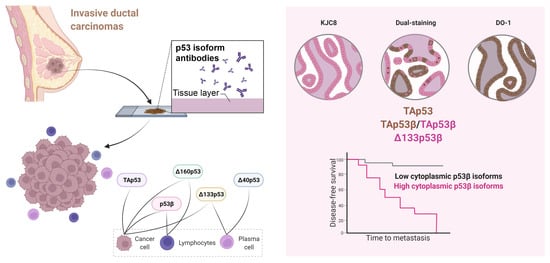
Cytoplasmic p53β Isoforms Are Associated with Worse Disease-Free Survival in Breast Cancer
Abstract
:1. Introduction
2. Results
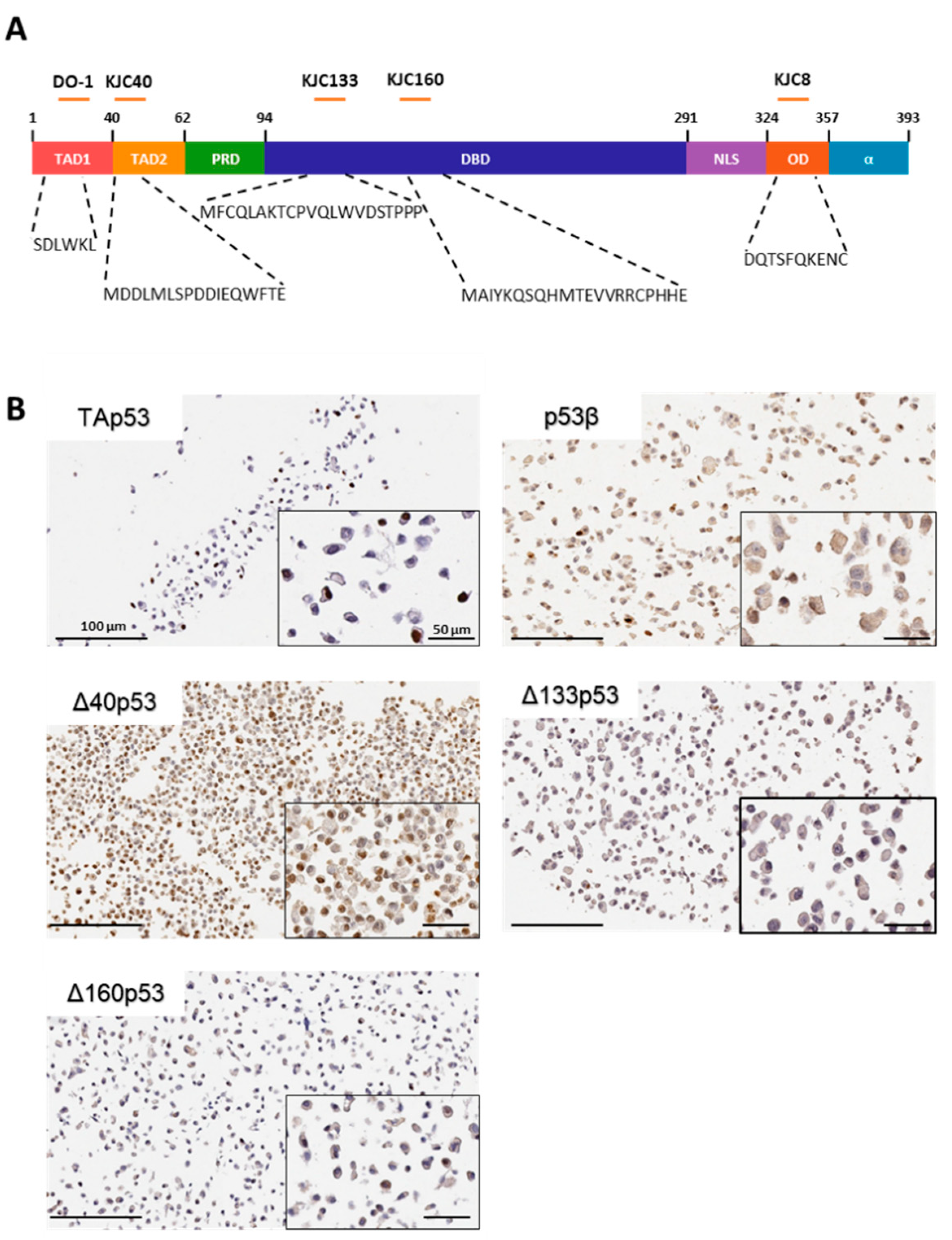
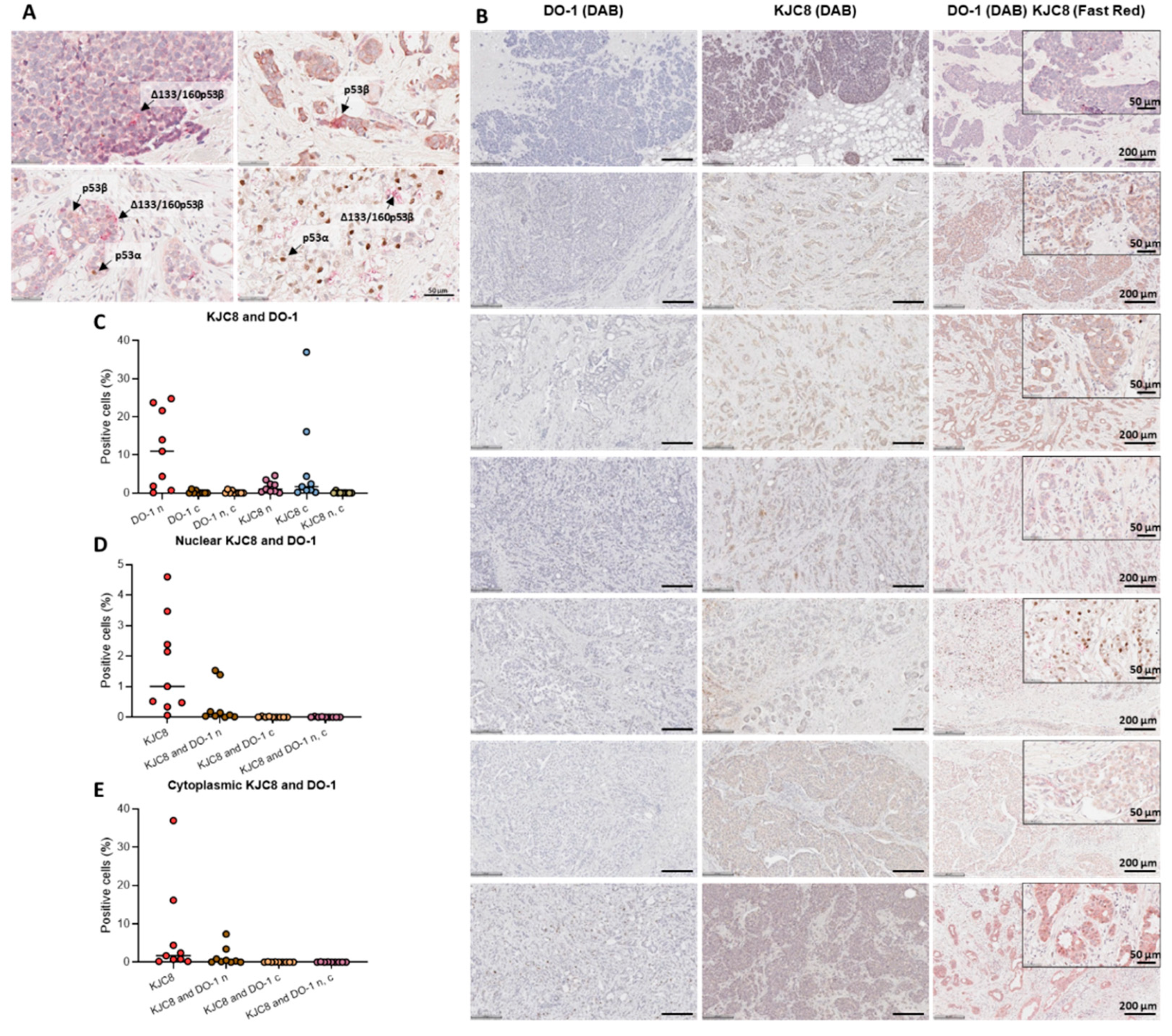
2.1. Characterisation of p53 Isoform Localisation in Breast Cancer Tissues
