Identification and Defensive Characterization of PmCYP720B11v2 from Pinus massoniana
Abstract
:1. Introduction
2. Results
2.1. Cloning and Sequence Analysis of PmCYP720B11v2 from P. massoniana
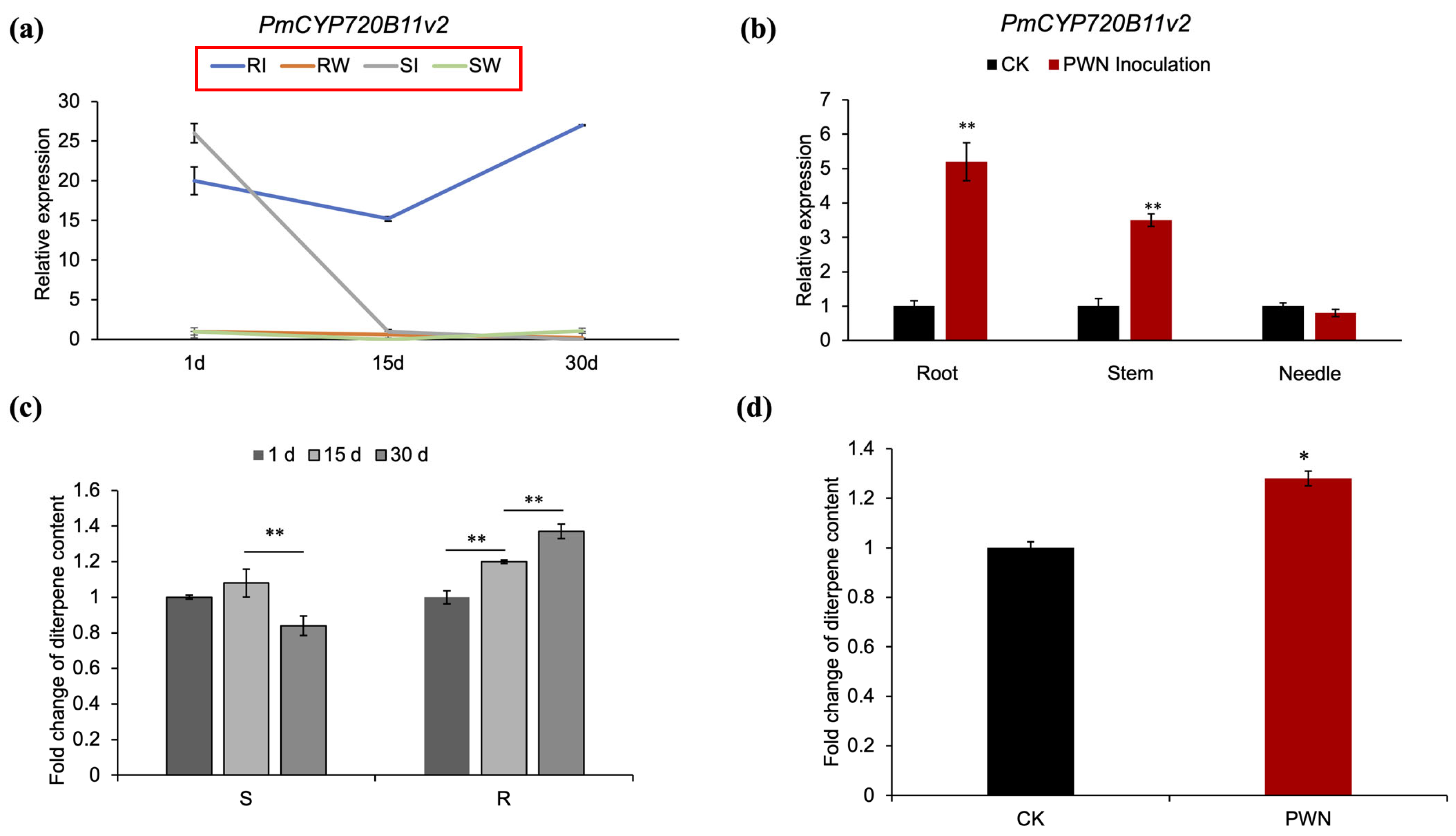
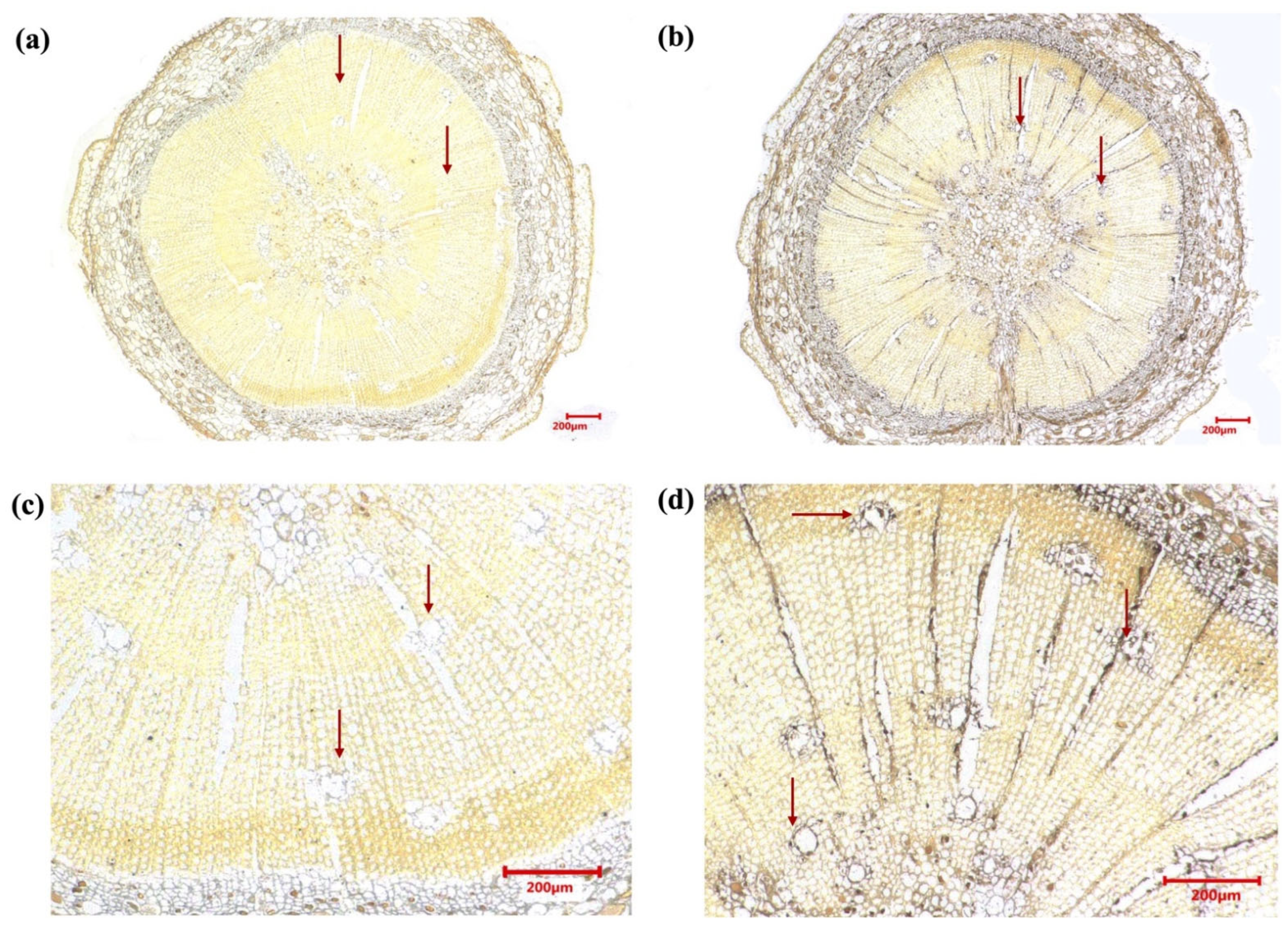
2.2. Tissue-Specific Expression Patterns and Localization of the PmCYP720B11v2 Gene in Resistant P. massoniana
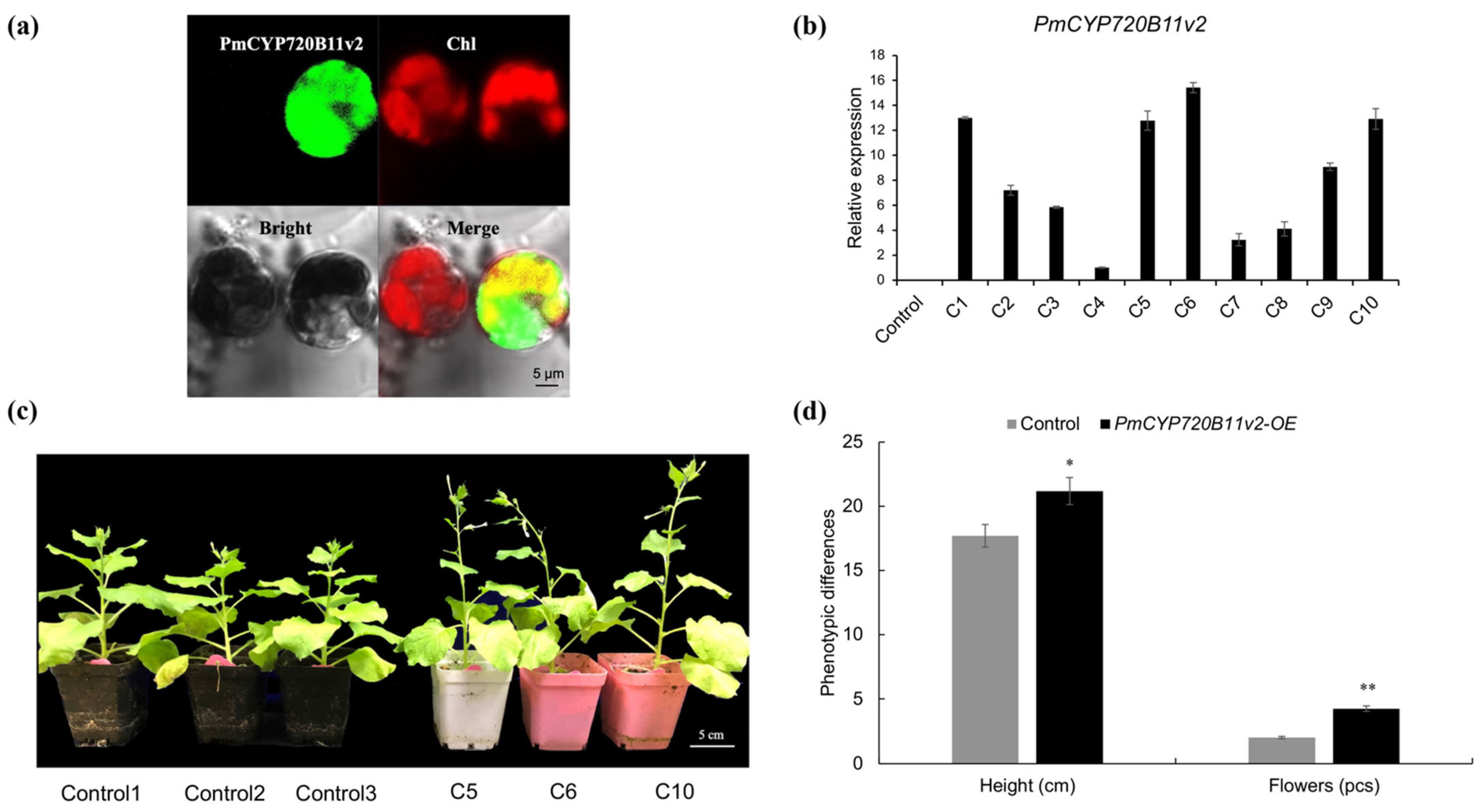
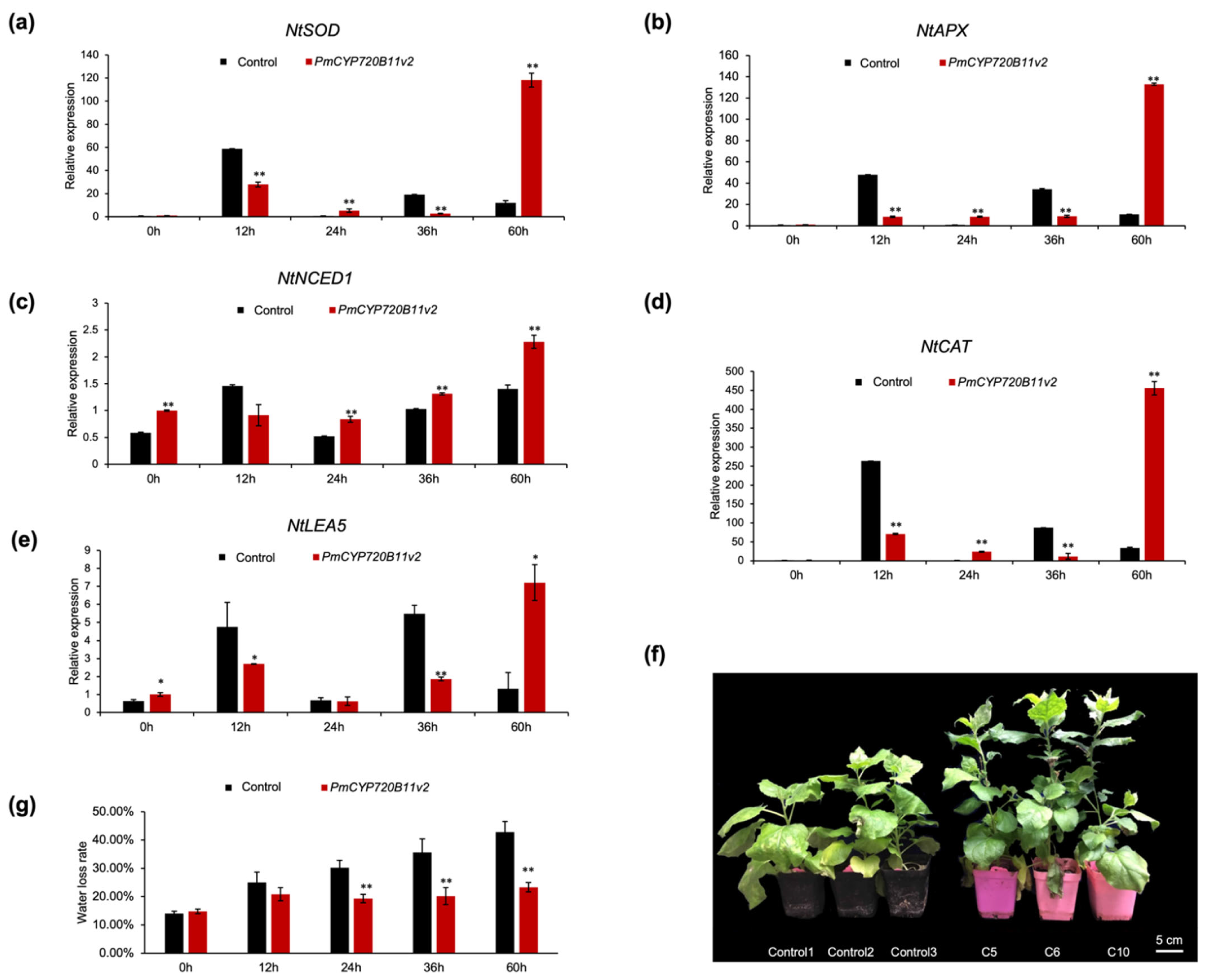
2.3. Overexpression of PmCYP720B11v2 Enhanced Growth and Drought Stress Tolerance in Tobacco
3. Discussion
3.1. Tissue-Specific Expression of PmCYP720B11v2 in P. massoniana in Response to PWN Infection
3.2. Overexpression of PmCYP720B11v2 Boosts Growth and Drought Stress Tolerance in Tobacco
4. Materials and Methods
4.1. Nematode Culture
4.2. Plant Material and Experimental Design
4.3. Identification of the PmCYP720B Family
4.4. Gene Cloning and Sequence Analysis
4.5. Real-Time Quantitative PCR (qRT–PCR) Analysis
4.6. Expression and Purification of Recombinant PmCYP720B11v2 Enzyme
4.7. Polyclonal Antibody Preparation
4.8. Immunohistochemistry
4.9. Subcellular Localization of the PmCYP720B11v2 Gene
4.10. Generation of PmCYP720B11v2 Transgenic Lines in Tobacco
4.11. Oleoresin Terpene Extraction from P. massoniana
4.12. GC–MS Analysis
4.13. Drought Stress Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamata, N.; Takeuchi, Y.; Gonthier, P.; Nicolotti, G. Pine wilt disease and other nematode diseases. J. Geophys. Res.-Atmos. 2013, 113, 84. [Google Scholar] [CrossRef] [Green Version]
- Valadas, V.; Oliveira, S.; Espada, M.; Laranjo, M.; Barbosa, P. The pine wood nematode, Bursaphelenchus xylophilus, in Portugal: Possible introductions and spread routes of a serious biological invasion revealed by molecular methods. Nematology 2012, 14, 899–911. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, J. Pinewood Nematode Bursaphelenchus xylophilus (Steiner and Buhrer) nickle. In Biological Invasions and Its Management in China; Springer: Singapore, 2017. [Google Scholar] [CrossRef]
