Proteomic and Phosphoproteomic Analyses Reveal a Complex Network Regulating Pollen Abortion and Potential Candidate Proteins in TCMS Wheat
Abstract
:1. Introduction
2. Results
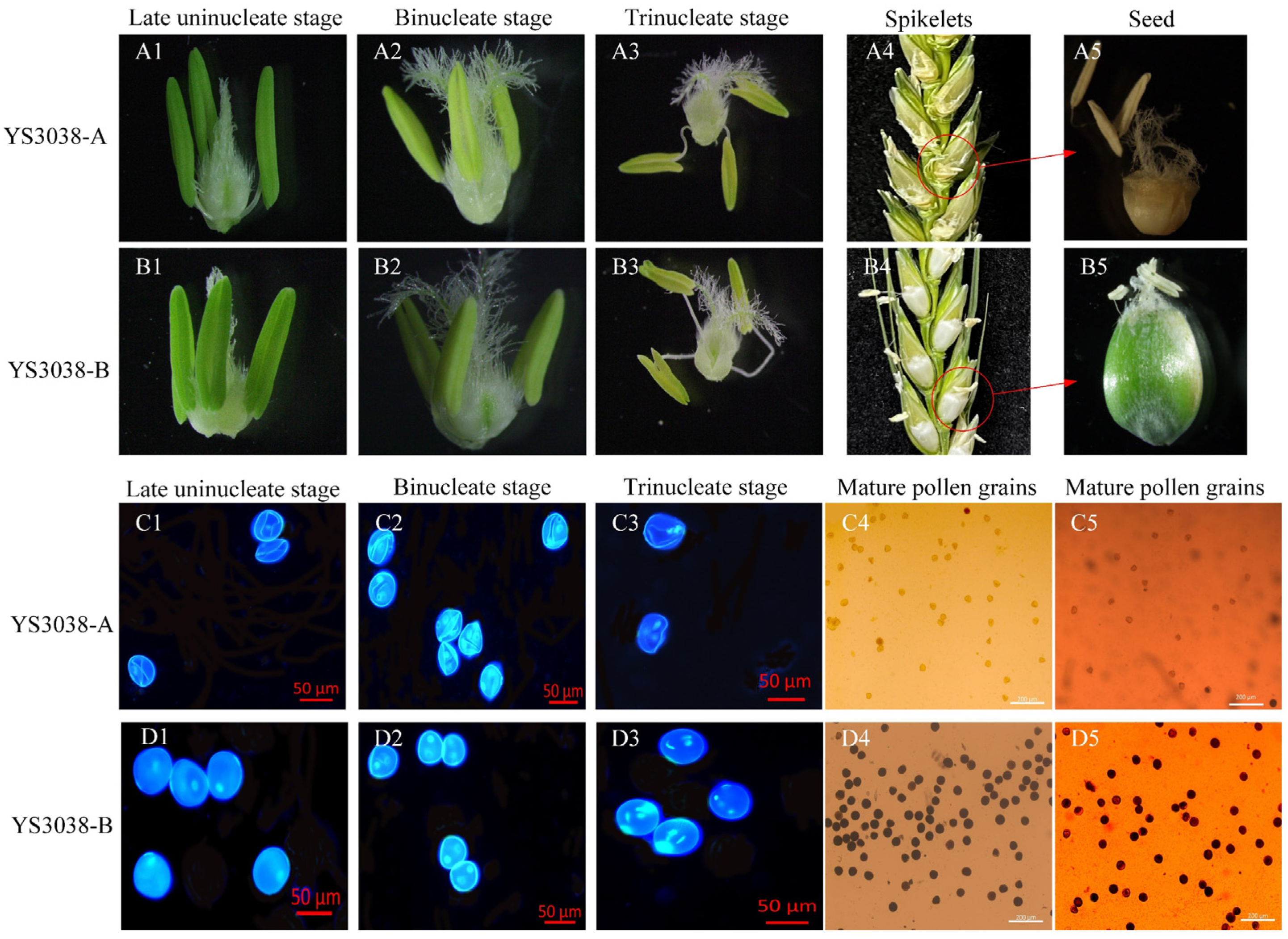
2.1. Phenotypic Characterization and Cytological Observations
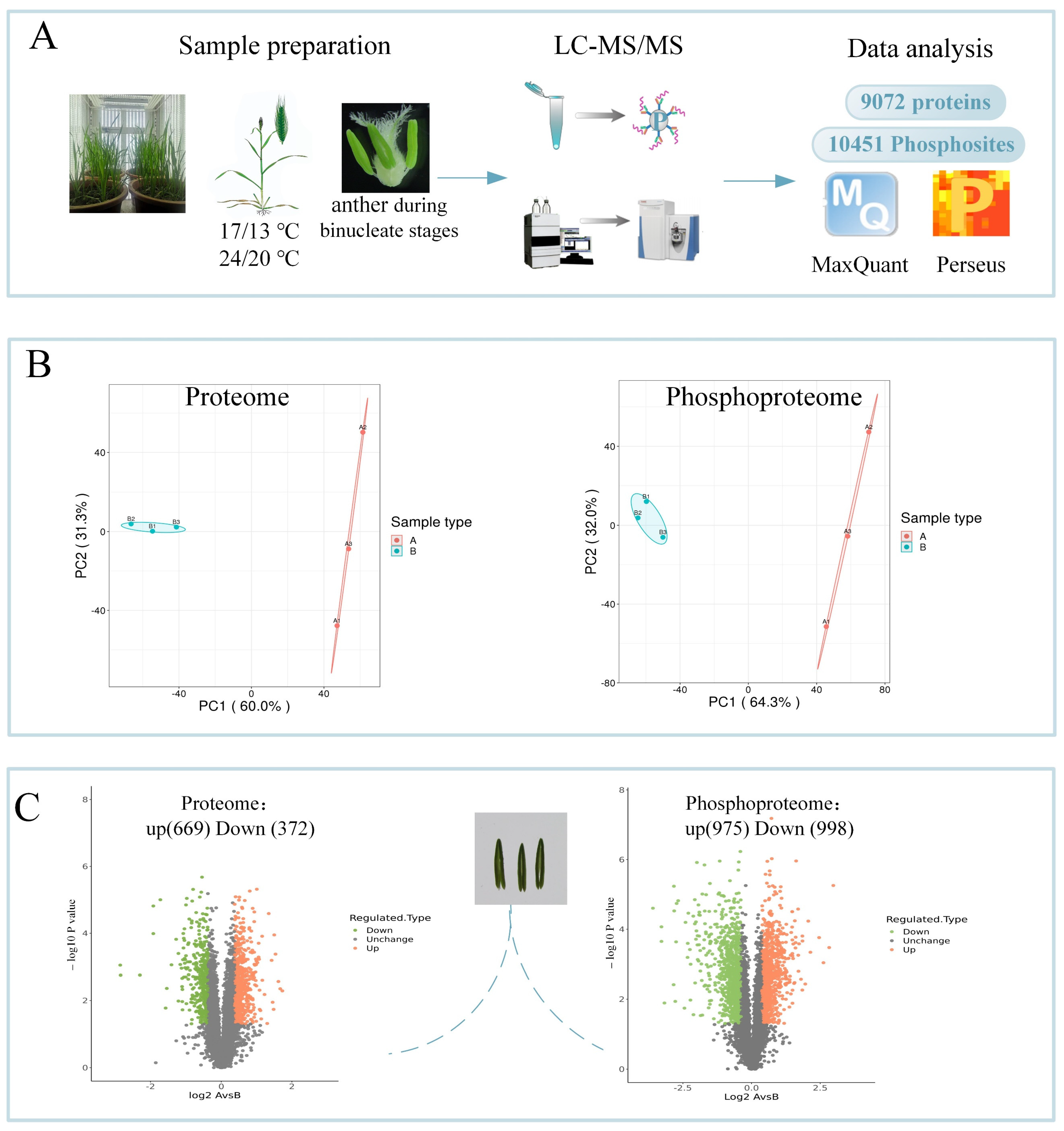
2.2. Proteomic and Phosphoproteomic Identification
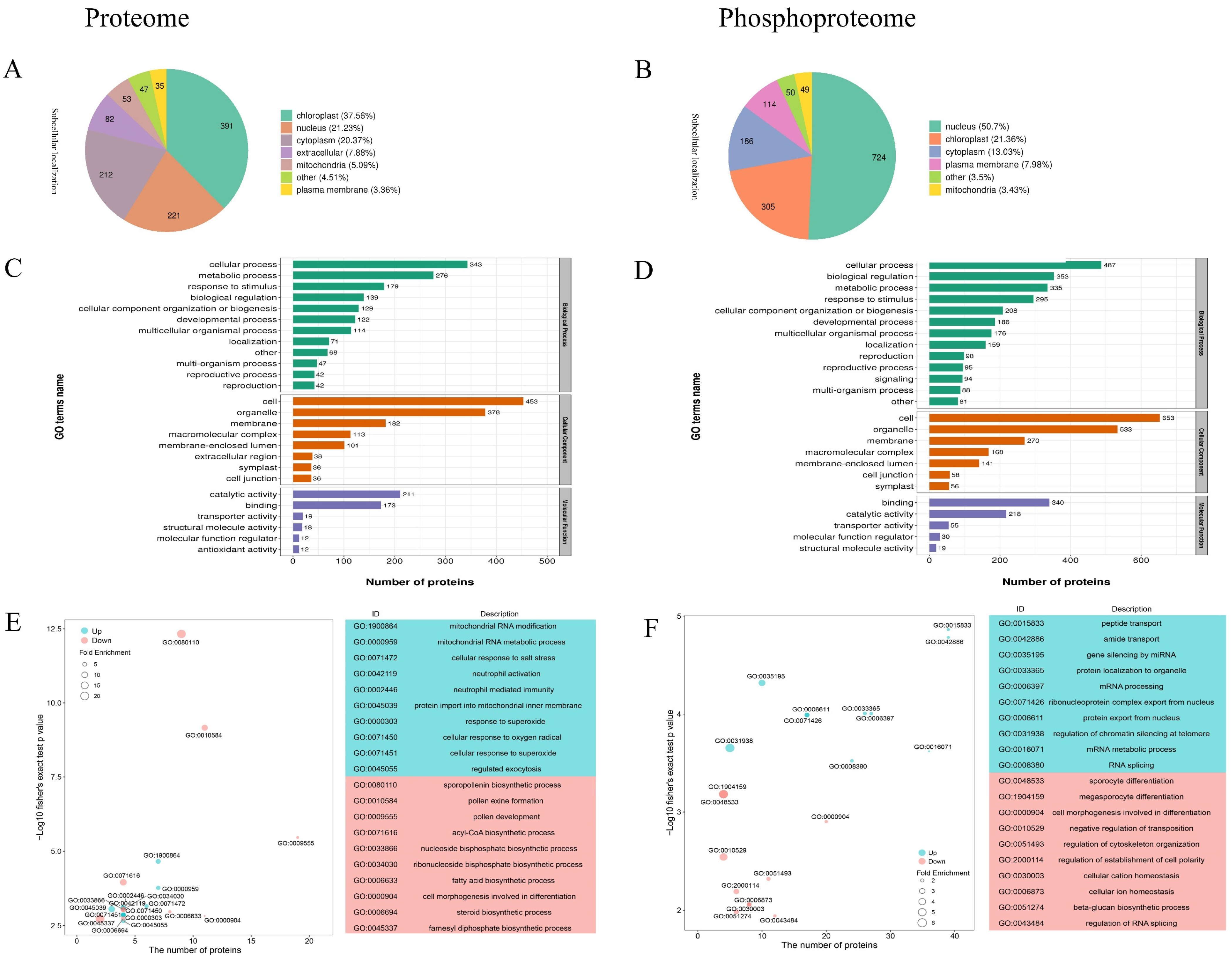
2.3. Subcellular Localization and Functional Classification
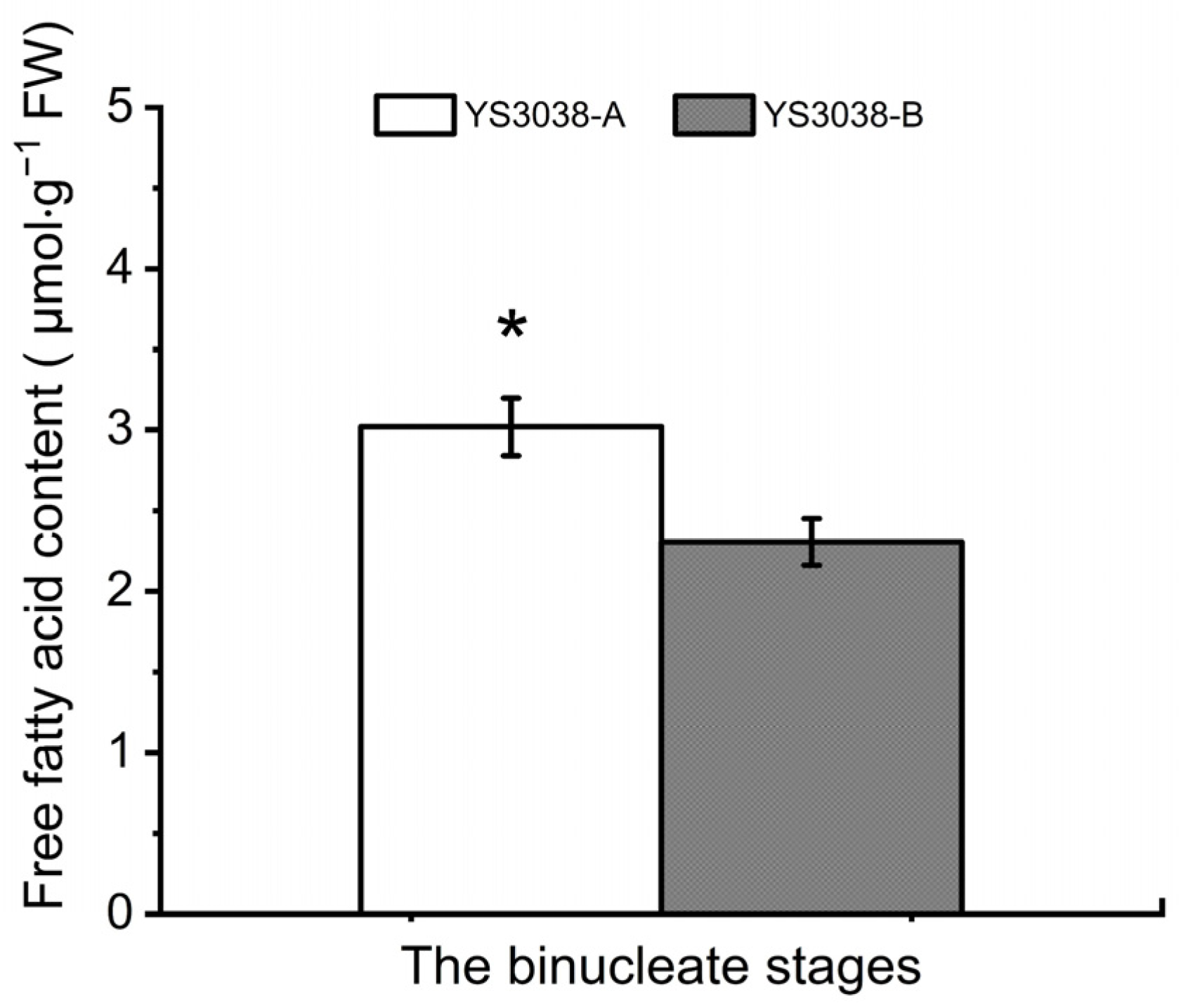
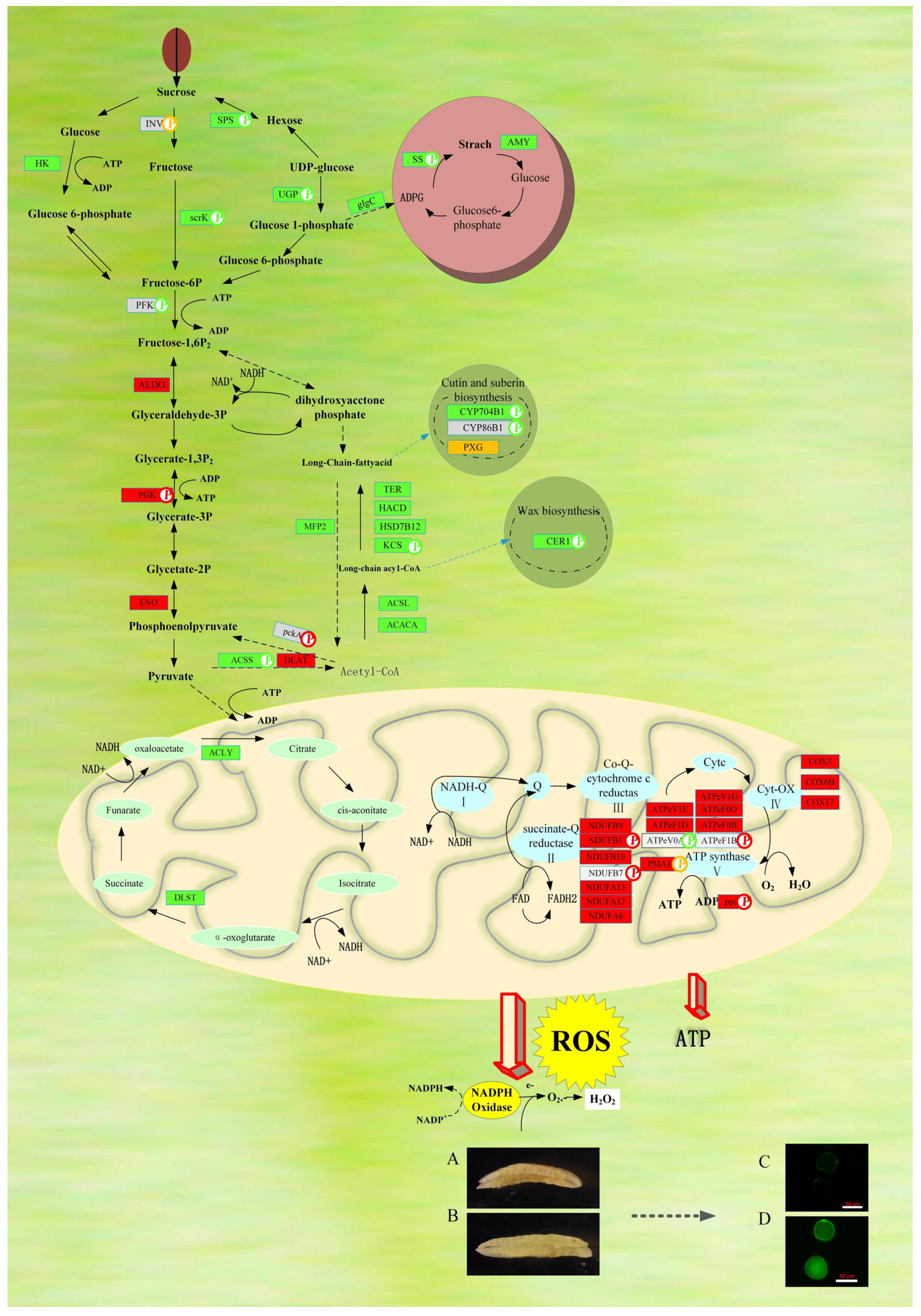
2.4. Differentially Expressed Proteins Highlight Fatty-Acid-Related Metabolic Pathways
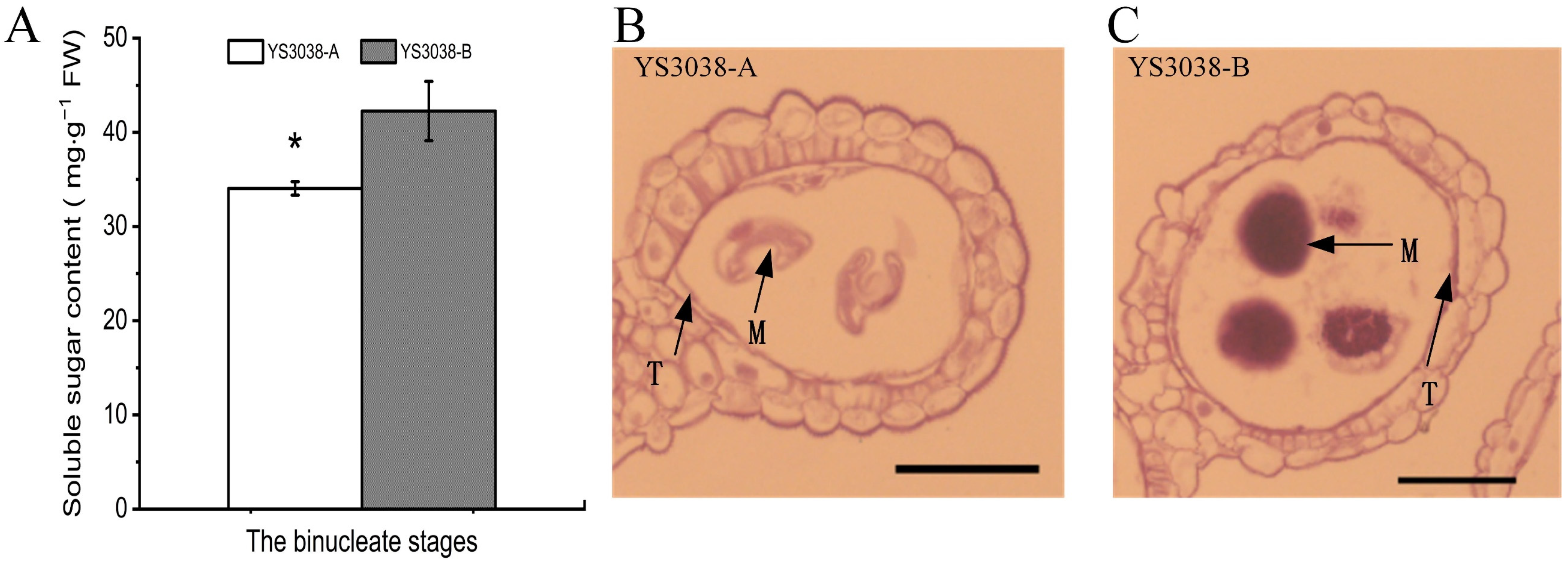
2.5. Abnormal Carbohydrate Metabolism in YS3038-A Anthers
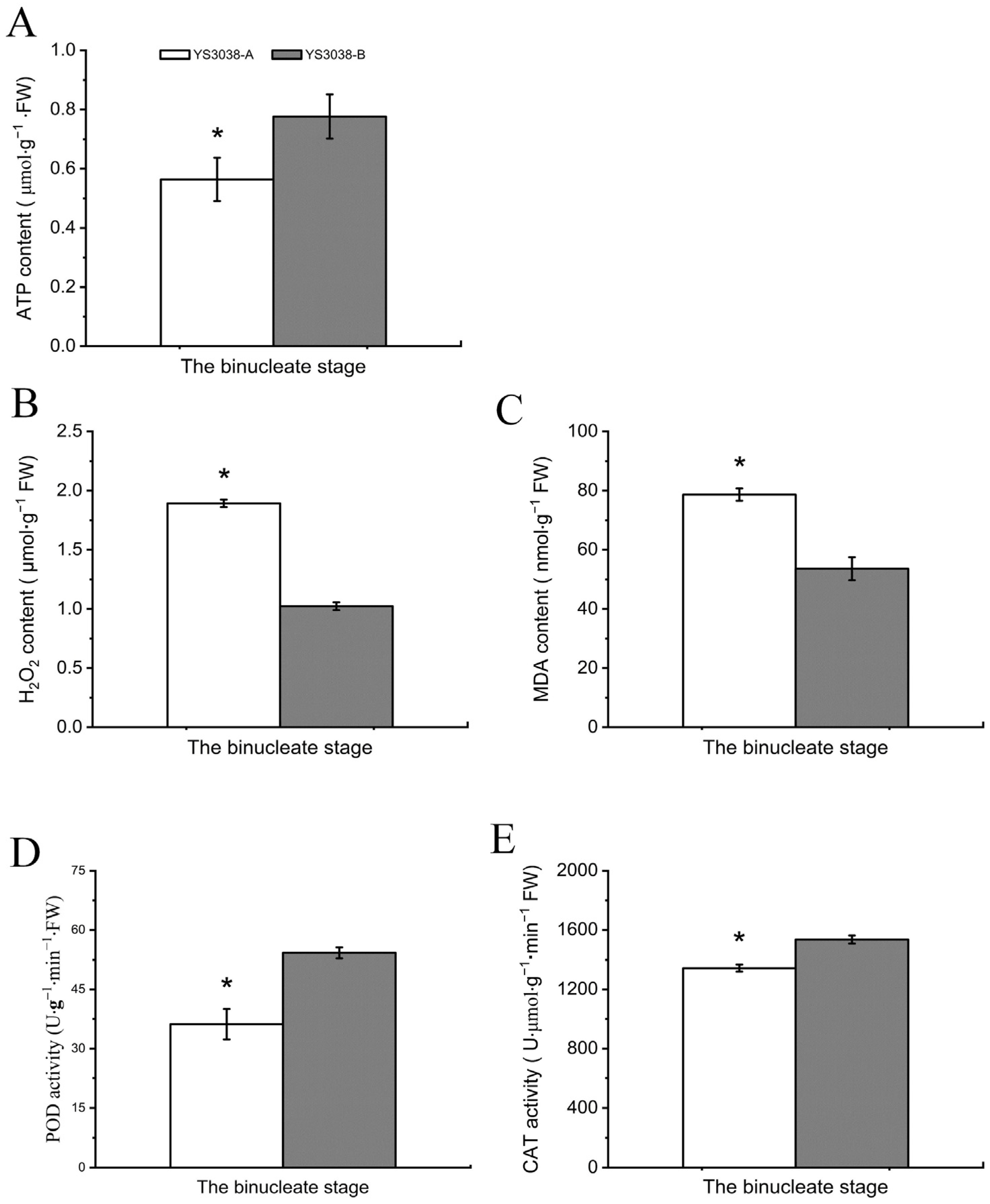
2.6. DAPs and DAPPS Interfere with the Electron Transport Chain, Reactive Oxygen Species Accumulation, and ATP Synthesis
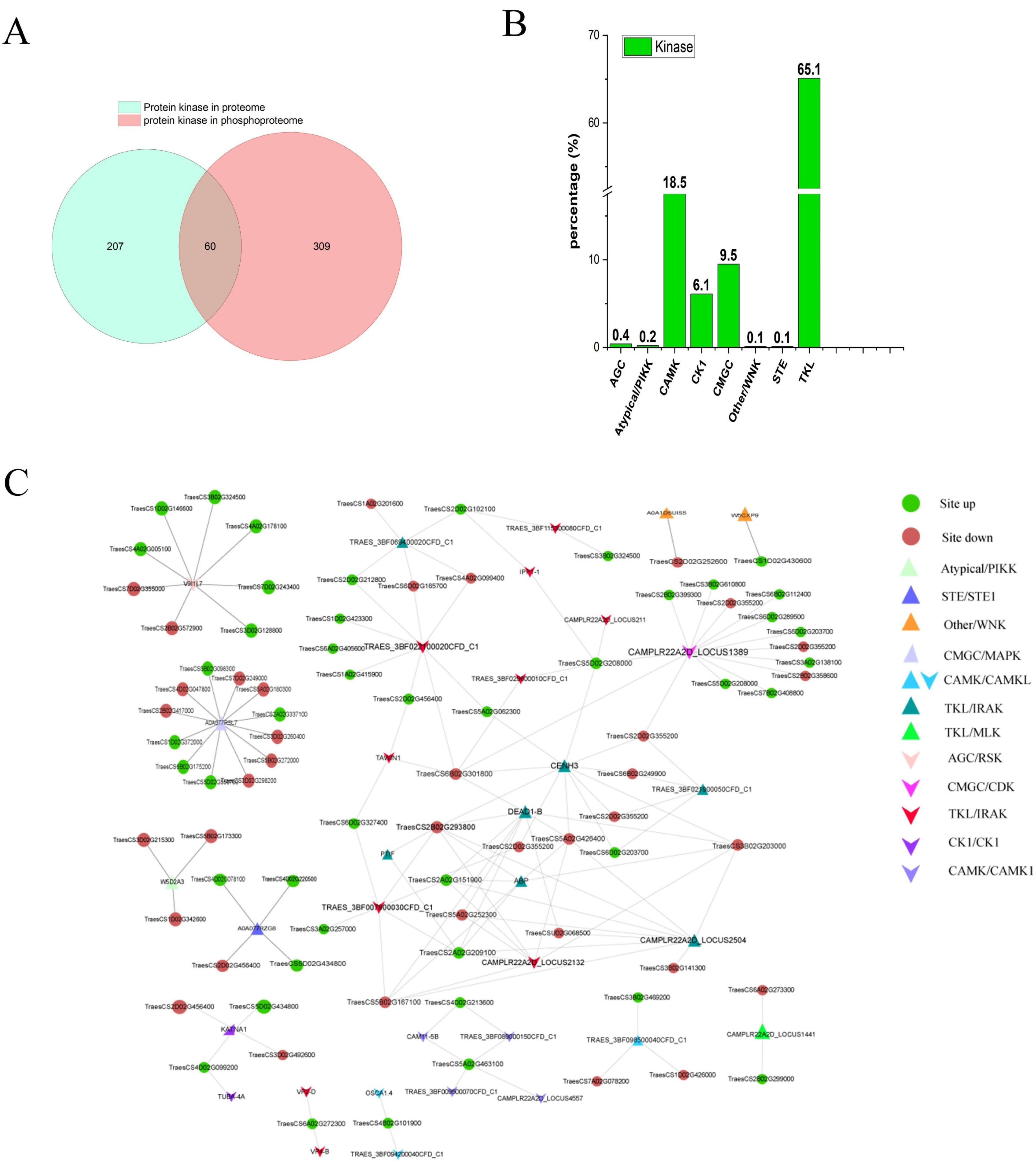
2.7. Phosphoproteomics Reveals the Protein Kinase Regulatory Network of YS3038
2.8. Other Sterility Candidate Proteins Identified among DAPs and DAPPs
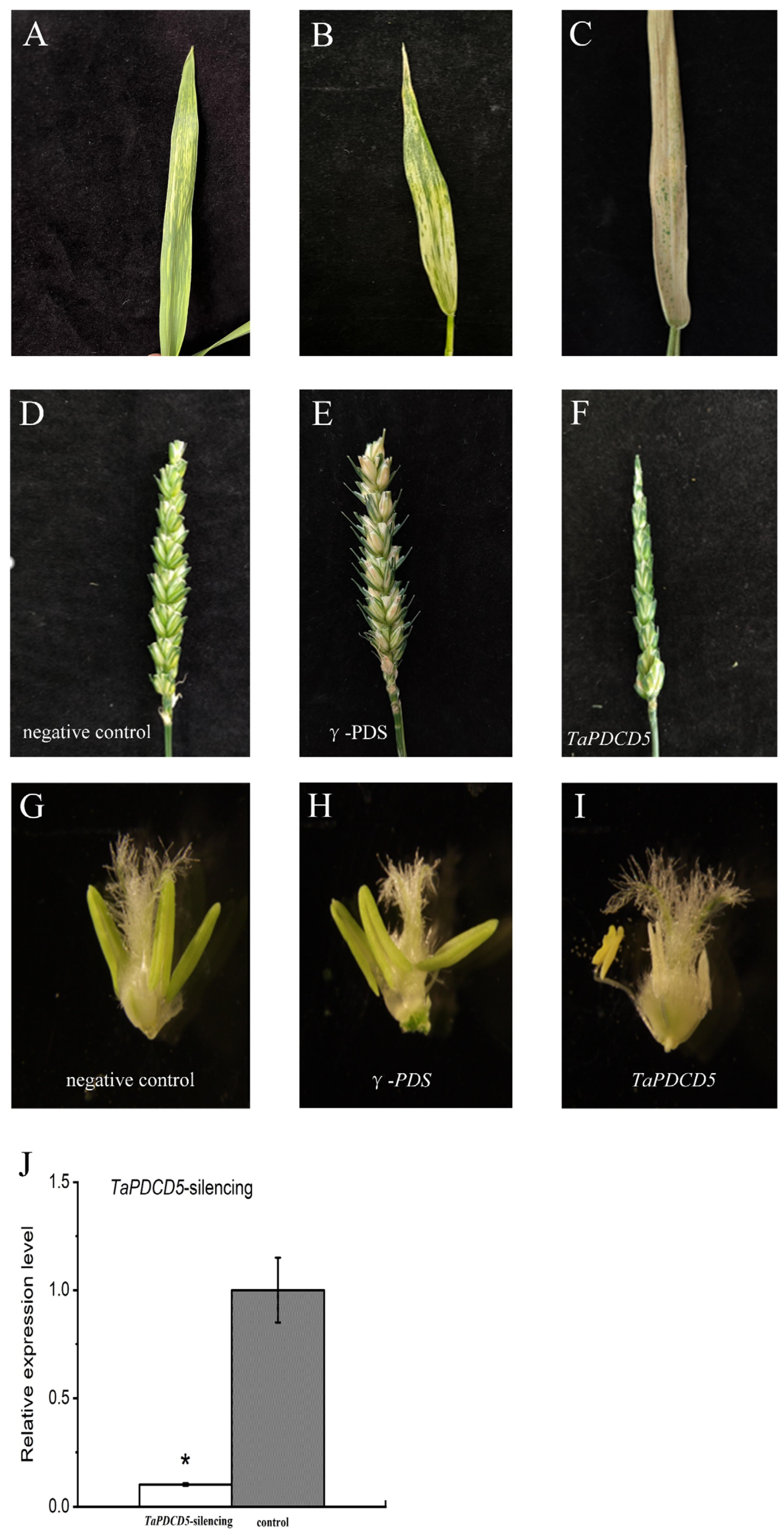
2.9. Functional Verification of TaPDCD5 via BSMV-VIGS
3. Discussion
3.1. A regulatory Network of Energy Metabolism a Candidate for TCMS in Wheat
