR4, MCC1 [41,42]), photosynthesis and respiration-related genes (such as AGT, TIM, etc. [43,44]), and stress response processes under stress conditions (such as ALDH7B4, AMP1, PAO5, etc. [45,46,47]). The protein interaction network analysis of TaM24 indicated that the TaM24 gene family was directly or indirectly involved in various biological activities of plants.
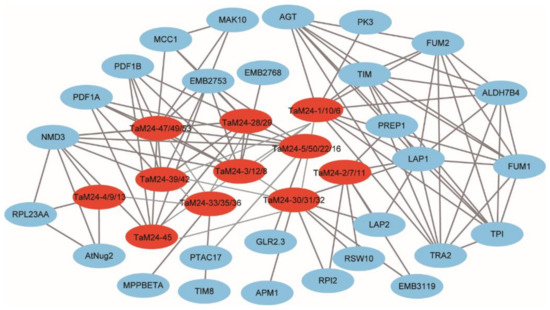
Figure 6.
Genes with other wheat proteins using Cytoscape. The red ellipse represents the TaM24 genes and the blue ellipse represents other genes. The gray line represents the interaction between the proteins.
2.7. Cis-Acting Element Analysis of Promoter Region
The promoter nucleic acid sequences of all TaM24 genes were submitted to the Plant CARE database, and many cis-acting elements were found (Figure 7, Table S8). In total, 92 types of cis-acting elements were identified in TaM24 genes, and each gene contained several TATA-boxes and CAAT-boxes, indicating that they could be transcribed normally. Further analysis showed that the environmental response elements contained in TaM24 genes accounted for about 23% (1206/5197), followed by hormone response elements accounting for about 13% (757/5197), and light response and plant growth related elements accounting for 11% and 6%, respectively. The elements related to stress response, drought, and osmotic stress were predominant among environmental response elements. For hormone response, most cis-acting elements were MeJA- and ABA-response elements. These results indicate that TaM24 genes are likely to be involved in the abiotic stress response of plants. In addition, root- and meristem-specific response elements accounted for most plant growth elements. We also visualized several common and important cis-acting elements. We found that almost all TaM24 genes have environmental stress-related elements (MYB, MYC, STRE) and hormone-responsive elements (ABRE, CGTCA, TGACG-motif), and it is speculated that TaM24 gene members may participate in abiotic stress resistance by responding to these cis-acting elements in plants. However, the cis-acting elements related to plant growth and development only appear in the promoters of some genes, especially the M24III subfamily.
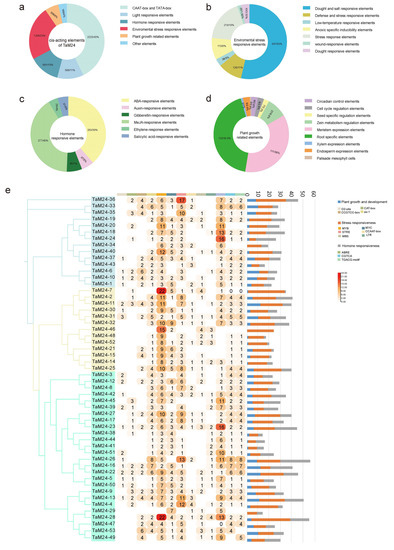
Figure 7.
The analysis results of TaM24 genes' promoter sequence (2000 bp upstream of genes). (a) The percentage distribution of cis-regulator elements in the promoters of TaM24 genes. (b) The percentage distribution of environmental stress related cis-elements. (c) The percentage distribution of hormone responsive cis-elements. (d) The percentage distribution of plant growth related cis-elements. (e) Statistics of common abiotic stress elements in TM24 genes. The ordinate represents different genes and the abscissa represents different cis-acting element sequences.
2.8. Analysis of Expression Patterns of TaM24 Genes
To further explore the expression of TaM24 genes in different tissues and developmental stages, we downloaded data from the wheat expression database (Table S9) and made an expression heatmap (Figure 8). About 69% (37/53) of genes were expressed in at least one stage of development, with an expression range of 1-8 Log2tpm (Log2tpmmax). Unexpressed genes may have undergone functional differentiation and redundant expression. From these data, we determined that the expression of TaM24 genes has tissue and space-time specificity. The expression levels of some genes are concentrated in the leaf/stem/shoot and spike development stages of wheat, for example, TaM24-2, 7, 11, and a large part of the M24III subfamily. Genes that are highly expressed in specific tissues during a specific period are critical for the formation of specific organs in wheat. Interestingly, TaM24-39 of the M24I subfamily; TaM24-25, 24, and 21 of the M24II subfamily; and TaM24-3, TaM24-8, 9, 13, 4, and 47 of the M24III subfamily have relatively higher expression over all development stages, indicating that they are vital for the growth and development of wheat. In addition, those genes that do not span the entire developmental stage of this tissue are presumed to be critical for the morphological development of specific organs in wheat.
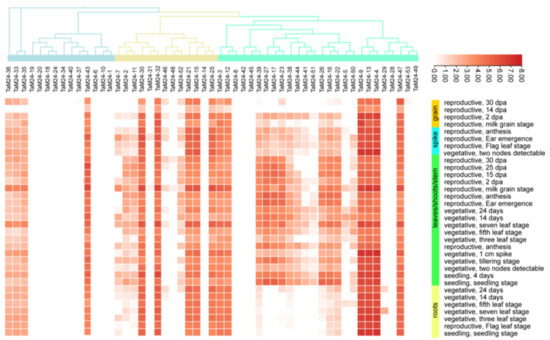
Figure 8.
The expression of TaM24 genes during wheat developmental stages of different tissues (based on log2tpm). TaM24 gene expression levels were downloaded from the wheat expression database. The heatmap shows the TaM24 genes expression levels: abscissa represents different genes and ordinate represents different growth periods of different tissues. Different colors represent different expression values: red: higher expression; white: lower expression.