- Ye, J.R. Epidemic Status of Pine Wilt Disease in China and ItsPrevention and Control Techniques and Counter Measures. Sci. Silvae Sin. 2019, 55, 1–10. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Wang, J.; You, Y.; Yang, Y.; Shi, Z.; Huang, X.; Zheng, L.; Li, Z.; Ming, A.; et al. Mixed-species plantation with Pinus massoniana and Castanopsis hystrix accelerates C loss in recalcitrant coniferous litter but slows C loss in labile broadleaf litter in southern China. For. Ecol. Manag. 2018, 422, 207–213. [Google Scholar] [CrossRef]
- Zhao, B.G.; Li, R.G. The Role of Bacteria Associated with the Pine Wood Nematode in Pathogenicity and Toxin-Production Related to Pine Wilt. In Pine Wilt Disease; Springer: Tokyo, Japan, 2008; pp. 250–259. [Google Scholar] [CrossRef]
- Bohlmann, J. Pine terpenoid defences in the mountain pine beetle epidemic and in other conifer pest interactions: Specialized enemies are eating holes into a diverse, dynamic and durable defence system. Tree Physiol. 2012, 32, 943–945. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Q.; Zhou, Z.; Yin, H.; Xie, Y.; Wei, Y. Two terpene synthases in resistant Pinus massoniana contribute to defence against Bursaphelenchus xylophilus. Plant Cell Environ. 2020, 44, 257–274. [Google Scholar] [CrossRef]
- Martin, D.; Bohlmann, J. Molecular Biochemistry and Genomics of Terpenoid Defenses in Conifers. Recent Adv. Phytochem. 2005, 39, 29–56. [Google Scholar] [CrossRef]
- Zulak, K.G.; Bohlmann, J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant Biol. 2010, 52, 86–97. [Google Scholar] [CrossRef]
- Dezene, P.W.; Bohlmann, J. The Role of Terpene Synthases in the Direct and Indirect Defense of Conifers Against Insect Herbivory and Fungal Pathogens. In Multigenic and Induced Systemic Resistance in Plants; Tuzun, S., Bent, E., Eds.; Springer: Boston, MA, USA, 2006; pp. 296–313. [Google Scholar] [CrossRef]
- Keeling, C.; Bohlmann, J. Diterpene resin acids in conifers. Phytochemistry 2006, 67, 2415–2423. [Google Scholar] [CrossRef]
- Boone, C.K.; Aukema, B.H.; Bohlmann, J.; Carroll, A.L.; Raffa, K.F. Efficacy of tree defense physiology varies with bark beetle population density: A basis for positive feedback in eruptive species. Can. J. For. Res. 2011, 41, 1174–1188. [Google Scholar] [CrossRef]