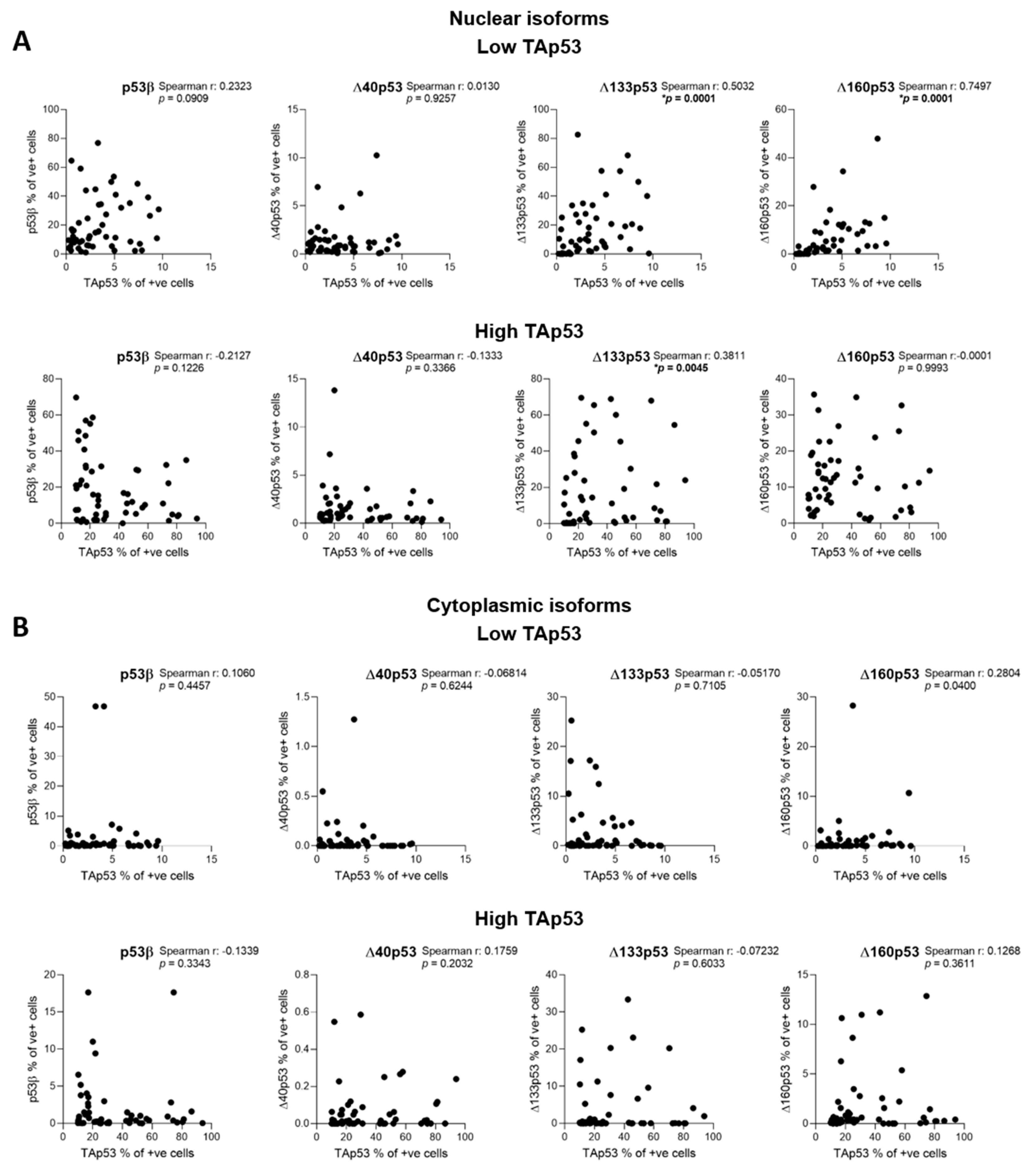
2.2. Characterisation of p53 Isoform Expression in Breast Cancer Tissues
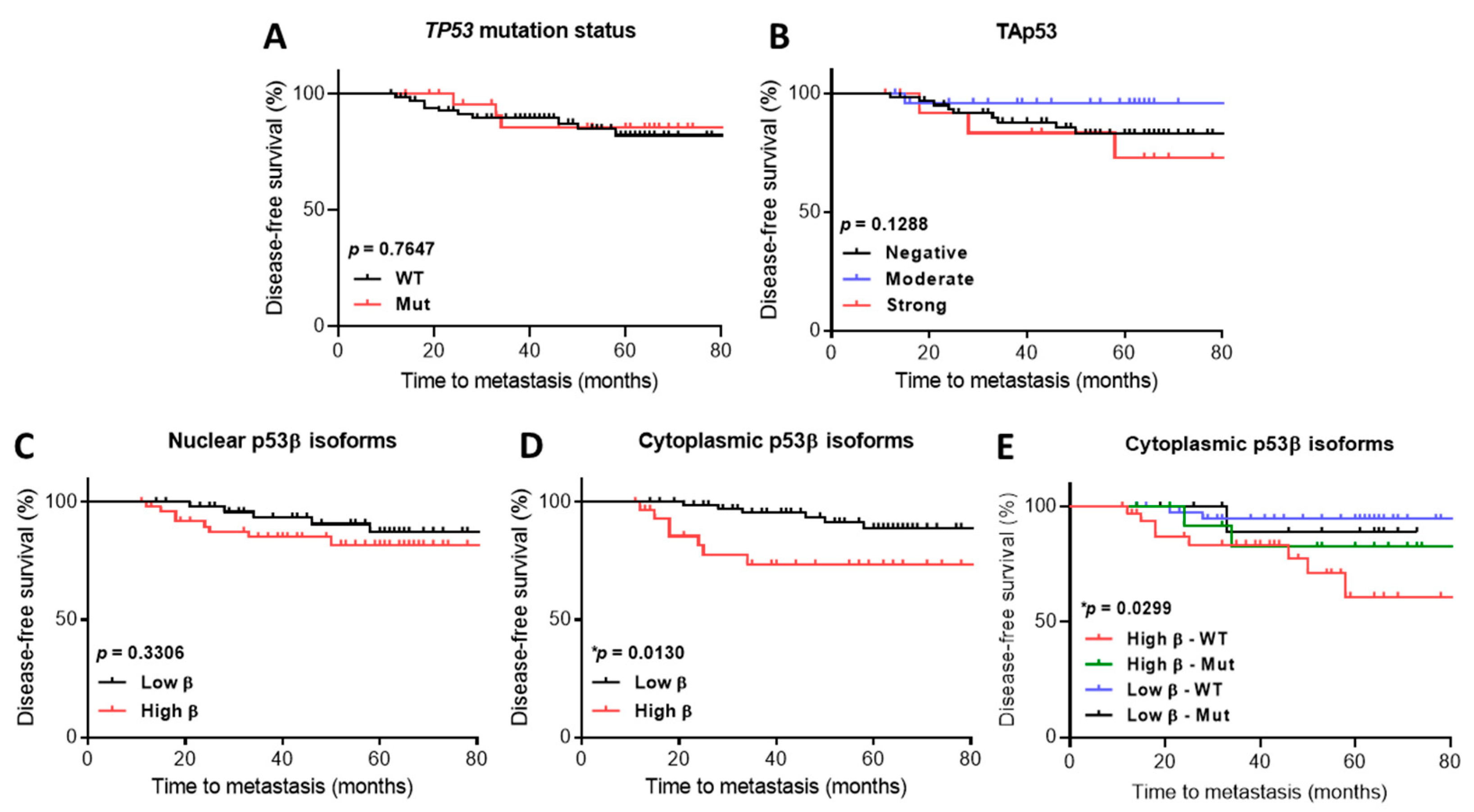
2.3. The association of p53 Isoform Expression with Tp53 Mutation Status and Clinicopathological Features
2.4. p53 Isoform Expression Association with Disease-Free Survival