Previously, we found in the analysis of cis-acting elements and the protein-protein interaction network that TaM24 genes may be involved in abiotic stress responses in wheat. Therefore, in order to explore whether the expression of TaM24 genes will be affected by adversity stress, we also downloaded the relevant data of TaM24 genes from the wheat expression database (Figure S4, Table S9). We found that some genes showed changes in gene expression levels after being subjected to drought, heat and cold stress. Therefore, we speculate that these genes may be involved in abiotic stress in plants. In order to explore the expression pattern of TaM24 genes under environmental stress and hormonal stress, we randomly selected four genes (TaM24-2, TaM24-9, TaM24-12, TaM24-50) with a variety of abiotic stress cis-acting elements and high gene expression as the research objects.
We used RT-qPCR to determine the expression patterns of these four genes in drought, NaCl, ABA, and MeJA responses (Figure 9). We found that the expression of all four genes changed under the stress treatments, but the degree and regularity of the changes varied. Under drought stress, the expression levels of TaM24-2, TaM24-9, TaM24-12, and TaM24-50 were up-regulated, and reached the peak at 12 h, 9 h, 24 h, and 12 h respectively (Figure 9a). In ABA treatment, the expression levels of all four genes began to decline after reaching the peak, and then returned to the initial state (Figure 9b). Under NaCl stress, the expression levels of TaM24-9 and TaM24-12 were up-regulated more than five-fold, reaching the highest values at 12 and 6 h, respectively (Figure 9c). Following MeJA treatment, the transcript levels of TaM24-2 and TaM24-9 were up-regulated more than four-fold, reaching the highest values at 3 and 1 h, respectively (Figure 9d). Based on the cis-acting element analysis of TaM24 genes members, gene expression analysis, and results of previous studies, we chose TaM24-9 for the next experiment [15,16,17].

Figure 9.
The real-time quantitative PCR analyses of four TaM24 genes under drought (a), ABA (b), NaCl (c), and MeJA (d) treatments. The mean and SD were calculated from three biological replicates. An ANOVA test demonstrated that there were significant differences (* p < 0.05, ** p < 0.01).
2.9. Overexpression of TaM24-9 Affects Seed Germination under Drought or NaCl Stress in Arabidopsis
To detect the effect of TaM24-9 genes on the seed germination rate under drought or salt stress, we chose TaM24-9 overexpressing Arabidopsis lines (OE-1, OE-2, OE-3). The OE lines, wild-type Arabidopsis (WT), and corresponding mutant Arabidopsis (ebp1/g2) were evenly seeded on a Murashige and Skoog (MS) medium with no additives or containing 9% polyethylene glycol 6000 (9% PEG6000), 12% PEG6000, 100 mM NaCl, and 150 mM NaCl. We found that WT, overexpression Arabidopsis lines, and mutant Arabidopsis (ebp1/g2) on MS medium were not significantly different (Figure 10 and Table S10). This suggested that TaM24-9 may not affect plant growth under normal growth conditions. On MS supplemented with PEG6000, the seed germination rates of all strains were inhibited. From the germination stage, the germination rates of seeds from each overexpression line were significantly higher than the WT (65.7%) and mutant Arabidopsis (ebp1/g2) (56%) within 60 h. On MS medium containing NaCl, the germination rates of seeds of WT, mutant Arabidopsis (ebp1/g2), and overexpression Arabidopsis were significantly lower than on MS, and the difference was more obvious with increased salt content. Compared with the TaM24-9 overexpression strain, the germination speed and rate of WT and mutant Arabidopsis (ebp1/g2) seeds were slower (Figure 11 and Table S10).
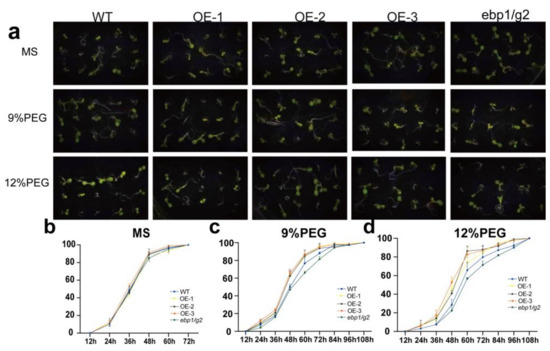
Figure 10.
Germination assay of wild-type (WT), TaM24-9 overexpression of Arabidopsis, and mutant Arabidopsis (ebp1/g2) seeds under PEG6000 treatment. (a) The phenotypes of WT, TaM24-9 overexpression Arabidopsis, and mutant Arabidopsis (ebp1/g2) seeds under 9% and 12% PEG6000 treatments. (b) The germination rates of WT, TaM24-9 overexpression Arabidopsis, and mutant Arabidopsis (ebp1/g2) seeds at different time points on MS medium. (c) The germination rates under 9% PEG6000 treatment. (d) The germination rates under 12% PEG6000 treatment.
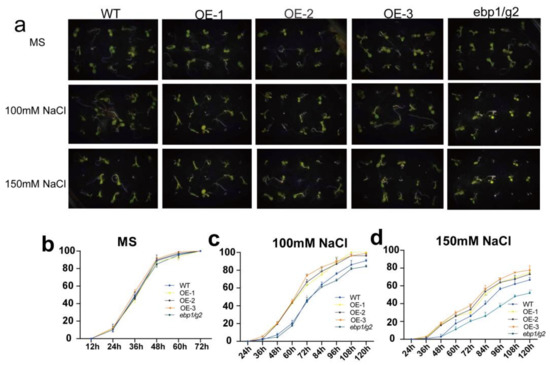
Figure 11.