- McKay, S.A.B.; Hunter, W.L.; Godard, K.-A.; Wang, S.X.; Martin, D.M.; Bohlmann, J.; Plant, A.L. Insect Attack and Wounding Induce Traumatic Resin Duct Development and Gene Expression of (—)-Pinene Synthase in Sitka Spruce. Plant Physiol. 2003, 133, 368–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.; Tholl, D.; Gershenzon, J.; Bohlmann, J. Methyl Jasmonate Induces Traumatic Resin Ducts, Terpenoid Resin Biosynthesis, and Terpenoid Accumulation in Developing Xylem of Norway Spruce Stems. Plant Physiol. 2002, 129, 1003–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, B.; Madilao, L.L.; Ralph, S.; Bohlmann, J. Insect-Induced Conifer Defense. White Pine Weevil and Methyl Jasmonate Induce Traumatic Resinosis, de Novo Formed Volatile Emissions, and Accumulation of Terpenoid Synthase and Putative Octadecanoid Pathway Transcripts in Sitka Spruce. Plant Physiol. 2005, 137, 369–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeling, C.; Bohlmann, J. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006, 170, 657–675. [Google Scholar] [CrossRef] [PubMed]
- Celedon, J.M.; Bohlmann, J. Oleoresin defenses in conifers: Chemical diversity, terpene synthases and limitations of oleoresin defense under climate change. New Phytol. 2019, 224, 1444–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bathe, U.; Tissier, A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry 2019, 161, 149–162. [Google Scholar] [CrossRef]
- Calla, B. Signatures of selection and evolutionary relevance of cytochrome P450s in plant-insect interactions. Curr. Opin. Insect Sci. 2020, 43, 92–96. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Xia, X.J.; Zhou, Y.H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [Green Version]
- Udawat, P.; Jha, R.K.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a Cytosolic Abiotic Stress Responsive Universal Stress Protein (SbUSP) Mitigates Salt and Osmotic Stress in Transgenic Tobacco Plants. Front. Plant Sci. 2016, 7, 518. [Google Scholar] [CrossRef] [Green Version]