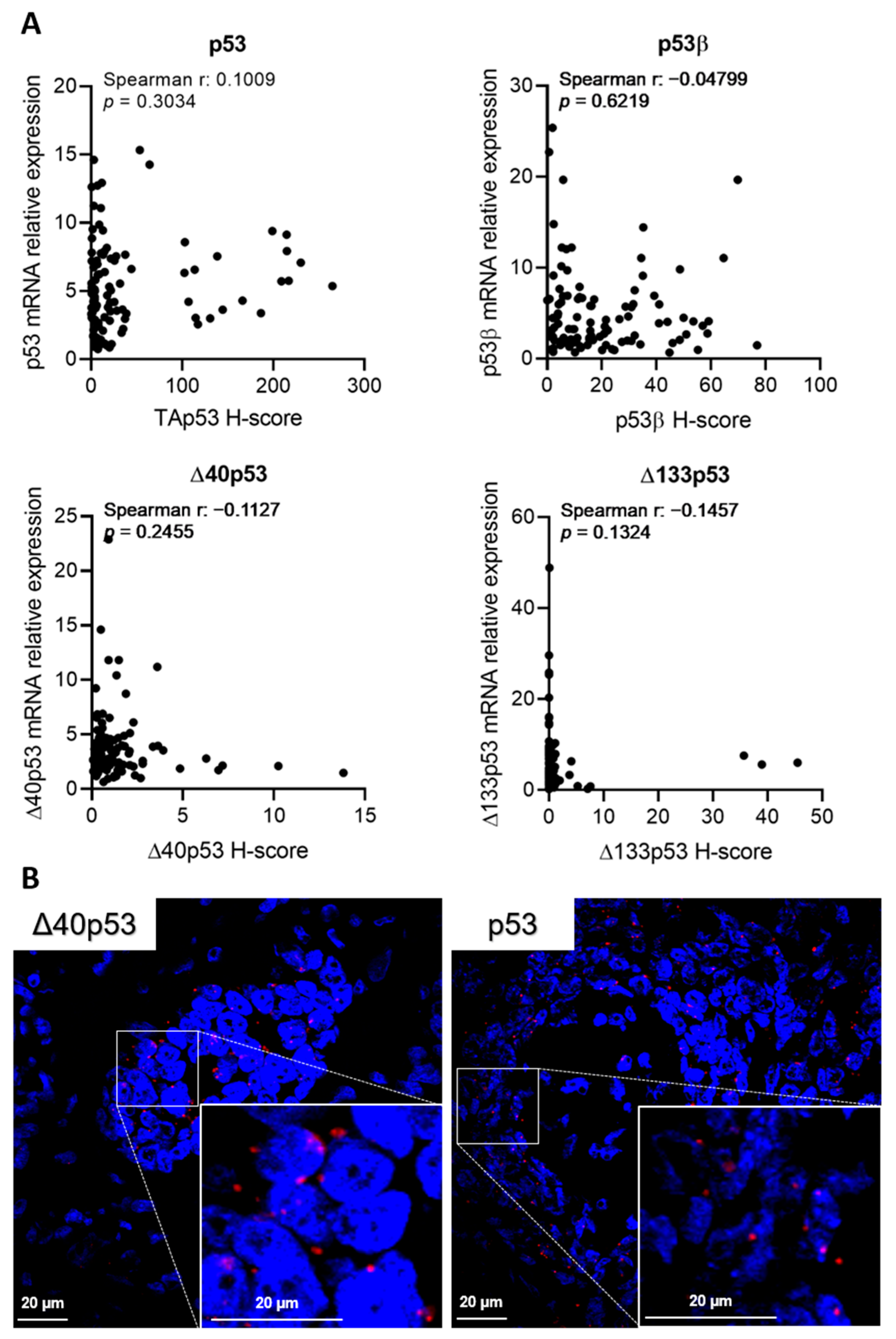
2.5. p53 Isoform mRNA Levels Do Not Correlate with Their Protein Product in Breast Cancer
3. Discussion
4. Materials and Methods
4.1. Subcloning of p53 Isoform Plasmid Vectors
4.2. Transfection of p53 Isoforms Plasmids into Breast Cancer Cells and Cell Pellet Preparation
4.3. Study Cohort
4.4. Antibodies
4.5. Immunohistochemistry
4.6. In Situ Hybridisation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer (IARC); WHO. Globocan 2020. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/20-Breast-fact-sheet.pdf (accessed on 16 September 2021).
