Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix
Abstract
1. Introduction
2. Results
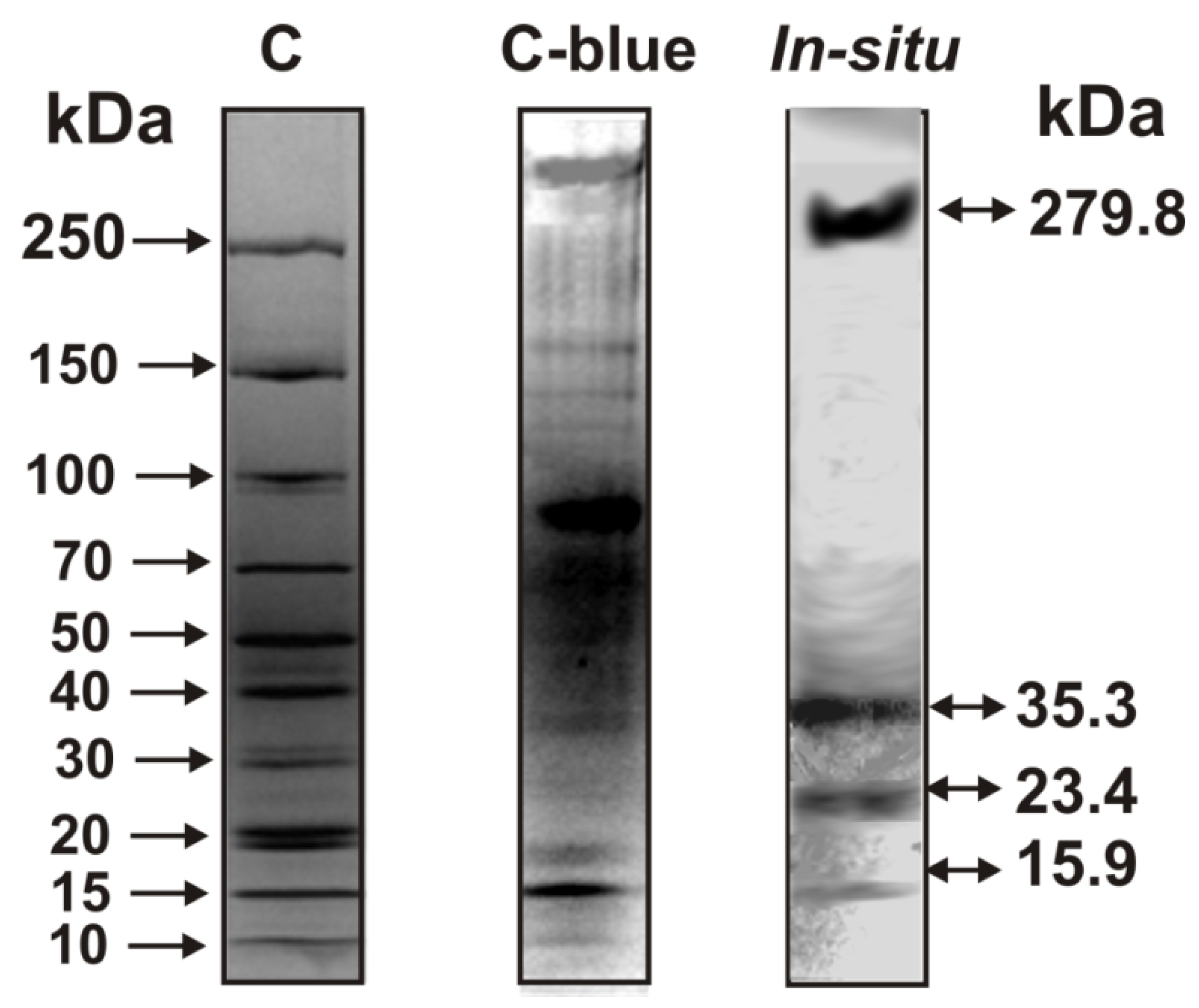
2.1. Isolation and Analysis of Sea Cucumber Protein Complex
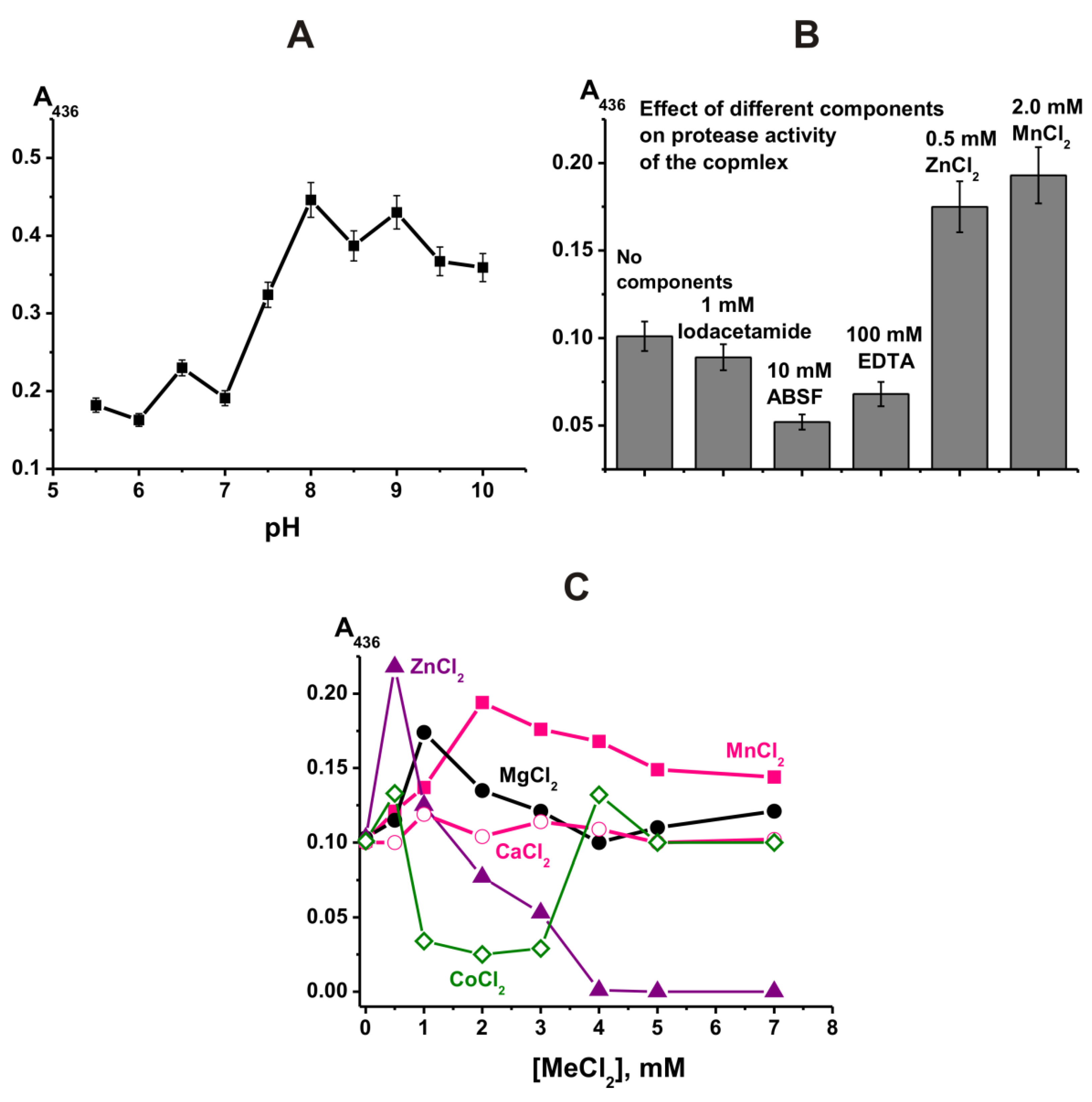
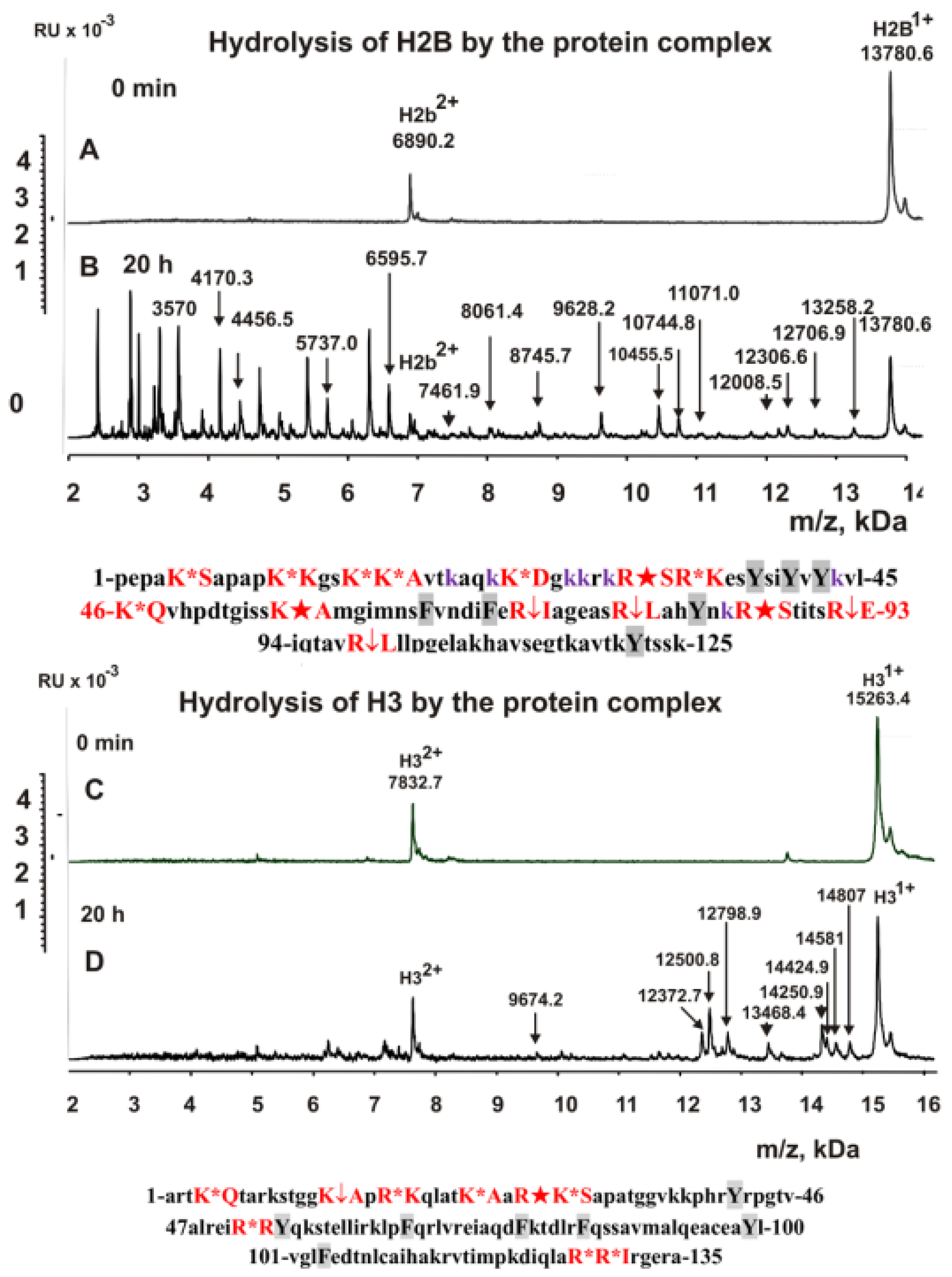
2.2. Protease Activities of the Complex
2.3. Type of Metal-Independent Proteases of the Complex
2.4. DNase Activities of the Complex
2.5. In Situ Assay of DNase Activity
2.6. ATPase Activity of the Complex
3. Discussion
4. Materials and methods
4.1. Chemicals
4.2. Purification of Stable Complexes by Gel Filtration
4.3. Protease Activity of the Complex
4.4. DNase Activity Assay
4.5. In Situ Analysis of DNase Activity
4.6. ATPase Activity of the Complex
4.7. MALDI-TOF Mass Spectrometry Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hyman, L.H. The Invertebrates: Echinodermata. The Coelome Bilateria; McGraw-Hill Book Co.: New York, NY, USA, 1955; p. 763. [Google Scholar]
- Dolmatov, I.Y.; Mashanov, V.S. Regeneration in Holothurians; Dalnauka: Vladivostok, Russia, 2007; pp. 1–212. [Google Scholar]
- Dolmatov, I.Y. Regeneration of the digestive system in holothurians. Zh. Obshch. Biol. 2009, 70, 319–330. [Google Scholar]
- Dolmatov, I.Y. Regeneration of the aquapharyngeal complex in the holothurian Eupentacta fraudatrix (Holothuroidea, Dendrochirota). In Keys for Regeneration; Taban, C.H., Boilly, B., Eds.; Karger: Basel, Germany, 1992; Volume 23, pp. 40–50. [Google Scholar]
- Levin, V.S.; Dalnevostochnyi Trepang. Biologiya, Dobycha, Vosproizvodstvo (The Far East Trepang Biology, Catching, Reproduction); St. Petersburg, Goland, Russia. 2000. Available online: https://search.rsl.ru/ru/record/01000663732 (accessed on 18 May 2022).