- Kodaira, K.-S.; Qin, F.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2 Zinc-Finger Proteins AZF1 and AZF2 Negatively Regulate Abscisic Acid-Repressive and Auxin-Inducible Genes under Abiotic Stress Conditions. Plant Physiol. 2011, 157, 742–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zeng, J.; Li, Y.; Rong, X.; Sun, J.; Sun, T.; Li, M.; Wang, L.; Feng, Y.; Chai, R.; et al. Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 2015, 6, 615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Hou, Y.; Xu, Y.; Peng, R.; et al. Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum. Genes 2019, 10, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamiru, M.; Undan, J.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H.; et al. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Ding, J.; Lee, D.; Lu, X.; Feng, Y.-Q.; Song, W. Overexpression of SoCYP85A1, a Spinach Cytochrome p450 Gene in Transgenic Tobacco Enhances Root Development and Drought Stress Tolerance. Front. Plant Sci. 2017, 8, 1909. [Google Scholar] [CrossRef] [Green Version]
- Ro, D.-K.; Arimura, G.-I.; Lau, S.Y.W.; Piers, E.; Bohlmann, J. Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc. Natl. Acad. Sci. USA 2005, 102, 8060–8065. [Google Scholar] [CrossRef] [Green Version]
- Geisler, K.; Jensen, N.B.; Yuen, M.M.; Madilao, L.; Bohlmann, J. Modularity of Conifer Diterpene Resin Acid Biosynthesis: P450 Enzymes of Different CYP720B Clades Use Alternative Substrates and Converge on the Same Products. Plant Physiol. 2016, 171, 152–164. [Google Scholar] [CrossRef]
- Hamberger, B.; Ohnishi, T.; Hamberger, B.; Séguin, A.; Bohlmann, J. Evolution of Diterpene Metabolism: Sitka Spruce CYP720B4 Catalyzes Multiple Oxidations in Resin Acid Biosynthesis of Conifer Defense against Insects. Plant Physiol. 2011, 157, 1677–1695. [Google Scholar] [CrossRef] [Green Version]
- Robert, J.A.; Madilao, L.L.; White, R.; Yanchuk, A.; King, J.; Bohlmann, J. Terpenoid metabolite profiling in Sitka spruce identifies association of dehydroabietic acid, (+)-3-carene, and terpinolene with resistance against white pine weevil. Botany 2010, 88, 810–820. [Google Scholar] [CrossRef]
