Characterization of the Key Bibenzyl Synthase in Dendrobium sinense
Abstract
:1. Introduction
2. Results
2.1. Total Contents of Bibenzyls in Different Tissues of D. sinense
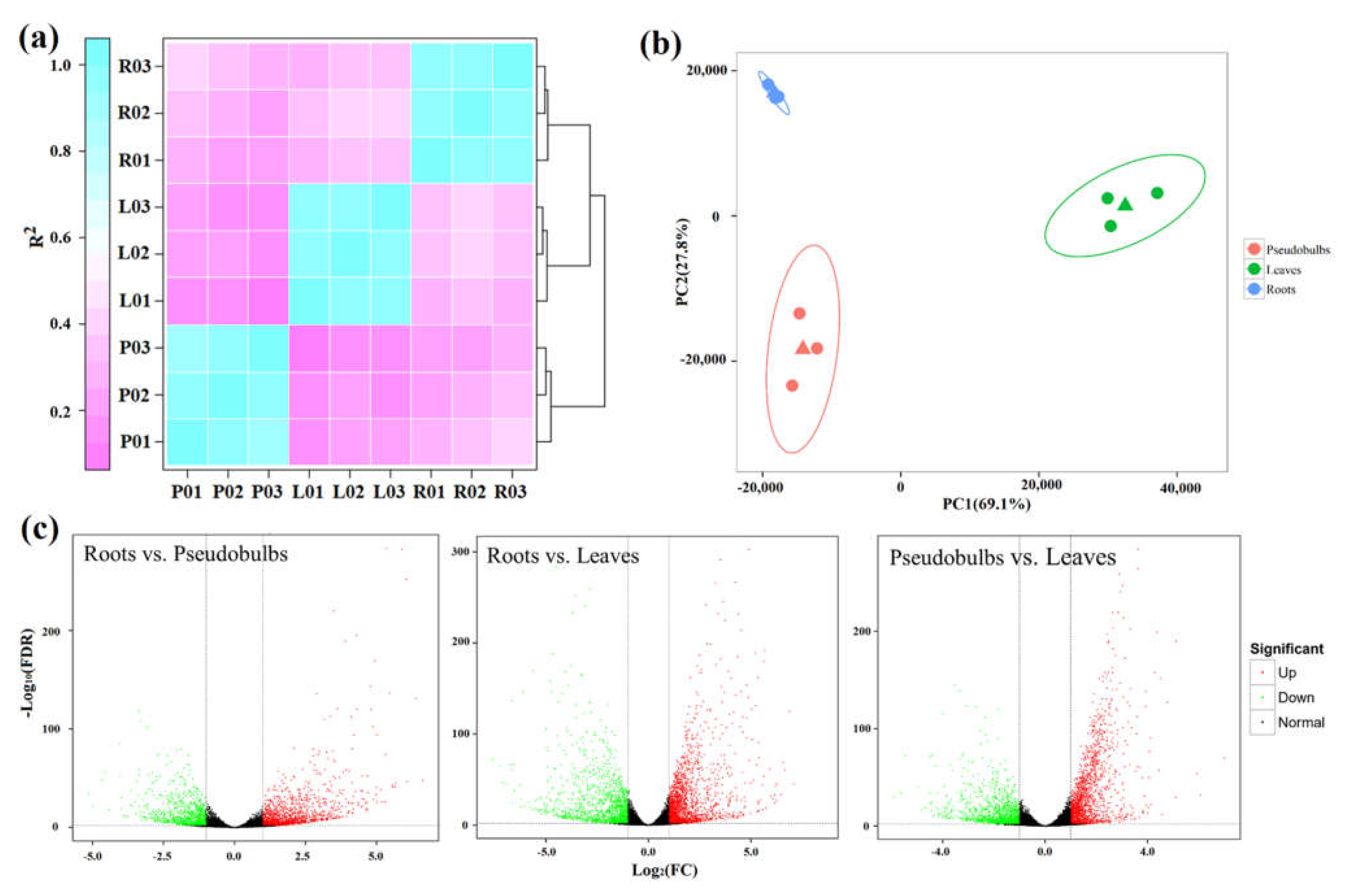
2.2. Transcriptome Sequencing and Analysis
2.3. High-Throughput Analysis of Gene Expression
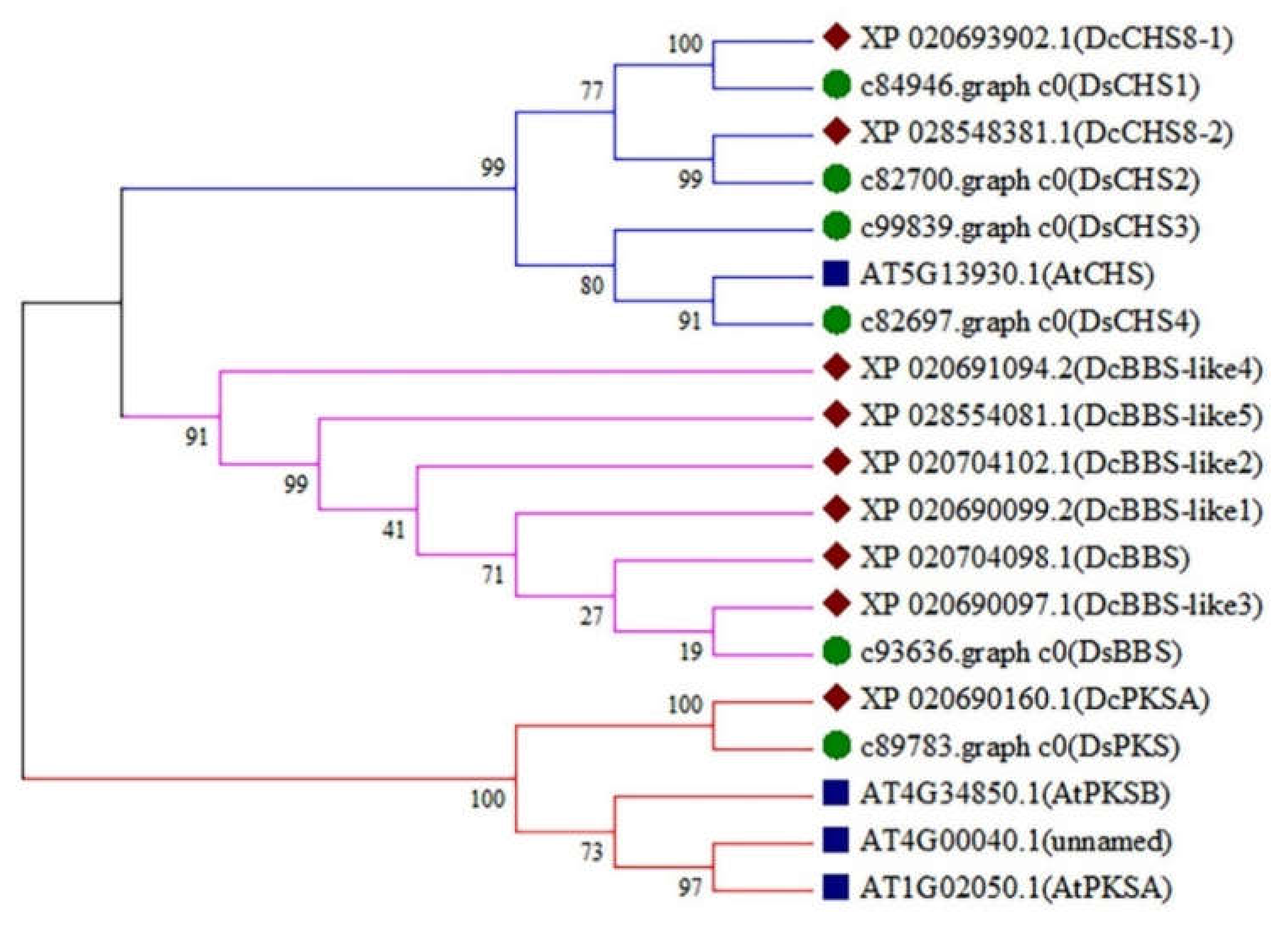
2.4. Identification of the Type III PKS Genes in D. sinense
2.5. Expression Analysis
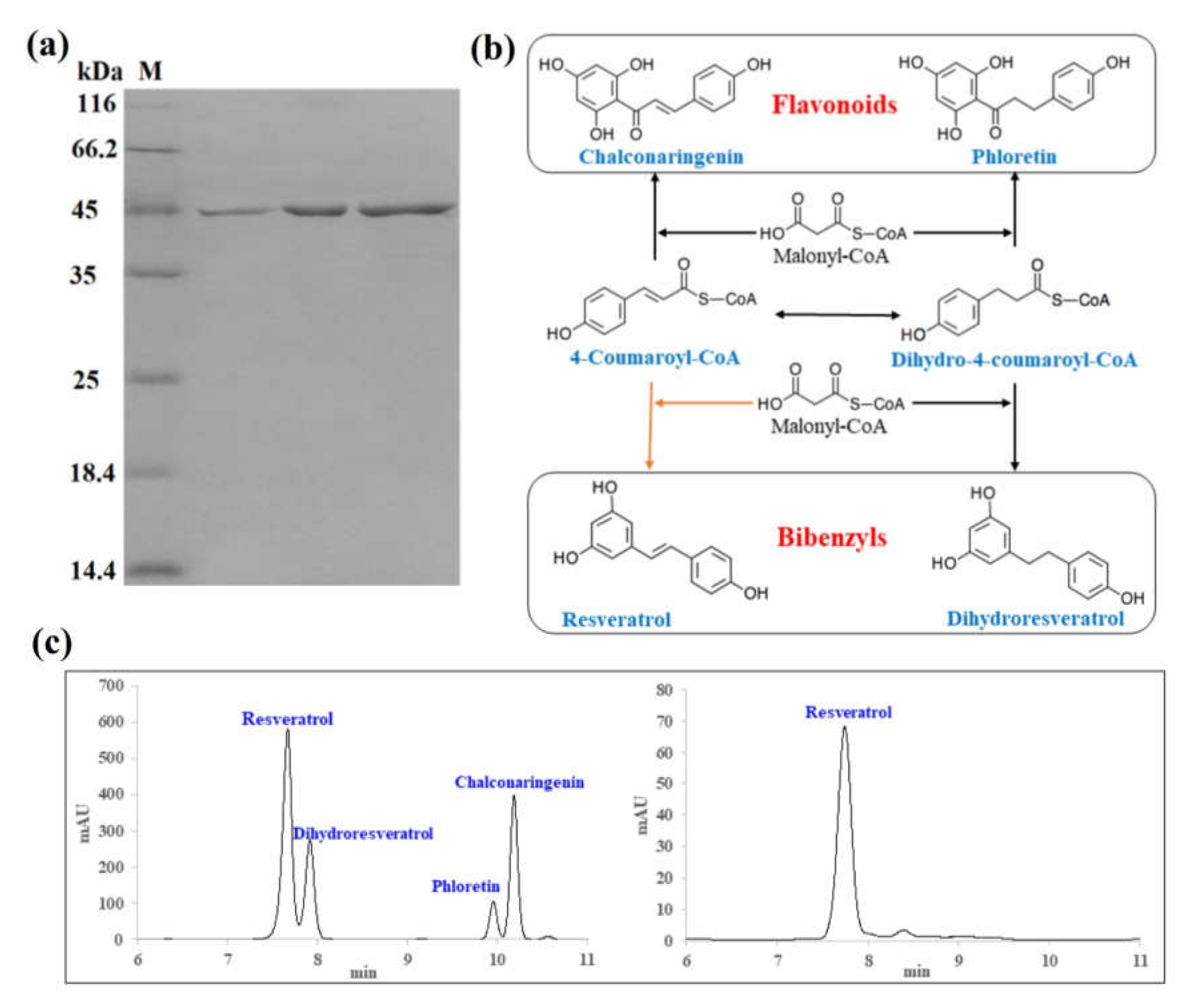
2.6. High Expression of Recombinant DsBBS Protein
2.7. Enzyme Activity Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of Bibenzyl Content
4.3. Transcriptome Preparation and Sequencing
4.4. Functional Annotation and Expression Analysis
4.5. Identification of Type III PKS Genes and Phylogeny Tree
4.6. RT-qPCR Analysis
4.7. Gene Cloning and Recombinant Vector Construction
4.8. Protein Purification and Enzymatic Assay
4.9. HPLC Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, C.; Chen, J.; Huang, W.; Song, X.; Niu, J. Transcriptomics and metabolomics reveal purine and phenylpropanoid metabolism response to drought stress in Dendrobium sinense, an endemic orchid species in Hainan Island. Front. Genet. 2021, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Sze, S.C.W.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biot. 2012, 93, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Khoonrit, P.; Mirdogan, A.; Dehlinger, A.; Mekboonsonglarp, W.; Likhitwitayawuid, K.; Priller, J.; Böttcher, C.; Sritularak, B. Immune modulatory effect of a novel 4, 5-dihydroxy-3, 3, 4-trimethoxybibenzyl from Dendrobium lindleyi. PLoS ONE 2020, 15, e0238509. [Google Scholar] [CrossRef] [PubMed]
- Pengpaeng, P.; Sritularak, B.; Chanvorachote, P. Dendrofalconerol A sensitizes anoikis and inhibits migration in lung cancer cells. J. Nat. Med. 2015, 69, 178–190. [Google Scholar] [CrossRef]