3.2. Protein Kinase-Related DAPs and DAPPs
3.3. Transcription Factors
3.4. Epigenetic Differences
4. Materials and Methods
4.1. Plant Materials
4.2. Total Protein Extraction and Trypsin Digestion
4.3. TMT Labeling and Phosphopeptide Enrichment
4.4. LC-MS/MS Analysis
4.5. Database Search and Data Analysis
4.6. Phenotypic Characterization and Cytological Observations
4.7. Determination of Physiological Indexes of Anthers
4.8. Quantitative Real-Time PCR
4.9. Functional Verification of Candidate Genes via BSMV-VIGS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Longin, C.F.H.; Muhleisen, J.; Maurer, H.P.; Zhang, H.; Gowda, M.; Reif, J.C. Hybrid breeding in autogamous cereals. Theor. Appl. Genet. 2012, 125, 1087–1096. [Google Scholar] [CrossRef]
- Melonek, J.; Duarte, J.; Martin, J.; Beuf, L.; Murigneux, A.; Varenne, P.; Comadran, J.; Specel, S.; Levadoux, S.; Bernath-Levin, K.; et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat. Commun. 2021, 12, 1036. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- Liu, Y.J.; Li, D.; Gong, J.; Wang, Y.B.; Chen, Z.B.; Pang, B.S.; Chen, X.C.; Gao, J.G.; Yang, W.B.; Zhang, F.T.; et al. Comparative transcriptome and DNA methylation analysis in temperature-sensitive genic male sterile wheat BS366. BMC Genom. 2021, 22, 911. [Google Scholar] [CrossRef]