- Zerbe, P.; Bohlmann, J. Plant diterpene synthases: Exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 2015, 33, 419–428. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, Y.; Xuelian, C.; Hao, Y.; Yongcheng, W.; Zhou, Z. Transcriptomic Profiling Reveals Differentially Expressed Genes Associated with Pine Wood Nematode Resistance in Masson Pine (Pinus massoniana Lamb.). Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.A.; Croteau, R.B. Resin-based defenses in conifers. Trends Plant Sci. 1999, 4, 184–190. [Google Scholar] [CrossRef]
- Kolosova, N.; Bohlmann, J. Conifer Defense Against Insects and Fungal Pathogens. In Growth and Defence in Plants; Springer: Berlin/Heidelberg, Germany, 2012; pp. 85–109. [Google Scholar] [CrossRef]
- Yahyaa, M.; Tholl, D.; Cormier, G.; Jensen, R.; Simon, P.W.; Ibdah, M. Identification and Characterization of Terpene Synthases Potentially Involved in the Formation of Volatile Terpenes in Carrot (Daucus carota L.) Roots. J. Agric. Food Chem. 2015, 63, 4870–4878. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng./Biotechnol. 2015, 148, 63. [Google Scholar] [CrossRef]
- Burke, J.L.; Carroll, A.L. The influence of variation in host tree monoterpene composition on secondary attraction by an invasive bark beetle: Implications for range expansion and potential host shift by the mountain pine beetle. For. Ecol. Manag. 2016, 359, 59–64. [Google Scholar] [CrossRef]
- Martin, D.M.; Fäldt, J.; Bohlmann, J. Functional Characterization of Nine Norway Spruce TPS Genes and Evolution of Gymnosperm Terpene Synthases of the TPS-d Subfamily. Plant Physiol. 2004, 135, 1908–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeling, C.I.; Weisshaar, S.; Ralph, S.G.; Jancsik, S.; Hamberger, B.; Dullat, H.K.; Bohlmann, J. Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol. 2011, 11, 43. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.C.; Keeling, C.I.; Bohlmann, J. The cytochrome P450 CYP6DE1 catalyzes the conversion of α-pinene into the mountain pine beetle aggregation pheromone trans-verbenol. Sci. Rep. 2019, 9, 1477. [Google Scholar] [CrossRef]
- Chiu, C.C.; Keeling, C.I.; Bohlmann, J. Cytochromes P450 Preferentially Expressed in Antennae of the Mountain Pine Beetle. J. Chem. Ecol. 2018, 45, 178–186. [Google Scholar] [CrossRef]