- Alberts, B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 1998, 92, 291–294. [Google Scholar] [CrossRef]
- Eubel, H.; Braun, H.P.; Millar, A.H. Blue-native PAGE in plants: A tool in analysis of protein-protein interactions. Plant Methods 2005, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, L.; Liang, G. Identification and analysis of multi-protein complexes in placenta. PLoS ONE 2013, 138, e62988. [Google Scholar] [CrossRef]
- Soboleva, S.E.; Dmitrenok, P.S.; Verkhovod, T.D.; Buneva, V.N.; Sedykh, S.E.; Nevinsky, G.A. Very stable high molecular mass multiprotein complex with DNase and amylase activities in human milk. J. Mol. Recognit. 2015, 28, 20–34. [Google Scholar] [CrossRef]
- Burkova, E.E.; Dmitrenok, P.S.; Sedykh, S.E.; Buneva, V.N.; Soboleva, S.E.; Nevinsky, G.A. Extremely stable soluble high molecular mass multi-protein complex with DNase activity in human placental tissue. PLoS ONE 2014, 9, e111234. [Google Scholar] [CrossRef][Green Version]
- Burkova, E.E.; Dmitrenok, P.S.; Bulgakov, D.V.; Ermakov, E.A.; Buneva, V.N.; Soboleva, S.E.; Nevinsky, G.A. Identification of major proteins of a very stable high molecular mass multi-protein complex of human placental tissue possessing nine different catalytic activities. Biochem. Anal. Biochem. 2018, 7, 351. [Google Scholar] [CrossRef]