Germination assay of wild-type (WT), TaM24-9 overexpression Arabidopsis, and mutant Arabidopsis (ebp1/g2) seeds under NaCl treatment. (a) The phenotypes of WT, TaM24-9 overexpression Arabidopsis, and mutant Arabidopsis (ebp1/g2) seeds under 100 mM and 150 mM NaCl treatments. (b) The germination rates of WT, TaM24-9 overexpression Arabidopsis, and mutant Arabidopsis (ebp1/g2) seeds at different time points on MS medium. (c) The germination rates under 100 mM NaCl treatment. (d) The germination rates under 150 mM NaCl treatment.
2.10. Overexpression of TaM24-9 Affects Growth State of Arabidopsis at the Seedling Stage under Drought and NaCl Stress
Through the result of the germination rate, we found that TaM24-9 can still maintain a higher germination rate than WT and mutant Arabidopsis (ebp1/g2) under drought and salt stress. To explore whether TaM24-9 is also involved in plant stress resistance at the seedling stage, we stressed Arabidopsis seedlings. WT, mutant Arabidopsis (ebp1/g2), and TaM24-9 overexpression Arabidopsis lines, which were cultured on MS medium, were first transferred to MS medium with 12% PEG6000 and MS medium with 100 mM NaCl. Then we evaluated the phenotypic changes of these strains after seven days. For WT cultured on MS medium, the root length of TaM24-9 overexpression Arabidopsis strains did not change significantly, and the root length of mutant Arabidopsis (ebp1/g2) was slightly shorter than WT and TaM24-9 overexpression Arabidopsis. (Figure 12 and Table S11). However, the growth of each strain was significantly limited on the MS medium supplemented with PEG6000 or NaCl, and the growth state of the overexpression strain was better than that of the WT and mutant Arabidopsis (ebp1/g2) (Figure 12 and Table S11).
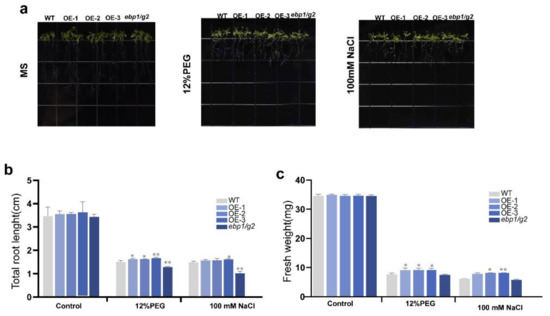
Figure 12.
The overexpression of TaM24-9 enhanced the tolerance to drought and salt stresses in Arabidopsis. (a) Morphological differences between wild-type (WT), TaM24-9 overexpression Arabidopsis, and mutant Arabidopsis (ebp1/g2) at seedling stages under PEG6000 treatment and NaCl. (b) Statistical analysis of root length and (c) fresh weight. * p < 0.05, ** p < 0.01.
After that, we conducted drought and salt resistance treatments in the seedling stage in the soil, followed by rehydration. We calculated respective survival rates and measured the corresponding proline (Pro) and malondialdehyde (MDA) contents. We observed phenotypic changes and found that all Arabidopsis strains were stressed after drought treatment. After rehydration treatment, the survival rate of TaM24-9 overexpression lines was higher than that of WT and corresponding mutant Arabidopsis (ebp1/g2) The amount of MDA in the strains overexpressing TaM24-9 was significantly lower than that in WT and mutant Arabidopsis (ebp1/g2), while on the contrary, the content of Pro was higher (Figure S5 and Table S11). We found that after NaCl treatment, the growth status of all strains was severely disrupted. Relatively speaking, after rehydration, TaM24-9 overexpression strains survived better (Figure S6 and Table S11). Similarly, the amount of Pro in overexpression strains was significantly higher than that of WT and the content of MDA was significantly lower than that of WT.
3. Discussion
Peptidase M24 proteins superfamily members have multiple biological functions in animals, plants, and microorganisms, and are an emerging gene family. In plants, the functions of most members of the peptidase M24 superfamily are not known in detail, but studies have shown that these genes are involved in a variety of biological processes, such as participating in the protein synthesis process [25,29,48,49,50,51], ensuring the normal development of plant organs [14,28,32,52,53], regulating the formation of special structures in legumes [54], abiotic response and biotic stress [15,16,34,54]. Therefore, this study aimed to identify and classify the members of the peptidase M24 in wheat, explore the role of TaM24 genes in drought and salt stress in Arabidopsis.
In this work, we identified 53 peptidase M24 members from the wheat genome and compared the number of peptidase M24 genes superfamily members identified in other species of monocotyledons and dicotyledons (Figure 1 and Table S2). For example, there were 16 putative peptidase M24 genes in rice and 12 in Arabidopsis. We found that apart from wheat, the number of peptidase M24 genes in other species is basically the same, whether it is a monocotyledon or dicotyledon, indicating that this gene superfamily is conserved in plant evolution. So, we speculate that the peptidase M24 superfamily did not undergo a gene deletion phenomenon. We counted the number of TaM24 on the D chromosome of wheat as 15, which was roughly the same as that AeM24. As is known to all that Ae. tauschii derived subgenome D chromosomes of wheat, therefore, the source of wheat peptidase M24 genes is likely related to the increase of chromosome number.
During chromosome mapping, we found that the distribution of peptidase M24 on wheat chromosomes is uneven, with most distributed in chromosomes 1 and 2. At the same time, we found that most of the collinearity regions also correspond to chromosomes 1 and 2. Gene duplication of peptidase M24 occurred due to fragment duplication or chromosome doubling. Additionally, the ratio of Ka/KS was less than 0.5, indicating that peptidase M24 was under positive purification selection during evolution, and suggesting that the gene is essential in the process of species evolution.