- Cai, C.-H.; Tan, C.-Y.; Chen, H.-Q.; Wang, H.; Mei, W.-L.; Song, X.-Q.; Dai, H.-F. Chemical constituents from Dendrobium sinense (II). Guihaia 2020, 40, 1368–1374. [Google Scholar]
- Chen, X.-J.; Mei, W.-L.; Cai, C.-H.; Guo, Z.-K.; Song, X.-Q.; Dai, H.-F. Four new bibenzyl derivatives from Dendrobium sinense. Phytochem. Lett. 2014, 9, 107–112. [Google Scholar] [CrossRef]
- Nandy, S.; Dey, A. Bibenzyls and bisbybenzyls of bryophytic origin as promising source of novel therapeutics: Pharmacology, synthesis and structure-activity. DARU J. Pharm. Sci. 2020, 28, 701–734. [Google Scholar] [CrossRef]
- He, L.; Su, Q.; Bai, L.; Li, M.; Liu, J.; Liu, X.; Zhang, C.; Jiang, Z.; He, J.; Shi, J. Recent research progress on natural small molecule bibenzyls and its derivatives in Dendrobium species. Eur. J. Med. Chem. 2020, 204, 112530. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, J.-K.; Wang, J.; Wang, N.-L.; Kurihara, H.; Kitanaka, S.; Yao, X.-S. Bioactive bibenzyl derivatives and fluorenones from Dendrobium nobile. J. Nat. Prod. 2007, 70, 24–28. [Google Scholar] [CrossRef]
- Costa, M.A.; Collins, R.E.; Anterola, A.M.; Cochrane, F.C.; Davin, L.B.; Lewis, N.G. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 2003, 64, 1097–1112. [Google Scholar] [CrossRef]
- Chezem, W.R.; Clay, N.K. Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 2016, 131, 26–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate: Coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.S.; Pearson, L.A.; Neilan, B.A. Genome mining and evolutionary analysis reveal diverse type III polyketide synthase pathways in cyanobacteria. Genome Biol. Evol. 2021, 13, evab056. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-N.; Wang, L.; Sun, B.; Gao, S.; Cheng, A.-X.; Lou, H.-X. Functional characterization of a chalcone synthase from the liverwort Plagiochasma appendiculatum. Plant Cell Rep. 2015, 34, 233–245. [Google Scholar] [CrossRef]