- Soboleva, S.E.; Burkova, E.E.; Dmitrenok, P.S.; Bulgakov, D.V.; Menzorova, N.I.; Buneva, V.N.; Nevinsky, G.A. Extremely stable high molecular mass soluble multiprotein complex from eggs of sea urchin Strongylocentrotus intermedius with phosphatase activity. J. Mol. Recognit. 2018, 31, e2753. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Kostrikina, I.A.; Dmitrenok, P.S.; Soboleva, S.E.; Nevinsky, G.A. Very Stable Two Mega Dalton High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix. Molecules 2021, 26, 5703. [Google Scholar] [CrossRef]
- Zaksas, N.P.; Timofeeva, A.M.; Dmitrenok, P.S.; Soboleva, S.E.; Nevinsky, G. Comparison of the Content of Several Elements in Seawater, Sea Cucumber Eupentacta fraudatrix and Its High-Molecular-Mass Multiprotein Complex. Molecules 2022, 27, 1958. [Google Scholar] [CrossRef]
- Milcovich, G.; Antunes, F.; Golob, S.; Farra, R.; Grassi, M.; Voinovich, D.; Grassi, G.; Asaro, F.J. Thermo-responsive hydrogels from cellulose-based polyelectrolytes and catanionic vesicles for biomedical application. Biomed. Mater. Res A 2016, 104, 1668–1679. [Google Scholar] [CrossRef]
- Fersht, A. Enzyme Structure and Mechanism, 2nd ed.; W. H. Freeman & Co.: New York, NY, USA, 1985. [Google Scholar]
- Alcala-Torano, R.; Walther, M.; Sommer, D.J.; Park, C.K.; Ghirlanda, G. Rational design of a hexameric protein assembly stabilized by metal chelation. Biopolymers 2018, 109, e23233. [Google Scholar] [CrossRef]
- Kocyła, A.; Tran, J.B.; Krężel, A. Galvanization of Protein-Protein Interactions in a Dynamic Zinc Interactome. Trends Biochem. Sci. 2021, 46, 64–79. [Google Scholar] [CrossRef]
- Butcher, S.E. The spliceosome and its metal ions. Met. Ions Life Sci. 2011, 9, 235–251. [Google Scholar]