Analysis of GO and PPI helps with the understanding of the ability of proteins in organisms. For cellular components, TaM24 genes are important components of the intracellular organelle (GO:0043229), organelle (GO:0043226), and cytoplasm (GO:0005737). In terms of molecular function, the main function of TaM24 genes is in activity of peptidase activity (GO:0008233), hydrolase activity (GO:0016787), and DNA/RNA binding (GO:0005488) (image not showing). Therefore, we speculate that the TaM24 genes are also involved in plant growth and stress response through enzymatic activity or nucleic acid binding protein. For biological processes, TaM24 members are mainly involved in modifying proteins by breaking peptide bonds, such as the protein metabolic process (GO:0019538) and the proteolysis process (GO:0006508). We speculate that the TaM24 regulates the physiological process of plants by directly or indirectly participating in the process of protein metabolism. Based on the existing annotation information, we speculate that members of TaM24 may participate in plant stress resistance by promoting the synthesis, translation, processing, and modification of target proteins. The TaM24 proteins were found to interact with cell wall synthesis-related genes (NMD3, RSW10 [39,40]) and stress response processes under stress conditions (ALDH7B4, AMP1, PAO5 [45,46,47]) during protein network prediction.
Our analysis of cis-elements in the promoter region of peptidase M24 superfamily genes in wheat showed that the most responsive hormone elements were ABA sensitive and MeJA sensitive cis-elements (ABRE, CGTCA, TGACG-motif, etc.). Among the environmental stress response elements, the most important ones in wheat are those related to drought and salt stress (MYB, MYC, STRE, etc.). Many signals of plants under environmental stress are closely related to hormone transmission [55,56]. These results suggest that the peptidase M24 superfamily of peptidases may be involved in responding to drought and salt stress by relying on the ABA or MeJA pathways. In order to verify our hypothesis, four genes were selected for qRT-PCR verification. The results verified that these four genes responded to drought, salt, MeJA and ABA. Although all four genes changed under environmental stress and hormone treatment, their expression patterns were not exactly the same. Due to the complex composition of the peptidase M24 proteins family, we speculate that their expression differences indicate their involvement in different anti-stress transmission pathways.
The most intuitive reflection of plant stress is phenotype changes, such as decreased germination rate, lodging, leaf curl, the emergence of plaque on leaves, or a decreased seed setting rate [7,13,57]. The main reason for these phenotypic changes is to affect the growth and development of plants by changing plant physiology and metabolism. In this study, we cloned TaM24-9 and overexpressed it in Arabidopsis. We subjected overexpression Arabidopsis, WT, and mutant Arabidopsis (ebp1/g2) to drought stress and salt stress. Under stress conditions, plants showed decreased yield, reduced height, increased MDA content, decreased enzyme activity, and even plant death. In this study, we observed that Arabidopsis transformed with TaM24-9 had stronger resistance to drought and salt, because it had better germination rates under drought and salt stress. GO and PPI analysis found that TaM24-9 can be combined with DNA, RNA, 60 s ribosomal subunit [58], cell wall formation related genes NMD3 [40] etc. Therefore, we speculate that the higher germination rate of transgenic plants may promote ribosome biogenesis. In previous studies, it was that the homologous protein of TaM24-9 in maize and potato also confirmed that it can improve the cell cycle activity of TaM24-9 overexpression plants [26,59]. Under drought stress, we found that the root system of the overexpressed lines was more complex, and the peptidase M24 genes superfamily has root-specific cis-acting elements. Plant can promote plant root activity under drought and salt stress [12,60,61]. The survival of transgenic Arabidopsis is higher when soil-cultured Arabidopsis is subjected to drought and salt stress treatments. In addition, the content of MDA is an important index to measure the stress degree of plants. The greater the stress, the higher the content of MDA [60,62]. On the contrary, for Pro, a soluble substance in cells, the higher its content, the stronger its protection of plants. Plants can increase the content of Pro in response to salt and drought stress [15,63]. The TaM24-9 overexpression plants had higher Pro content and lower MDA content. This result is consistent with studies of TaM24-9 homologous proteins found in Hevea brasiliensis [15], Arabidopsis [17], and Ammopiptanthus mongolicus [16]. In summary, we believe that the TaM24-9 gene is crucial in plant growth, development, and stress response.
4. Materials and Methods
4.1. Identification of Peptidase M24 Gene Family Members in Wheat
We downloaded the genome sequence and genome annotation information of wheat from the Ensembl plants database (http://plants.ensembl.org/index.html; accessed on 29 August 2021) [64]. A hidden Markov model (HMM) of the typical domain of peptidase M24 was obtained from the Pfam database (https://pfam.xfam.org/; accessed on 29 August 2021) [65]. The Pfam ID used to identify the member of the peptidase M24 genes in wheat is PF00557. The protein sequences of other species were obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/pz/portal.html; accessed on 29 August 2021) [66]. The protein sequences of wheat were constructed into a local database, and the protein sequences containing peptidase M24 domain in wheat were screened by HMMR 3.0 software on the HMM model (E value < E−10) [67]. We submitted these sequences to the NCBI protein Batch CD-search database (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi; accessed on 29 August 2021) and the SMART database (https://smart.embl-heidelberg.de/; accessed on 29 August 2021) [68,69], and next removed sequences without the significant conserved domain of peptidase M24. ExPASY (https://www.expasy.org/; accessed on 30 August 2021) [70] was used to predict the physicochemical parameters of the peptidase M24 proteins, including molecular weight (Mw) and theoretical pI. We submitted the protein sequences to WOLF PSORT (https://wolfpsort.hgc.jp/; accessed on 30 August 2021) [71] for protein subcellular localization prediction.