- Liu, H.; Sun, Z.; Hu, L.; Li, C.; Wang, X.; Yue, Z.; Han, Y.; Yang, G.; Ma, K.; Yin, G. Comparative Transcriptome Analysis of Male Sterile Anthers Induced by High Temperature in Wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 727966. [Google Scholar] [CrossRef]
- Song, X.Y.; Zhang, L.L.; Zeng, J.L.; Qian, H.H.; Li, H.B.; He, B.R. Development of thermo-sensitive cytoplasmic male sterile (TCMS) lines of wheat characterized by complete male sterility at lower-temperatures and partially restored fertility at higher-temperatures. Euphytica 2013, 192, 393–399. [Google Scholar] [CrossRef]
- Geng, X.; Ye, J.; Yang, X.; Li, S.; Zhang, L.; Song, X. Identification of Proteins Involved in Carbohydrate Metabolism and Energy Metabolism Pathways and Their Regulation of Cytoplasmic Male Sterility in Wheat. Int. J. Mol. Sci. 2018, 19, 324. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Shi, F.; Zhou, L.; Zhou, Y.; Liu, Z.; Ji, R.; Feng, H. iTRAQ-based proteomic analysis of fertile and sterile flower buds from a genetic male sterile line ‘AB01’ in Chinese cabbage (Brassica campestris L. ssp. pekinensis). J. Proteom. 2019, 204, 103395. [Google Scholar] [CrossRef]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef]