- Castellano, I.; Migliaccio, O.; Ferraro, G.; Maffioli, E.; Marasco, D.; Merlino, A.; Zingone, A.; Tedeschi, G.; Palumbo, A. Biotic and environmental stress induces nitration and changes in structure and function of the sea urchin major yolk protein toposome. Sci. Rep. 2018, 8, 4610. [Google Scholar] [CrossRef]
- Pinsino, A.; Matranga, V. Sea urchin immune cells as sentinels of environmental stress. Dev. Comp. Immunol. 2015, 49, 198–205. [Google Scholar] [CrossRef]
- Charney, J.; Tomarelli, R.M. A colorimetric method for the determination of the proteolytic activity of duodenal juice. J. Biol. Chem. 1947, 171, 501–505. [Google Scholar] [CrossRef]
- Solov’eva, N.I. Matrix metalloproteinases and their biological functions. Bioorg. Khim. 1998, 24, 245–255. [Google Scholar]
- Verma, R.P.; Hansch, C. Matrix metalloproteinases (MMPs): Chemical-biological functions and (Q)SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef]
- Whittaker, M.; Floyd, C.D.; Brown, P.; Gearing, A.J. Design and therapeutic application of matrix metalloproteinase inhibitors. Chem. Rev. 1999, 99, 2735–2776. [Google Scholar] [CrossRef]
- Baranova, S.V.; Dmitrienok, P.S.; Ivanisenko, N.V.; Buneva, V.N.; Nevinsky, G.A. Antibodies to H1 histone from the sera of HIV-infected patients recognize and catalyze site-specific degradation of this histone. J. Mol. Recognit. 2017, 30, e2588. [Google Scholar] [CrossRef]
- Baranova, S.V.; Dmitrienok, P.S.; Ivanisenko, N.V.; Buneva, V.N.; Nevinsky, G.A. Antibodies to H2a and H2b histones from the sera of HIV-infected patients catalyze site-specific degradation of these histones. Mol. Biosyst. 2017, 13, 1090–1101. [Google Scholar] [CrossRef]