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdon, J.C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcel, V.; Perrier, S.; Aoubala, M.; Ageorges, S.; Groves, M.J.; Diot, A.; Fernandes, K.; Tauro, S.; Bourdon, J.C. Delta160p53 is a novel N-terminal p53 isoform encoded by Delta133p53 transcript. FEBS Lett. 2010, 584, 4463–4468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffens Reinhardt, L.; Zhang, X.; Wawruszak, A.; Groen, K.; De Iuliis, G.N.; Avery-Kiejda, K.A. Good Cop, Bad Cop: Defining the Roles of Delta40p53 in Cancer and Aging. Cancers 2020, 12, 1659. [Google Scholar] [CrossRef]
- Ghosh, A.; Stewart, D.; Matlashewski, G. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell Biol. 2004, 24, 7987–7997. [Google Scholar] [CrossRef] [Green Version]
- Courtois, S.; Verhaegh, G.; North, S.; Luciani, M.G.; Lassus, P.; Hibner, U.; Oren, M.; Hainaut, P. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 2002, 21, 6722–6728. [Google Scholar] [CrossRef] [Green Version]
- Ray, P.S.; Grover, R.; Das, S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006, 7, 404–410. [Google Scholar] [CrossRef] [Green Version]
- Candeias, M.M.; Powell, D.J.; Roubalova, E.; Apcher, S.; Bourougaa, K.; Vojtesek, B.; Bruzzoni-Giovanelli, H.; Fåhraeus, R. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 2006, 25, 6936–6947. [Google Scholar] [CrossRef] [Green Version]
- Solomon, H.; Bräuning, B.; Fainer, I.; Ben-Nissan, G.; Rabani, S.; Goldfinger, N.; Moscovitz, O.; Shakked, Z.; Rotter, V.; Sharon, M. Post-translational regulation of p53 function through 20S proteasome-mediated cleavage. Cell Death Differ. 2017, 24, 2187–2198. [Google Scholar] [CrossRef]
- Avery-Kiejda, K.A.; Zhang, X.D.; Adams, L.J.; Scott, R.J.; Vojtesek, B.; Lane, D.P.; Hersey, P. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin. Cancer Res. 2008, 14, 1659–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdon, J.C.; Khoury, M.P.; Diot, A.; Baker, L.; Fernandes, K.; Aoubala, M.; Quinlan, P.; Purdie, C.A.; Jordan, L.B.; Prats, A.C.; et al. p53 mutant breast cancer patients expressing p53gamma have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011, 13, R7. [Google Scholar] [CrossRef] [PubMed]
- Hofstetter, G.; Berger, A.; Fiegl, H.; Slade, N.; Zoric, A.; Holzer, B.; Schuster, E.; Mobus, V.J.; Reimer, D.; Daxenbichler, G.; et al. Alternative splicing of p53 and p73: The novel p53 splice variant p53delta is an independent prognostic marker in ovarian cancer. Oncogene 2010, 29, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Anensen, N.; Oyan, A.M.; Bourdon, J.C.; Kalland, K.H.; Bruserud, O.; Gjertsen, B.T. A distinct p53 protein isoform signature reflects the onset of induction chemotherapy for acute myeloid leukemia. Clin. Cancer Res. 2006, 12, 3985–3992. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.; Giannini, C.; Sarkaria, J.N.; Schroeder, M.; Rogers, J.; Mastroeni, D.; Scrable, H. p53 isoform profiling in glioblastoma and injured brain. Oncogene 2013, 32, 3165–3174. [Google Scholar] [CrossRef] [Green Version]
- Melo Dos Santos, N.; de Oliveira, G.A.P.; Ramos Rocha, M.; Pedrote, M.M.; Diniz da Silva Ferretti, G.; Pereira Rangel, L.; Morgado-Diaz, J.A.; Silva, J.L.; Rodrigues Pereira Gimba, E. Loss of the p53 transactivation domain results in high amyloid aggregation of the Delta40p53 isoform in endometrial carcinoma cells. J. Biol. Chem. 2019, 294, 9430–9439. [Google Scholar] [CrossRef] [Green Version]
- Hofstetter, G.; Berger, A.; Berger, R.; Zoric, A.; Braicu, E.I.; Reimer, D.; Fiegl, H.; Marth, C.; Zeimet, A.G.; Ulmer, H.; et al. The N-terminally truncated p53 isoform Delta40p53 influences prognosis in mucinous ovarian cancer. Int. J. Gynecol. Cancer 2012, 22, 372–379. [Google Scholar] [CrossRef]
- Oh, L.; Hainaut, P.; Blanchet, S.; Ariffin, H. Expression of p53 N-terminal isoforms in B-cell precursor acute lymphoblastic leukemia and its correlation with clinicopathological profiles. BMC Cancer 2020, 20, 110. [Google Scholar] [CrossRef] [Green Version]
- Avery-Kiejda, K.A.; Morten, B.; Wong-Brown, M.W.; Mathe, A.; Scott, R.J. The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis 2014, 35, 586–596. [Google Scholar] [CrossRef]
- Morten, B.C.; Wong-Brown, M.W.; Scott, R.J.; Avery-Kiejda, K.A. The presence of the intron 3 16 bp duplication polymorphism of p53 (rs17878362) in breast cancer is associated with a low Delta40p53:p53 ratio and better outcome. Carcinogenesis 2016, 37, 81–86. [Google Scholar] [CrossRef]