- Friederich, S.; Rueffer, M.; Asakawa, Y.; Zenk, M.H. Cytochromes P-450 catalyze the formation of marchantins A and C in Marchantia polymorpha. Phytochemistry 1999, 52, 1195–1202. [Google Scholar] [CrossRef]
- Reinecke, T.; Kindl, H. Characterization of bibenzyl synthase catalysing the biosynthesis of phytoalexins of orchids. Phytochemistry 1993, 35, 63–66. [Google Scholar] [CrossRef]
- Preisigmuller, R.; Gnau, P.; Kindl, H. The inducible 9, 10-dihydrophenanthrene pathway: Characterization and expression of bibenzyl synthase and S-adenosylhomocysteine hydrolase. Arch. Biochem. Biophys. 1995, 317, 201–207. [Google Scholar] [CrossRef]
- Cheng, Y.; He, D.; He, J.; Niu, G.; Gao, R. Effect of light/dark cycle on photosynthetic pathway switching and CO2 absorption in two Dendrobium species. Front. Plant Sci. 2019, 10, 659. [Google Scholar] [CrossRef]
- Tan, C.-Y.; Mei, W.-L.; Zhao, Y.-X.; Huang, S.-Z.; Kong, F.-D.; Yang, N.-N.; Song, X.-Q.; Dai, H.-F. Chemical Constituents from Dendrobium sinense. J. Trop. Subtrop. Bot. 2017, 25, 189. [Google Scholar]
- Chen, X.-J.; Mei, W.-L.; Zuo, W.-J.; Zeng, Y.-B.; Guo, Z.-K.; Song, X.-Q.; Dai, H.-F. A new antibacterial phenanthrenequinone from Dendrobium sinense. J. Asian Nat. Prod. Res. 2013, 15, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Shui, L.; Huo, K.; Chen, Y.; Zhang, Z.; Li, Y.; Niu, J. Integrated metabolome and transcriptome revealed the flavonoid biosynthetic pathway in developing Vernonia amygdalina leaves. PeerJ 2021, 9, e11239. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zhang, X.; Han, X.; Zhang, L.; Li, X.; Zhan, H.; Ma, J.; Luo, P.; Zhang, W.; Cui, L. A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 2015, 6, 770. [Google Scholar] [CrossRef] [Green Version]