- Baranova, S.V.; Dmitrenok, P.S.; Zubkova, A.D.; Ivanisenko, N.V.; Odintsova, E.S.; Buneva, V.N.; Nevinsky, G.A. Antibodies against H3 and H4 histones from the sera of HIV-infected patients catalyze site-specific degradation of these histones. J. Mol. Recognit. 2018, 31, e2703. [Google Scholar] [CrossRef]
- Baranovskii, A.G.; Buneva, V.N.; Nevinsky, G.A. Human deoxyribonucleases. Biochemistry 2004, 69, 587–601. [Google Scholar] [CrossRef]
- Suck, D. DNA recognition by DNase I. J. Mol. Recognit. 1994, 7, 65–70. [Google Scholar] [CrossRef]
- Keyel, P.A. DNases in health and disease. Dev. Biol. 2017, 429, 1–11. [Google Scholar] [CrossRef]
- Lauková, L.; Konečná, B.; Janovičová, Ľ.; Vlková, B.; Celec, P. Deoxyribonucleases and Their Applications in Biomedicine. Biomolecules 2020, 10, 1036. [Google Scholar] [CrossRef]
- Bernardi, G. The Enzymes; Boyer, P.D., Ed.; Academic Press: New York, NY, USA, 1971. [Google Scholar]
- Abramova, Z.I.; Kosareva, T.I.; Vinter, V.G. The interaction of the Ca2+-and Mg2+-dependent DNases of sea urchin embryos with DNA. Tsitologiia 1995, 37, 893–899. [Google Scholar]
- Menzorova, N.I.; Rasskazov, V.A. Effect of bivalent metal ions on enzymatic activity of Ca2+,Mg2+-dependent DNase from sea urchin Stronglyocentrotus intermedius embryos. Biokhimiia 1980, 45, 544–553. [Google Scholar]
- Sibirtsev, J.T.; Shastina, V.V.; Menzorova, N.I.; Makarieva, T.N.; Rasskazov, V.A. Ca2+, Mg2+-dependent DNase involvement in apoptotic effects in spermatozoa of sea urchin Strongylocentrotus intermedius induced by two-headed sphingolipid rhizochalin. Mar. Biotechnol. 2011, 13, 536–543. [Google Scholar] [CrossRef]