4.2. Phylogenetic Analysis and Classification of TaM24 Proteins
Multiple sequence alignments of M24 protein sequences were generated by Clustal Omega [72], and the approximate maximum likelihood phylogenetic tree was constructed by PhyML3.0 [73] with the LG (Le and Gascuel) model [74]. The Shimodaira-Hasegawa test [75] (1000 resamples) was used to compute local support values. The trees were visualized using Evolview (https://www.evolgenius.info/evolview/#login; accessed on 2 September 2021) [76].
4.3. Chromosomal Location and Identification of Homoeologs
The location information of TaM24 genes were obtained from a wheat genome annotation file, and the distribution map of TaM24 genes on chromosomes was drawn with Tbtools [77]. Homoeologous genes were identified using the Ensembl Plants database [64].
4.4. Gene Duplication and Evolutionary Selection Analysis
Gene duplication events were predicted according to a previously described process [36,63] according to the following standards based on aligned genes: (1) covers more than 80% of comparable nucleotide sequences; and (2) the identity of the aligned regions is > 80%. Tbtools software was then used to visualize and calculate the rates of synonymous (KS), (non-synonymous) Ka, and Ka/KS values of collinear block gene pairs [77].
4.5. Gene Structure and Conserved Motif Analysis
Conserved motifs in TaM24 proteins sequences were predicted by MEME (http://meme-suit/org/; accessed on 2 September 2021) [78]. According to the annotation file of the wheat genome, gene structure was analyzed and visualized by Tbtools [77].
4.6. Cis-Acting Element Analysis
According to the wheat genome annotation file, we used Tbtools [77] to extract the 2,000 bp upstream of the coding sequence (CDS) from the wheat genome file. In order to predict the cis-acting element in the promoter region, we submitted these sequences to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed on 6 September 2021) [79].
4.7. Gene Ontology (GO) Analysis and Protein Interaction Network (PPI) Analysis
The amino acid sequences of wheat peptidase M24 were submitted to the AGriGO database (http://systemsbiology.cau.edu.cn/agriGOv2/; accessed on 9 September 2021) [80], and the results were analyzed and visualized. The TaM24 proteins sequences were submitted to the Triticeae-GeneTribe website (http://wheat.cau.edu.cn/TGT/; accessed on 9 September 2021) [81] to obtain the orthologous pair between wheat and Arabidopsis, and then it was submitted to the String database (http://string-db.org/cgi; accessed on 9 September 2021) [82]. Finally, Protein-Protein interaction networks are visualized through Cytoscape software v3.7.1 [83].
4.8. Expression Analysis of TaM24 Genes
We downloaded the expression data of different tissues (root, leaf/shoots/steam, spikes, and grains) in different growth periods and abiotic stresses of wheat from the Wheat expression browser database (http://www.wheat-expression.com; accessed on 12 September 2021) [84]. We then used Tbtools to draw the expression profile heat map [77].
4.9. RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from wheat leaves sprouting for seven days using TRIGene (GenStar, Beijing, China). The FastKing RT Kit (With gDNase) (TIANGEN, Beijing, China) was used for cDNA synthesis. QuantStudio 3 Flex Real-Time PCR system (ThermoFisher, Foster City, CA, USA) and PerfectStart® Green qPCR SuperMix (TransGen, Beijing, China) were used for quantitative real-time PCR (qRT-PCR). The wheat β-actin gene (GenBank accession number AB181991.1) was used as an internal reference for all qRT-PCR analyses. Three repetitions were set for each sample. The relative expression levels of each gene were calculated based on the 2−ΔΔCt value. The primers used in this experiment are shown in Table S12.
4.10. Plant Materials, Growth Conditions, and Stress Treatments
Chinese Spring seeds cultured for about one week were treated for gene expression analysis. The culture conditions were 22 °C/20 °C (light for 16 h/dark for 8 h). Seven-day-old seedlings were treated with drought stress (no water supply), NaCl (100 mM), ABA (100 μm), and MeJA (100 μm) and samples were taken after treatment at 0 (CK), 0.5, 1, 3, 6, 9, 12, and 24 h. For all samples, three replicates were quickly placed in liquid nitrogen and stored at −80 °C.
In order to obtain overexpression strains, we connected the protein CDS of TaM24-9 to the pCAMBIA1302 vector. The correctly sequenced recombinant plasmid was transformed into Agrobacterium tumefaciens strain GV3101, and then transformed into the Arabidopsis Columbia type using the flower immersion method [85]. Mutant and overexpression information of Arabidopsis thaliana are shown in Figure S7. The TaM24-9 overexpression strains were cultured to the T3 generation. The growth conditions of Arabidopsis were the same as for wheat. We cultured the Arabidopsis seeds on MS medium containing PEG6000 (9% and 12%), NaCl (100 mM and 150 mM), and MS with no additives. We recorded the germination rate every 12 h.
The same growth state overexpression Arabidopsis seedlings were cultured vertically to the MS medium containing 12% PEG6000 or 100 mM NaCl, and the root length was recorded after seven days of growth. To test drought tolerance in the soil, we transferred Arabidopsis seedlings with the same growth state on MS to the soil. After three weeks of culture, the seedlings were subjected to drought stress (no water supply) for two weeks, and then rehydrated for three days. The survival rate and the contents of MDA and Pro were recorded. In order to investigate salt tolerance, after three weeks of culture, plants were irrigated with 100 mM NaCl for 3 days, and the survival rate and the contents of MDA and Pro were recorded. We recorded the survival and performed three independent biological replications.
4.11. Measurement of Malondialdehyde and Proline Contents
The levels of Pro and MDA in Arabidopsis leaves were determined by using the corresponding detection kits (Cominbio, Suzhou, China). Each sample measurement was repeated three times.
4.12. Statistical Analysis
The above experiments were repeated three times. We used GraphPad Prism 8.0 software and spss 22.0 software for statistical analysis. The data was calculated as mean ± standard deviation (SD) and performed ANOVA. The significance levels were defined as * (p < 0.05); and ** (p < 0.01).