- Yaqoob, N.; Yakovlev, I.A.; Krokene, P.; Kvaalen, H.; Solheim, H.; Fossdal, C.G. Defence-related gene expression in bark and sapwood of Norway spruce in response to Heterobasidion parviporum and methyl jasmonate. Physiol. Mol. Plant Pathol. 2012, 77, 10–16. [Google Scholar] [CrossRef]
- Pham, T.; Chen, H.; Dai, L.; Vu, T.Q.T. Isolation a P450 gene in Pinus armandi and its expression after inoculation of Leptographium qinlingensis and treatment with methyl jasmonate. Russ. J. Plant Physiol. 2016, 63, 111–118. [Google Scholar] [CrossRef]
- Fahn, A. Secretory tissues in vascular plants. New Phytol. 1988, 108, 229–257. [Google Scholar] [CrossRef]
- Eyles, A.; Bonello, P.; Ganley, R.; Mohammed, C. Induced resistance to pests and pathogens in trees. New Phytol. 2009, 185, 893–908. [Google Scholar] [CrossRef]
- Ro, D.-K.; Bohlmann, J. Diterpene resin acid biosynthesis in loblolly pine (Pinus taeda): Functional characterization of abietadiene/levopimaradiene synthase (PtTPS-LAS) cDNA and subcellular targeting of PtTPS-LAS and abietadienol/abietadienal oxidase (PtAO, CYP720B1). Phytochemistry 2006, 67, 1572–1578. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Lazar, G.; Goodman, H.M. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 103, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Luong, P.; Huq, E. The F-Box Protein MAX2 Functions as a Positive Regulator of Photomorphogenesis in Arabidopsis. Plant Physiol. 2007, 145, 1471–1483. [Google Scholar] [CrossRef] [Green Version]
- Stirnberg, P.; Furner, I.J.; Leyser, H.M.O. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007, 50, 80–94. [Google Scholar] [CrossRef]
- Nelson, D.R.; Schuler, M.A.; Paquette, S.M.; Werck-Reichhart, D.; Bak, S. Comparative Genomics of Rice and Arabidopsis. Analysis of 727 Cytochrome P450 Genes and Pseudogenes from a Monocot and a Dicot. Plant Physiol. 2004, 135, 756–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christ, B.; Süssenbacher, I.; Moser, S.; Bichsel, N.; Egert, A.; Müller, T.; Kräutler, B.; Hörtensteiner, S. Cytochrome P450 CYP89A9 Is Involved in the Formation of Major Chlorophyll Catabolites during Leaf Senescence in Arabidopsis. Plant Cell 2013, 25, 1868–1880. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a Soybean Cytochrome P450 Family Gene Involved in the Jasmonic Acid and Ethylene Signaling Pathway, Enhances Plant Resistance to Biotic and Abiotic Stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef] [Green Version]