- Vu, L.D.; Zhu, T.; Verstraeten, I.; van de Cotte, B.; International Wheat Genome Sequencing, C.; Gevaert, K.; De Smet, I. Temperature-induced changes in the wheat phosphoproteome reveal temperature-regulated interconversion of phosphoforms. J. Exp. Bot. 2018, 69, 4609–4624. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Zhang, Z.; Long, H.; Zhang, Z.; Hong, Y.; Zhang, X.; You, C.; Liang, W.; Ma, H.; Lu, P. Proteomic and phosphoproteomic analyses reveal extensive phosphorylation of regulatory proteins in developing rice anthers. Plant J. 2015, 84, 527–544. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Luo, D.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, F.; Li, R. Development of High Yielding Glutinous Cytoplasmic Male Sterile Rice (Oryza sativa L.) Lines through CRISPR/Cas9 Based Mutagenesis of Wx and TGW6 and Proteomic Analysis of Anther. Agronomy 2018, 8, 290. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Zhang, Z.; You, C.; Zhang, X.; Lu, J.; Ma, H. Abundant protein phosphorylation potentially regulates Arabidopsis anther development. J. Exp. Bot. 2016, 67, 4993–5008. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Li, R.; Zhou, R. Comparative phosphoproteomic analysis reveals differentially phosphorylated proteins regulate anther and pollen development in kenaf cytoplasmic male sterility line. Amino Acids 2018, 50, 841–862. [Google Scholar] [CrossRef]