- Abe, I. Biosynthesis of medicinally important plant metabolites by unusual type III polyketide synthases. J. Nat. Med. 2020, 74, 639–646. [Google Scholar] [CrossRef]
- Jiang, C.; Schommer, C.K.; Kim, S.Y.; Suh, D.-Y. Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 2006, 67, 2531–2540. [Google Scholar] [CrossRef]
- Suh, D.-Y.; Fukuma, K.; Kagami, J.; Yamazaki, Y.; Shibuya, M.; Ebizuka, Y.; Sankawa, U. Identification of amino acid residues important in the cyclization reactions of chalcone and stilbene synthases. Biochem. J. 2000, 350, 229–235. [Google Scholar] [CrossRef]
- Wang, X.; Yam, T.W.; Meng, Q.; Zhu, J.; Zhang, P.; Wu, H.; Wang, J.; Zhao, Y.; Song, X. The dual inoculation of endophytic fungi and bacteria promotes seedlings growth in Dendrobium catenatum (Orchidaceae) under in vitro culture conditions. Plant Cell Tissue Organ Cult. 2016, 126, 523–531. [Google Scholar] [CrossRef]
- Fang, J.; Houlin, X.; Ping, H.; Xiaoyan, H.; Zijun, N.; Jianjun, T.; Tingmo, Z. Determination of Bibenzyl Compounds Content in Dendrobinm denneanum by UV-Visible Spectrophotometry. J. Chengdu Univ. TCM 2014, 37, 7–10. [Google Scholar]
- Liu, Y.; Wang, Z.; Pang, S.; Zhao, W.; Kang, L.; Zhang, Y.; Zhang, H.; Yang, J.; Wang, Z.; Lu, P. Evaluation of dynamic developmental processes and the molecular basis of the high body fat percentage of different proglottid types of Moniezia expansa. Parasites Vectors 2019, 12, 390. [Google Scholar] [CrossRef]
- Gu, H.; Chen, S.; Zhang, M.; Wen, Y.; Li, B. Differences in the expression profiles of lncRNAs and mRNAs in partially injured anterior cruciate ligament and medial collateral ligament of rabbits. PeerJ 2022, 10, e12781. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Niu, J.; Bi, Q.; Deng, S.; Chen, H.; Yu, H.; Wang, L.; Lin, S. Identification of AUXIN RESPONSE FACTOR gene family from Prunus sibirica and its expression analysis during mesocarp and kernel development. BMC Plant Biol. 2018, 18, 21. [Google Scholar] [CrossRef] [Green Version]