- An, J.-P.; Li, R.; Qu, F.-J.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. Ectopic expression of an apple cytochrome P450 gene MdCYPM1 negatively regulates plant photomorphogenesis and stress response in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 483, 1–9. [Google Scholar] [CrossRef]
- Kikuchi, T.; Jones, J.T.; Aikawa, T.; Kosaka, H.; Ogura, N. A family of glycosyl hydrolase family 45 cellulases from the pine wood nematode Bursaphelenchus xylophilus. FEBS Lett. 2004, 572, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Chen, F.; Luo, Y.-Q.; Wang, Z.; Xie, B.-Y. First isolation of pine wood nematode from Pinus tabuliformis forests in China. For. Pathol. 2012, 43, 59–66. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Q.; Zhou, Z.; Yin, H.; Xie, Y. Overexpression of geranyl diphosphate synthase (PmGPPS1) boosts monoterpene and diterpene production involved in the response to pine wood nematode invasion. Tree Physiol. 2021, 42, 411–424. [Google Scholar] [CrossRef]
- Chang, F.C.; Lu, C.M.; Sha, S. The Plant Biology Experiment; Nanjing Normal University Press: Nanjing, China, 2007; pp. 209–225. ISBN 978-7-81101-650-5. [Google Scholar]
- Finn, R.D.; Mistry, J.; Schuster-Böckler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef] [Green Version]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Gascuel, O. Neighbor-Joining Revealed. Mol. Biol. Evol. 2006, 23, 1997–2000. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Chen, Y.C.; Gao, M.; Yin, H.F.; Wu, L.W.; Wang, Y.D. Overexpression of geranyl diphosphate synthase small subunit 1 (LcGPPS.SSU1) T enhances the monoterpene content and biomass. Ind. Crops Prod. 2019, 143, 11926. [Google Scholar] [CrossRef]
- Chen, R.; He, X.; Chen, J.; Gu, T.; Liu, P.; Xu, T.; Teale, S.A.; Hao, D. Traumatic Resin Duct Development, Terpenoid Formation, and Related Synthase Gene Expression in Pinus massoniana Under Feeding Pressure of Monochamus alternatus. J. Plant Growth Regul. 2018, 38, 897–908. [Google Scholar] [CrossRef]