- Powell, D.J.; Hrstka, R.; Candeias, M.; Bourougaa, K.; Vojtesek, B.; Fahraeus, R. Stress-dependent changes in the properties of p53 complexes by the alternative translation product p53/47. Cell Cycle 2008, 7, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Grover, R.; Ray, P.S.; Das, S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 2008, 7, 2189–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourougaa, K.; Naski, N.; Boularan, C.; Mlynarczyk, C.; Candeias, M.M.; Marullo, S.; Fåhraeus, R. Endoplasmic Reticulum Stress Induces G2 Cell-Cycle Arrest via mRNA Translation of the p53 Isoform p53/47. Mol. Cell 2010, 38, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Sharathchandra, A.; Katoch, A.; Das, S. IRES mediated translational regulation of p53 isoforms. Wiley Interdiscip. Rev. RNA 2014, 5, 131–139. [Google Scholar] [CrossRef]
- Zhang, X.; Groen, K.; Morten, B.C.; Steffens Reinhardt, L.; Campbell, H.G.; Braithwaite, A.W.; Bourdon, J.C.; Avery-Kiejda, K.A. The effect of p53 and its N-terminally truncated isoform, Δ40p53, on breast cancer migration and invasion. Mol. Oncol. 2021, 16, 447–465. [Google Scholar] [CrossRef]
- Ungewitter, E.; Scrable, H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010, 24, 2408–2419. [Google Scholar] [CrossRef] [Green Version]
- Dookeran, K.A.; Dignam, J.J.; Ferrer, K.; Sekosan, M.; McCaskill-Stevens, W.; Gehlert, S. p53 as a marker of prognosis in African-American women with breast cancer. Ann. Surg. Oncol. 2010, 17, 1398–1405. [Google Scholar] [CrossRef]
- Yang, P.; Du, C.W.; Kwan, M.; Liang, S.X.; Zhang, G.J. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci. Rep. 2013, 3, 2246. [Google Scholar] [CrossRef] [Green Version]
- Bankhead, P.; Fernández, J.A.; McArt, D.G.; Boyle, D.P.; Li, G.; Loughrey, M.B.; Irwin, G.W.; Harkin, D.P.; James, J.A.; McQuaid, S.; et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab. Investig. 2018, 98, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhang, D.; Qin, X.; Owzar, K.; McCann, J.J.; Kastan, M.B. DNA-Damage-Induced Alternative Splicing of p53. Cancers 2021, 13, 251. [Google Scholar] [CrossRef]
- Campbell, H.; Fleming, N.; Roth, I.; Mehta, S.; Wiles, A.; Williams, G.; Vennin, C.; Arsic, N.; Parkin, A.; Pajic, M.; et al. ∆133p53 isoform promotes tumour invasion and metastasis via interleukin-6 activation of JAK-STAT and RhoA-ROCK signalling. Nat. Commun. 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadea, G.; Arsic, N.; Fernandes, K.; Diot, A.; Joruiz, S.M.; Abdallah, S.; Meuray, V.; Vinot, S.; Anguille, C.; Remenyi, J.; et al. TP53 drives invasion through expression of its Δ133p53β variant. eLife 2016, 5, e14734. [Google Scholar] [CrossRef] [PubMed]
- Arsic, N.; Ho-Pun-Cheung, A.; Lopez-Crapez, E.; Assenat, E.; Jarlier, M.; Anguille, C.; Colard, M.; Pezet, M.; Roux, P.; Gadea, G. The p53 isoform delta133p53ß regulates cancer cell apoptosis in a RhoB-dependent manner. PLoS ONE 2017, 12, e0172125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazantseva, M.; Eiholzer, R.A.; Mehta, S.; Taha, A.; Bowie, S.; Roth, I.; Zhou, J.; Joruiz, S.M.; Royds, J.A.; Hung, N.A.; et al. Elevation of the TP53 isoform Δ133p53β in glioblastomas: An alternative to mutant p53 in promoting tumor development. J. Pathol. 2018, 246, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Kazantseva, M.; Mehta, S.; Eiholzer, R.A.; Gimenez, G.; Bowie, S.; Campbell, H.; Reily-Bell, A.L.; Roth, I.; Ray, S.; Drummond, C.J.; et al. The Δ133p53β isoform promotes an immunosuppressive environment leading to aggressive prostate cancer. Cell Death Dis. 2019, 10, 631. [Google Scholar] [CrossRef]
- Arsic, N.; Gadea, G.; Lagerqvist, E.L.; Busson, M.; Cahuzac, N.; Brock, C.; Hollande, F.; Gire, V.; Pannequin, J.; Roux, P. The p53 isoform Delta133p53beta promotes cancer stem cell potential. Stem Cell Rep. 2015, 4, 531–540. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Qi, R.; Tang, Y.; Wang, W.; Wei, G.; Nussinov, R.; Ma, B. Conformational stability and dynamics of the cancer-associated isoform Δ133p53β are modulated by p53 peptides and p53-specific DNA. FASEB J. 2019, 33, 4225–4235. [Google Scholar] [CrossRef]
- Arsic, N.; Slatter, T.; Gadea, G.; Villain, E.; Fournet, A.; Kazantseva, M.; Allemand, F.; Sibille, N.; Seveno, M.; de Rossi, S.; et al. Δ133p53β isoform pro-invasive activity is regulated through an aggregation-dependent mechanism in cancer cells. Nat. Commun. 2021, 12, 5463. [Google Scholar] [CrossRef]
- Tadijan, A.; Precazzini, F.; Hanžić, N.; Radić, M.; Gavioli, N.; Vlašić, I.; Ozretić, P.; Pinto, L.; Škreblin, L.; Barban, G.; et al. Altered Expression of Shorter p53 Family Isoforms Can Impact Melanoma Aggressiveness. Cancers 2021, 13, 5231. [Google Scholar] [CrossRef]
- Kim, S.; An, S.S.A. Role of p53 isoforms and aggregations in cancer. Medicine 2016, 95, e3993. [Google Scholar] [CrossRef]