- Porowińska, D.; Wujak, M.; Roszek, K.; Komoszyński, M. Prokaryotic expression systems. Postepy Hig. I Med. Dosw. (Online) 2013, 67, 119–129. [Google Scholar] [CrossRef]



| Genes | ID | ORF | ||
|---|---|---|---|---|
| Start | Stop | Length (nt|aa) | ||
| DsCHS1 | c82697.graph_c0 | 75 | > 764 | - |
| DsCHS2 | c82700.graph_c0 | 87 | 1274 | 1188|395 |
| DsCHS3 | c84946.graph_c0 | 55 | 1227 | 1173|390 |
| DsPKS | c89783.graph_c0 | 558 | 1571 | 1014|337 |
| DsBBS | c93636.graph_c0 | 101 | 1273 | 1173|390 |
| DsCHS4 | c99839.graph_c0 | 41 | > 838 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wang, Y.; Liang, C.; Liu, L.; Song, X.; Zhao, Y.; Wang, J.; Niu, J. Characterization of the Key Bibenzyl Synthase in Dendrobium sinense. Int. J. Mol. Sci. 2022, 23, 6780. https://doi.org/10.3390/ijms23126780
Chen Y, Wang Y, Liang C, Liu L, Song X, Zhao Y, Wang J, Niu J. Characterization of the Key Bibenzyl Synthase in Dendrobium sinense. International Journal of Molecular Sciences. 2022; 23(12):6780. https://doi.org/10.3390/ijms23126780
Chicago/Turabian StyleChen, Yan, Yu Wang, Chongjun Liang, Liyan Liu, Xiqiang Song, Ying Zhao, Jia Wang, and Jun Niu. 2022. "Characterization of the Key Bibenzyl Synthase in Dendrobium sinense" International Journal of Molecular Sciences 23, no. 12: 6780. https://doi.org/10.3390/ijms23126780
APA StyleChen, Y., Wang, Y., Liang, C., Liu, L., Song, X., Zhao, Y., Wang, J., & Niu, J. (2022). Characterization of the Key Bibenzyl Synthase in Dendrobium sinense. International Journal of Molecular Sciences, 23(12), 6780. https://doi.org/10.3390/ijms23126780