- Shukla, P.; Gautam, R.; Singh, N.K.; Ahmed, I.; Kirti, P.B. A proteomic study of cysteine protease induced cell death in anthers of male sterile tobacco transgenic plants. Physiol. Mol. Biol. Plants 2019, 25, 1073–1082. [Google Scholar] [CrossRef]
- Han, Y.; Gao, Y.; Zhao, Y.; Zhang, D.; Zhao, C.; Xin, F.; Zhu, T.; Jian, M.; Ding, Q.; Ma, L. Energy metabolism involved in fertility of the wheat TCMS line YS3038. Planta 2019, 250, 2159–2171. [Google Scholar] [CrossRef]
- Araki, S.; Le, N.T.; Koizumi, K.; Villar-Briones, A.; Nonomura, K.I.; Endo, M.; Inoue, H.; Saze, H.; Komiya, R. miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat. Commun. 2020, 11, 3115. [Google Scholar] [CrossRef]
- Hedgcoth, C.; el-Shehawi, A.M.; Wei, P.; Clarkson, M.; Tamalis, D. A chimeric open reading frame associated with cytoplasmic male sterility in alloplasmic wheat with Triticum timopheevi mitochondria is present in several Triticum and Aegilops species, barley, and rye. Curr. Genet. 2002, 41, 357–365. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, Q.; Zhang, L.; Li, Z.; Wang, C.; Zhang, G. Comparative Proteomic Analysis of Developmental Changes in P-Type Cytoplasmic Male Sterile and Maintainer Anthers in Wheat. Int. J. Mol. Sci. 2021, 22, 2012. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, M.; Thilges, K.; Cho, M.J.; Cigan, A.M. MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum L.) and combining mutations in all three homeologs causes male sterility. PLoS ONE 2017, 12, e0177632. [Google Scholar] [CrossRef]
- Singh, M.; Albertsen, M.C.; Cigan, A.M. Male Fertility Genes in Bread Wheat (Triticum aestivum L.) and Their Utilization for Hybrid Seed Production. Int. J. Mol. Sci. 2021, 22, 8157. [Google Scholar] [CrossRef]
- Shi, J.; Tan, H.; Yu, X.H.; Liu, Y.; Liang, W.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.; Kai, G.; et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Li, Y.; Li, Y.; Ma, Y.; Zhao, Y.; Wang, C.; Chi, H.; Chen, M.; Ding, Y.; Guo, X.; et al. Proteomic analysis reveals that sugar and fatty acid metabolisms play a central role in sterility of the male-sterile line 1355A of cotton. J. Biol. Chem. 2019, 294, 7057–7067. [Google Scholar] [CrossRef]
- Harris, J.R.; Boekema, E.J. Membrane Protein Complexes: Structure and Function; Sub-Cellular Biochemistry 87 Series; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Tiwari, B.S.; Belenghi, B.; Levine, A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 2002, 128, 1271–1281. [Google Scholar] [CrossRef] [Green Version]