- De Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilborg, A.; Wilhelm, M.T.; Wiman, K.G. Regulation of tumor suppressor p53 at the RNA level. J. Mol. Med. 2010, 88, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Sharathchandra, A.; Lal, R.; Khan, D.; Das, S. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol. 2012, 9, 1429–1439. [Google Scholar] [CrossRef] [Green Version]
- Weingarten-Gabbay, S.; Khan, D.; Liberman, N.; Yoffe, Y.; Bialik, S.; Das, S.; Oren, M.; Kimchi, A. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 2014, 33, 611–618. [Google Scholar] [CrossRef]
- Swiatkowska, A.; Dutkiewicz, M.; Zydowicz-Machtel, P.; Szpotkowska, J.; Janecki, D.M.; Ciesiołka, J. Translational Control in p53 Expression: The Role of 5’-Terminal Region of p53 mRNA. Int. J. Mol. Sci. 2019, 20, 5382. [Google Scholar] [CrossRef] [Green Version]
- Haronikova, L.; Olivares-Illana, V.; Wang, L.; Karakostis, K.; Chen, S.; Fåhraeus, R. The p53 mRNA: An integral part of the cellular stress response. Nucleic Acids Res. 2019, 47, 3257–3271. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.; Markovic, S.N.; Scrable, H. Dominant Effects of D40p53 on p53 Function and Melanoma Cell Fate. J. Investig. Dermatol. 2013, 34, 791–800. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Rall-Scharpf, M.; Bourdon, J.-C.; Wiesmüller, L.; Biber, S. p53 isoforms differentially impact on the POLι dependent DNA damage tolerance pathway. Cell Death Dis. 2021, 12, 941. [Google Scholar] [CrossRef] [PubMed]
- Levandowski, C.B.; Jones, T.; Gruca, M.; Ramamoorthy, S.; Dowell, R.D.; Taatjes, D.J. The Δ40p53 isoform inhibits p53-dependent eRNA transcription and enables regulation by signal-specific transcription factors during p53 activation. PLoS Biol. 2021, 19, e3001364. [Google Scholar] [CrossRef] [PubMed]
- Agupitan, A.D.; Neeson, P.; Williams, S.; Howitt, J.; Haupt, S.; Haupt, Y. P53: A Guardian of Immunity Becomes Its Saboteur through Mutation. Int. J. Mol. Sci. 2020, 21, 3452. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Mandinova, A.; Aaronson, S.A.; Lee, S.W. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat. Rev. Immunol. 2016, 16, 741–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legscha, K.J.; Antunes Ferreira, E.; Chamoun, A.; Lang, A.; Awwad, M.H.S.; Ton, G.; Galetzka, D.; Guezguez, B.; Hundemer, M.; Bourdon, J.C.; et al. Δ133p53α enhances metabolic and cellular fitness of TCR-engineered T cells and promotes superior antitumor immunity. J. Immunother. Cancer. 2021, 9, e001846. [Google Scholar] [CrossRef]
- Mondal, A.M.; Horikawa, I.; Pine, S.R.; Fujita, K.; Morgan, K.M.; Vera, E.; Mazur, S.J.; Appella, E.; Vojtesek, B.; Blasco, M.A.; et al. p53 isoforms regulate aging- and tumor-associated replicative senescence in T lymphocytes. J. Clin. Investig. 2013, 123, 5247–5257. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Mondal, A.M.; Horikawa, I.; Nguyen, G.H.; Kumamoto, K.; Sohn, J.J.; Bowman, E.D.; Mathe, E.A.; Schetter, A.J.; Pine, S.R.; et al. p53 isoforms Δ133p53 and p53β are endogenous regulators of replicative cellular senescence. Nat. Cell Biol. 2009, 11, 1135–1142. [Google Scholar] [CrossRef]
- Mohammed, Z.M.; Going, J.J.; Edwards, J.; Elsberger, B.; McMillan, D.C. The relationship between lymphocyte subsets and clinico-pathological determinants of survival in patients with primary operable invasive ductal breast cancer. Br. J. Cancer 2013, 109, 1676–1684. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Fu, P.; Yao, M.; Chen, Y.; Du, L. Breast cancer stem cells phenotype and plasma cell-predominant breast cancer independently indicate poor survival. Pathol. Res. Pract. 2016, 212, 294–301. [Google Scholar] [CrossRef]
- Yeong, J.; Lim, J.C.T.; Lee, B.; Li, H.; Chia, N.; Ong, C.C.H.; Lye, W.K.; Putti, T.C.; Dent, R.; Lim, E.; et al. High Densities of Tumor-Associated Plasma Cells Predict Improved Prognosis in Triple Negative Breast Cancer. Front. Immunol. 2018, 9, 1209. [Google Scholar] [CrossRef]
- Schmidt, M.; Böhm, D.; von Törne, C.; Steiner, E.; Puhl, A.; Pilch, H.; Lehr, H.A.; Hengstler, J.G.; Kölbl, H.; Gehrmann, M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008, 68, 5405–5413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanker, L.C.; Rody, A.; Holtrich, U.; Pusztai, L.; Ruckhaeberle, E.; Liedtke, C.; Ahr, A.; Heinrich, T.M.; Sänger, N.; Becker, S.; et al. Prognostic evaluation of the B cell/IL-8 metagene in different intrinsic breast cancer subtypes. Breast Cancer Res. Treat 2013, 137, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Alistar, A.; Chou, J.W.; Nagalla, S.; Black, M.A.; D’Agostino, R., Jr.; Miller, L.D. Dual roles for immune metagenes in breast cancer prognosis and therapy prediction. Genome Med. 2014, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Kazantseva, M.; Mehta, S.; Eiholzer, R.A.; Hung, N.; Wiles, A.; Slatter, T.L.; Braithwaite, A.W. A mouse model of the Δ133p53 isoform: Roles in cancer progression and inflammation. Mamm. Genome 2018, 29, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Eiholzer, R.A.; Mehta, S.; Kazantseva, M.; Drummond, C.J.; McKinney, C.; Young, K.; Slater, D.; Morten, B.C.; Avery-Kiejda, K.A.; Lasham, A.; et al. Intronic TP53 Polymorphisms Are Associated with Increased Δ133TP53 Transcript, Immune Infiltration and Cancer Risk. Cancers 2020, 12, 2472. [Google Scholar] [CrossRef]