- Shin, J.M.; Munson, K.; Vagin, O.; Sachs, G. The gastric HK-ATPase: Structure, function and inhibition. Pflug. Arch. 2009, 457, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 7th ed.; W. H. Freeman and Company: New York, NY, USA, 2012. [Google Scholar]
- Sherman, D.J.; Lazarus, M.B.; Murphy, L.; Liu, C.; Walker, S.; Ruiz, N.; Kahne, D. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl. Acad. Sci. USA 2014, 111, 4982–4987. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timofeeva, A.M.; Kostrikina, I.A.; Dmitrenok, P.S.; Soboleva, S.E.; Nevinsky, G.A. Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix. Int. J. Mol. Sci. 2022, 23, 6677. https://doi.org/10.3390/ijms23126677
Timofeeva AM, Kostrikina IA, Dmitrenok PS, Soboleva SE, Nevinsky GA. Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix. International Journal of Molecular Sciences. 2022; 23(12):6677. https://doi.org/10.3390/ijms23126677
Chicago/Turabian StyleTimofeeva, Anna M., Irina A. Kostrikina, Pavel S. Dmitrenok, Svetlana E. Soboleva, and Georgy A. Nevinsky. 2022. "Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix" International Journal of Molecular Sciences 23, no. 12: 6677. https://doi.org/10.3390/ijms23126677
APA StyleTimofeeva, A. M., Kostrikina, I. A., Dmitrenok, P. S., Soboleva, S. E., & Nevinsky, G. A. (2022). Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix. International Journal of Molecular Sciences, 23(12), 6677. https://doi.org/10.3390/ijms23126677