5. Conclusions
A total of 53 peptidase M24 genes were identified in the wheat genome, and these genes were analyzed comprehensively and systematically by chromosome mapping, gene function and protein interaction network prediction, and cis acting element analysis. Our results show that TaM24-9, a member of the M24III subfamily, helped Arabidopsis plants resist drought and salt stress. This study not only presents reference information for the evolutionary mechanism of peptidase M24 in wheat, but also provides an important stress resistance gene for the genetic improvement of wheat.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23136904/s1.
Author Contributions
D.-H.M. and X.-H.Z. conceived and designed the experiments. L.-Y.Y. performed the experiments and wrote the paper. J.-G.G. and Y.-X.H. contributed to bioinformatics analysis. L.-L.Z. collected the previous studies. Y.L. and X.-X.X. designed primers and performed the qRT-PCR. X.Z. measured the phenotypic data of the plant. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by Key research and development program of Shaanxi Province (2022NY-186) and the Programme of Introducing Talents of Innovative Discipline to Universities (Project 111) from the State Administration of Foreign Experts Affairs (#B18042) “Crop breeding for disease resistance and genetic improvement”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Shiferaw, B.; Smale, M.; Braun, H.-J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013, 5, 291–317. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.K.; Balyan, H.S.; Sharma, S.; Kumar, R. Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 1569–1602. [Google Scholar] [CrossRef] [PubMed]
- Tyczewska, A.; Woźniak, E.; Gracz, J.; Kuczyński, J.; Twardowski, T. Towards Food Security: Current State and Future Prospects of Agrobiotechnology. Trends Biotechnol. 2018, 36, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyev, T.I.; Talai, J.M.; Huseynova, I.M.; Aliyev, J.A. Effect of drought stress on some physiological parameters, yield, yield components of durum (Triticum durum desf.) and bread (Triticum aestivum L.) wheat genotypes. Ekin J. Crop Breed. Genet. 2015, 1, 50–62. [Google Scholar]
- Noman, A.; Ali, Q.; Naseem, J.; Javed, M.T.; Kanwal, H.; Islam, W.; Aqeel, M.; Khalid, N.; Zafar, S.; Tayyeb, M.; et al. Sugar beet extract acts as a natural bio-stimulant for physio-biochemical attributes in water stressed wheat (Triticum aestivum L.). Acta Physiol. Plant. 2018, 40, 110. [Google Scholar] [CrossRef]
- Golfam, R.; Kiarostami, K.; Lohrasebi, T.; Hasrak, S.; Razavi, K. A review of drought stress on wheat (Triticum aestivum L.) starch. Farming Manag. 2021, 6, 47–57. [Google Scholar]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought Stress Tolerance in Wheat and Barley: Advances in Physiology, Breeding and Genetics Research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daryanto, S.; Wang, L.; Jacinthe, P.-A. Global Synthesis of Drought Effects on Maize and Wheat Production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef]
- Zhang, X.; Obringer, R.; Wei, C.; Chen, N.; Niyogi, D. Droughts in India from 1981 to 2013 and Implications to Wheat Production. Sci. Rep. 2017, 7, srep44552. [Google Scholar] [CrossRef] [Green Version]
- Flowers, T.J.; Flowers, S.A. Why does salinity pose such a difficult problem for plant breeders? Agric. Water Manag. 2005, 78, 15–24. [Google Scholar] [CrossRef]
- Wang, M.; Xia, G. The landscape of molecular mechanisms for salt tolerance in wheat. Crop J. 2018, 6, 42–47. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, J.-Y.; Wang, Y.-J.; Jiao, J.-L.; Wang, H.-Z. Silencing of ATG2 and ATG7 promotes programmed cell death in wheat via inhibition of autophagy under salt stress. Ecotoxicol. Environ. Saf. 2021, 225, 112761. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Han, S.; Zhong, S.; Wei, H.; Zhang, Y.; Zhao, Y.; Liu, B. Characterization and Fine Mapping of a Necrotic Leaf Mutant in Maize (Zea mays L.). J. Genet. Genom. 2013, 40, 307–314. [Google Scholar] [CrossRef]
- Cheng, H.; Chen, X.; Zhu, J.; Huang, H. Overexpression of a Hevea brasiliensis ErbB-3 Binding protein 1 Gene Increases Drought Tolerance and Organ Size in Arabidopsis. Front. Plant Sci. 2016, 7, 1703. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yu, G.; Sun, X.; Zhang, X.; Liu, J.; Pan, H. AcEBP1, an ErbB3-Binding Protein (EBP1) from halophyte Atriplex canescens, negatively regulates cell growth and stress responses in Arabidopsis. Plant Sci. 2016, 248, 64–74. [Google Scholar] [CrossRef]
- Li, C.; Liu, X.; Qiang, X.; Li, X.; Li, X.; Zhu, S.; Wang, L.; Wang, Y.; Liao, H.; Luan, S.; et al. EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol. 2018, 16, e2006340. [Google Scholar] [CrossRef] [Green Version]
- Bazan, J.F.; Weaver, L.H.; Roderick, S.L.; Huber, R.; Matthews, B.W. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc. Natl. Acad. Sci. USA 1994, 91, 2473–2477. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.Y.; Wang, X.W.; Rishi, A.K.; Lessor, T.; Xia, X.M.; A Gustafson, T.; Hamburger, A.W. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer 2000, 82, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Biswas, D.; Yu, Y.; Prall, M.; Formosa, T.; Stillman, D.J. The Yeast FACT Complex Has a Role in Transcriptional Initiation. Mol. Cell. Biol. 2005, 25, 5812–5822. [Google Scholar] [CrossRef] [Green Version]