- Choi, Y.I.; Jakhongir, M.; Choi, S.J.; Kim, L.; Park, I.; Han, J.Y.; Kim, J.M.; Chu, Y.C. High-quality cell block preparation from scraping of conventional cytology slide: A technical report on a modified cytoscrape cell block technique. Malays. J. Pathol. 2016, 38, 295–304. [Google Scholar]
- Morten, B.C.; Scott, R.J.; Avery-Kiejda, K.A. Comparison of the QuantiGene 2.0 Assay and Real-Time RT-PCR in the Detection of p53 Isoform mRNA Expression in Formalin-Fixed Paraffin-Embedded Tissues—A Preliminary Study. PLoS ONE 2016, 11, e0165930. [Google Scholar] [CrossRef]


| Patient Information | |
|---|---|
| Age (Years) | |
| Average (±SD) | 58.6 ± 15.1 |
| Median | 57.5 |
| Range | 28–90 |
| <40 | 10 |
| 40–50 | 28 |
| >50 | 70 |
| Grade | |
| 1 | 31 (28.1%) |
| 2 | 24 (22.2%) |
| 3 | 53 (49.1%) |
| Tumour size (mm) | |
| Average (±SD) | 29.6 ± 15.8 |
| Median | 25 |
| Range | 7–90 |
| ≤25 | 55 |
| >25 | 53 |
| No. of positive LNs | |
| 0 | 50 |
| 1–3 | 40 |
| >3 | 18 |
| Hormone receptor status | |
| ER positive | 80 (74.1%) |
| PR positive | 73 (67.6%) |
| HER2 positive | 18 (16.7%) |
| TNBC | 15 (13.9%) |
| Clinicopathological Features | Total | No Truncating TP53 Mutation | Truncating TP53 Mutation | Spearman r | p Value |
|---|---|---|---|---|---|
| n = 103 | n = 74 (72%) | n = 29 (28%) | |||
| Age (years), median = 58 | 0.2281 | 0.0205 * | |||
| Range = 28–90 | |||||
| <40 | 10 | 9 | 1 | ||
| 40–50 | 27 | 21 | 6 | ||
| >50 | 66 | 44 | 22 | ||
| Grade | −0.8275 | <0.0001 * | |||
| 1 or 2 | 50 | 21 | 29 | ||
| 3 | 53 | 53 | 0 | ||
| Tumour size (mm), median = 25 | −0.2130 | 0.0307 * | |||
| ≤25 | 53 | 35 | 18 | ||
| >25 | 50 | 39 | 11 | ||
| No. of positive LNs | −0.1270 | 0.2010 | |||
| 0 | 48 | 33 | 15 | ||
| 1–3 | 37 | 25 | 12 | ||
| >3 | 18 | 16 | 2 | ||
| ER | 0.3825 | <0.0001 * | |||
| Positive | 75 | 46 | 29 | ||
| Negative | 28 | 28 | 0 | ||
| PR | 0.3373 | 0.0005 * | |||
| Positive | 70 | 43 | 27 | ||
| Negative | 33 | 31 | 2 | ||
| HER2, (n = 102) | −0.2348 | 0.0176 * | |||
| Positive | 18 | 17 | 1 | ||
| Negative | 84 | 56 | 28 | ||
| TNBC | −0.2585 | 0.0084 * | |||
| Yes | 15 | 15 | 0 | ||
| No | 88 | 59 | 29 | ||
| Clinicopathological Features | Low Levels | High Levels | p Value |
|---|---|---|---|
| n = 54 | n = 54 | ||
| Age (years), median = 58 | 0.0361 * | ||
| Range = 28–90 | |||
| <40 | 9 | 1 | |
| 40–50 | 12 | 16 | |
| >50 | 37 | 33 | |
| Grade | 0.8474 | ||
| 1 or 2 | 28 | 27 | |
| 3 | 26 | 27 | |
| Tumour size (mm), median = 25 | 0.0039 * | ||
| ≤25 | 20 | 35 | |
| >25 | 34 | 19 | |
| No. of positive LNs | 0.3009 | ||
| 0 | 29 | 21 | |
| 1–3 | 17 | 23 | |
| >3 | 8 | 10 | |
| ER | 0.1877 | ||
| Positive | 43 | 37 | |
| Negative | 11 | 17 | |
| PR | 0.5374 | ||
| Positive | 38 | 35 | |
| Negative | 16 | 19 | |
| HER2, (n = 107) | 0.8005 | ||
| Positive | 10 | 9 | |
| Negative | 44 | 45 | |
| TNBC | 0.0123 * | ||
| Yes | 3 | 12 | |
| No | 51 | 42 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steffens Reinhardt, L.; Groen, K.; Morten, B.C.; Bourdon, J.-C.; Avery-Kiejda, K.A. Cytoplasmic p53β Isoforms Are Associated with Worse Disease-Free Survival in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 6670. https://doi.org/10.3390/ijms23126670
Steffens Reinhardt L, Groen K, Morten BC, Bourdon J-C, Avery-Kiejda KA. Cytoplasmic p53β Isoforms Are Associated with Worse Disease-Free Survival in Breast Cancer. International Journal of Molecular Sciences. 2022; 23(12):6670. https://doi.org/10.3390/ijms23126670
Chicago/Turabian StyleSteffens Reinhardt, Luiza, Kira Groen, Brianna C. Morten, Jean-Christophe Bourdon, and Kelly A. Avery-Kiejda. 2022. "Cytoplasmic p53β Isoforms Are Associated with Worse Disease-Free Survival in Breast Cancer" International Journal of Molecular Sciences 23, no. 12: 6670. https://doi.org/10.3390/ijms23126670
APA StyleSteffens Reinhardt, L., Groen, K., Morten, B. C., Bourdon, J.-C., & Avery-Kiejda, K. A. (2022). Cytoplasmic p53β Isoforms Are Associated with Worse Disease-Free Survival in Breast Cancer. International Journal of Molecular Sciences, 23(12), 6670. https://doi.org/10.3390/ijms23126670