- Han, C.P.; Kok, L.F.; Wang, P.H.; Wu, T.S.; Tyan, Y.S.; Cheng, Y.W.; Lee, M.Y.; Yang, S.F. Scoring of p16ink4a immunohistochemistry based on independent nuclear staining alone can sufficiently distinguish between endocervical and endometrial adenocarcinomas in a tissue microarray study. Mod. Pathol. 2009, 22, 797–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schirawski, J.; Planchais, S.; Haenni, A.-L. An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: A simple and reliable method. J. Virol. Methods 2000, 86, 85–94. [Google Scholar] [CrossRef]
- Kim, E.-J.; Kim, K.-J. Cloning, expression, purification, crystallization and X-ray crystallographic analysis of (S)-3-hydroxybutyryl-CoA dehydrogenase from Clostridium butyricum. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 485–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karanikas, C.; Walker, V.; Scaltsoyiannes, A.; Comte, G.; Bertrand, C. High vs. low yielding oleoresin Pinus halepensis Mill. trees GC terpenoids profiling as diagnostic tool. Ann. For. Sci. 2010, 67, 412. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Xie, Y.; Yin, H.; Zhou, Z.; Liu, Q. Identification and Defensive Characterization of PmCYP720B11v2 from Pinus massoniana. Int. J. Mol. Sci. 2022, 23, 6640. https://doi.org/10.3390/ijms23126640
Liu B, Xie Y, Yin H, Zhou Z, Liu Q. Identification and Defensive Characterization of PmCYP720B11v2 from Pinus massoniana. International Journal of Molecular Sciences. 2022; 23(12):6640. https://doi.org/10.3390/ijms23126640
Chicago/Turabian StyleLiu, Bin, Yini Xie, Huanhuan Yin, Zhichun Zhou, and Qinghua Liu. 2022. "Identification and Defensive Characterization of PmCYP720B11v2 from Pinus massoniana" International Journal of Molecular Sciences 23, no. 12: 6640. https://doi.org/10.3390/ijms23126640
APA StyleLiu, B., Xie, Y., Yin, H., Zhou, Z., & Liu, Q. (2022). Identification and Defensive Characterization of PmCYP720B11v2 from Pinus massoniana. International Journal of Molecular Sciences, 23(12), 6640. https://doi.org/10.3390/ijms23126640