- Stuwe, T.; Hothorn, M.; Lejeune, E.; Rybin, V.; Bortfeld, M.; Scheffzek, K.; Ladurner, A.G. The FACT Spt16 “peptidase” domain is a histone H3–H4 binding module. In Proceedings of the National Academy of Sciences 2008, Cambridge, UK, 22 January 2018; Volume 105, pp. 8884–8889. [Google Scholar]
- Hauser, F.; Strassner, J.; Schaller, A. Cloning, Expression, and Characterization of Tomato (Lycopersicon esculentum) Aminopeptidase P. J. Biol. Chem. 2001, 276, 31732–31737. [Google Scholar] [CrossRef] [Green Version]
- Murphy, A.S.; Hoogner, K.R.; Peer, W.A.; Taiz, L. Identification, Purification, and Molecular Cloning of N-1-Naphthylphthalmic Acid-Binding Plasma Membrane-Associated Aminopeptidases from Arabidopsis. Plant Physiol. 2002, 128, 935–950. [Google Scholar] [CrossRef] [Green Version]
- Duroux, M.; Houben, A.; Růžička, K.; Friml, J.; Grasser, K.D. The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 2004, 40, 660–671. [Google Scholar] [CrossRef]
- Zhang, W.-K.; Shen, Y.-G.; He, X.-J.; Du, B.-X.; Xie, Z.-M.; Luo, G.-Z.; Zhang, J.-S.; Chen, S.-Y. Characterization of a novel cell cycle-related gene from Arabidopsis. J. Exp. Bot. 2005, 56, 807–816. [Google Scholar] [CrossRef]
- Horváth, B.M.; Magyar, Z.; Zhang, Y.; Hamburger, A.W.; Bakó, L.; Visser, R.G.F.; Bachem, C.; Bögre, L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006, 25, 4909–4920. [Google Scholar] [CrossRef]
- Millar, A.H.; Nelson, C.J.; Li, L.; Taylor, N.L.; Stroher, E.; Petereit, J.J.P.P. The intermediate cleavage peptidase ICP55 modifies enzyme N-termini and alters protein stability in Arabidopsis mitochondria. Plant Physiol. 2015, 168, 415–427. [Google Scholar] [CrossRef] [Green Version]
- Lolas, I.B.; Himanen, K.; Gronlund, J.T.; Lynggaard, C.; Houben, A.; Melzer, M.; Van Lijsebettens, M.; Grasser, K.D. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010, 61, 686–697. [Google Scholar] [CrossRef]
- Ross, S.; Giglione, C.; Pierre, M.; Espagne, C.; Meinnel, T. Functional and Developmental Impact of Cytosolic Protein N-Terminal Methionine Excision in Arabidopsis. Plant Physiol. 2005, 137, 623–637. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.-J.; Shin, J.S.; Ok, S.H. Barley DNA-binding methionine aminopeptidase, which changes the localization from the nucleus to the cytoplasm by low temperature, is involved in freezing tolerance. Plant Sci. 2011, 180, 53–60. [Google Scholar] [CrossRef]
- Cao, P.; Song, J.; Zhou, C.; Weng, M.; Liu, J.; Wang, F.; Zhao, F.; Feng, D.; Wang, B. Characterization of multiple cold induced genes from Ammopiptanthus mongolicus and functional analyses of gene AmEBP1. Plant Mol. Biol. 2008, 69, 529–539. [Google Scholar] [CrossRef]
- Makam, S.N. The Arabidopsis Aminopeptidase-P (AtAPP1) is a Regulator of Auxin Signaling; Purdue University: West Lafayette, IN, USA, 2007. [Google Scholar]
- Sanjai, D. Arabidopsis Thaliana Aminopeptidase P1 (APP1) Functions as a Rate-Limiting Component of Auxin Signaling Transduction; Purdue University: West Lafayette, IN, USA, 2009. [Google Scholar]
- Balakireva, A.V.; Deviatkin, A.A.; Zgoda, V.G.; Kartashov, M.I.; Zhemchuzhina, N.S.; Dzhavakhiya, V.G.; Golovin, A.V.; Zamyatnin, J.A.A. Proteomics Analysis Reveals That Caspase-like and Metacaspase-like Activities Are Dispensable for Activation of Proteases Involved in Early Response to Biotic Stress in Triticum aestivum L. Int. J. Mol. Sci. 2018, 19, 3991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-wide identification of PYL gene family in wheat: Evolution, expression and 3D structure analysis. Genomics 2020, 113, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.-L.; Yan, J.; Fan, Y.; Li, Y.; Ruan, J.-J.; Wang, J.-Z.; Cheng, X.-B.; Cheng, J.-P. Genome-wide identification and phylogenetic relationships of the Hsp70 gene family of Aegilops tauschii, wild emmer wheat (Triticum dicoccoides) and bread wheat (Triticum aestivum). 3 Biotech 2021, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Bateman, A. How to use the MEROPS database and website to help understand peptidase specificity. Protein Sci. 2020, 30, 83–92. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2011, 40, D343–D350. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Elliott, J.E.; Williamson, R.E. Features of the primary wall CESA complex in wild type and cellulose-deficient mutants of Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 2627–2637. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.Q.; Zhang, A.H.; Zhang, Q.; Zhang, B.C.; Nan, J.; Li, X.; Liu, N.; Qu, H.; Lu, C.M.; Zhou, Y.H.; et al. Arabidopsis NMD3 Is Required for Nuclear Export of 60S Ribosomal Subunits and Affects Secondary Cell Wall Thickening. PLoS ONE 2012, 7, e35904. [Google Scholar] [CrossRef]
- Perrella, G.; Consiglio, M.F.; Aiese-Cigliano, R.; Cremona, G.; Sanchez-Moran, E.; Barra, L.; Errico, A.; Bressan, R.A.; Franklin, F.C.H.; Conicella, C. Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J. 2010, 62, 796–806. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 2005, 41, 845–858. [Google Scholar] [CrossRef]
- Liepman, A.H.; Vijayalakshmi, J.; Peisach, D.; Hulsebus, B.; Olsen, L.J.; Saper, M.A. Crystal Structure of Photorespiratory Alanine: Glyoxylate Aminotransferase 1 (AGT1) from Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1229. [Google Scholar] [CrossRef]
- Chen, M.; Thelen, J.J. The Plastid Isoform of Triose Phosphate Isomerase Is Required for the Postgerminative Transition from Heterotrophic to Autotrophic Growth in Arabidopsis. Plant Cell 2010, 22, 77–90. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Ye, T.; Wang, Y.; Chan, Z. Arabidopsis ALTERED MERISTEM PROGRAM 1 negatively modulates plant responses to abscisic acid and dehydration stress. Plant Physiol. Biochem. 2013, 67, 209–216. [Google Scholar] [CrossRef]
- Missihoun, T.; Hou, Q.; Mertens, D.; Bartels, D. Sequence and functional analyses of the aldehyde dehydrogenase 7B4 gene promoter in Arabidopsis thaliana and selected Brassicaceae: Regulation patterns in response to wounding and osmotic stress. Planta 2014, 239, 1281–1298. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Zhang, S.; Kojima, S.; Simm, S.; Berberich, T.; Kusano, T. Reducing Cytoplasmic Polyamine Oxidase Activity in Arabidopsis Increases Salt and Drought Tolerance by Reducing Reactive Oxygen Species Production and Increasing Defense Gene Expression. Front. Plant Sci. 2016, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Giglione, C.; Vallon, O.; Meinnel, T. Control of protein life-span by N-terminal methionine excision. EMBO J. 2003, 22, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Giglione, C.; Boularot, A.; Meinnel, T. Protein N-terminal methionine excision. Cell Mol. Life Sci. 2004, 61, 1455–1474. [Google Scholar] [CrossRef]
- Vögtle, F.-N.; Wortelkamp, S.; Zahedi, R.; Becker, D.; Leidhold, C.; Gevaert, K.; Kellermann, J.; Voos, W.; Sickmann, A.; Pfanner, N.; et al. Global Analysis of the Mitochondrial N-Proteome Identifies a Processing Peptidase Critical for Protein Stability. Cell 2009, 139, 428–439. [Google Scholar] [CrossRef]
- Singh, R.; Goyal, V.D.; Kumar, A.; Sabharwal, N.S.; Makde, R.D. Crystal structures and biochemical analyses of intermediate cleavage peptidase: Role of dynamics in enzymatic function. FEBS Lett. 2018, 593, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Are, V.N.; Ghosh, B.; Kumar, A.; Gadre, R.; Makde, R.D. Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum. Int. J. Biol. Macromol. 2016, 88, 36–43. [Google Scholar] [CrossRef]
- Frost, J.M.; Kim, M.Y.; Park, G.T.; Hsieh, P.-H.; Nakamura, M.; Lin, S.J.H.; Yoo, H.; Choi, J.; Ikeda, Y.; Kinoshita, T.; et al. FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4720–E4729. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-C.; Lei, Y.-W.; Liu, W.; Weng, L.; Lei, M.-J.; Hu, X.-H.; Dong, Z.; Luo, D.; Yang, J. LjCOCH interplays with LjAPP1 to maintain the nodule development in Lotus japonicus. Plant Growth Regul. 2018, 85, 267–279. [Google Scholar] [CrossRef]
- Kim, J.S.; Jeon, B.W.; Kim, J. Signaling Peptides Regulating Abiotic Stress Responses in Plants. Front. Plant Sci. 2021, 12, 704490. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.-T.; Zhao, M.-J.; Wang, C.-T.; Gao, Y.; Wang, Y.-X.; Liu, Y.-W.; Chen, M.; Chen, J.; Zhou, Y.-B.; Xu, Z.-S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 1–18. [Google Scholar] [CrossRef]
- Im, C.H.; Hwang, S.M.; Son, Y.S.; Heo, J.B.; Bang, W.Y.; Suwastika, I.N.; Shiina, T.; Bahk, J.D. Nuclear/Nucleolar GTPase 2 Proteins as a Subfamily of YlqF/YawG GTPases Function in Pre-60S Ribosomal Subunit Maturation of Mono- and Dicotyledonous Plants. J. Biol. Chem. 2011, 286, 8620–8632. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Sui, Z.; Liu, X.; Li, Y.; Li, H.; Xing, J.; Song, F.; Zhang, Y.; Sun, Q.; Ni, Z. Ectopic expression of a maize hybrid up-regulated gene, ErbB- 3 binding Protein 1 ( ZmEBP1), increases organ size by promoting cell proliferation in Arabidopsis. Plant Sci. 2016, 243, 23–34. [Google Scholar] [CrossRef]
- Wang, R.K.; Cao, Z.H.; Hao, Y.J. Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant. 2014, 150, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Richardson, T.; Chai, S.; Lynne McIntyre, C.; Rae, A.L.; Xue, G.-P. Drought-up-regulated TaNAC69-1 is a transcriptional repressor of TaSHY2 and TaIAA7, and enhances root length and biomass in wheat. Plant Cell Physiol. 2016, 57, 2076–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.-H.; Min, D.-H. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genom. 2021, 22, 1–19. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomic Data. In Plant genomics databases; 2016; Humana Press: New York, NY, USA; pp. 1–31. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [Green Version]
- Shimodaira, H.; Hasegawa, M. Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Song, W.; Xie, X.; Wang, Z.; Guan, P.; Peng, H.; Jiao, Y.; Ni, Z.; Sun, Q.; Guo, W. A Collinearity-Incorporating Homology Inference Strategy for Connecting Emerging Assemblies in the Triticeae Tribe as a Pilot Practice in the Plant Pangenomic Era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).