- Vercellino, I.; Sazanov, L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 2022, 23, 141–161. [Google Scholar] [CrossRef]
- Jiang, G.; Hassan, M.A.; Muhammad, N.; Arshad, M.; Chen, X.; Xu, Y.; Xu, H.; Ni, Q.; Liu, B.; Yang, W.; et al. Comparative Physiology and Transcriptome Analysis of Young Spikes in Response to Late Spring Coldness in Wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 811884. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y.; Fang, Z.; Zhang, Y.; Song, Q.; Hou, Z.; Sun, K.; Song, Y.; Li, Y.; Ma, D.; et al. Cytological and Proteomic Analysis of Wheat Pollen Abortion Induced by Chemical Hybridization Agent. Int. J. Mol. Sci. 2019, 20, 1615. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Li, J.; Ma, L.; Wang, H.; Zhou, H.; Ni, E.; Jiang, D.; Liu, Z.; Zhuang, C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc. Natl. Acad. Sci. USA 2019, 116, 7549–7558. [Google Scholar] [CrossRef] [Green Version]
- Pradet-Balade, B.; Boulmé, F.; Beug, H.; Müllner, E.; Garcia-Sanz, J.A. Translation control: Bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001, 26, 225–229. [Google Scholar] [CrossRef]
- Wang, Y.; Zha, X.; Zhang, S.; Qian, X.; Dong, X.; Sun, F.; Yang, J. Down-regulation of the OsPDCD5 gene induced photoperiod-sensitive male sterility in rice. Plant Sci. 2010, 178, 221–228. [Google Scholar] [CrossRef]
- Li, X.; Gao, X.; Wei, Y.; Deng, L.; Ouyang, Y.; Chen, G.; Li, X.; Zhang, Q.; Wu, C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 2011, 23, 1416–1434. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Wu, J.; Wei, C.; Li, K.; He, G.; Attla, K.; Qian, X.; Yang, J. Interaction between programmed cell death 5 and calcineurin B-like interacting protein kinase 23 in Oryza sativa. Plant Sci. 2006, 170, 1150–1155. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, H.; Cho, J.I.; Nguyen, C.D.; Moon, S.; Park, J.E.; Park, H.R.; Huh, J.H.; Jung, K.H.; Guiderdoni, E.; et al. Deficiency of rice hexokinase HXK5 impairs synthesis and utilization of starch in pollen grains and causes male sterility. J. Exp. Bot. 2020, 71, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Zhao, X.; Shao, Z.; Wei, Z.; Wang, Y.; Zhu, L.; Zhao, J.; Sun, M.; He, R.; He, G. Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 2007, 19, 847–861. [Google Scholar] [CrossRef] [Green Version]
- Mu, H.; Ke, J.; Liu, W.; Zhuang, C.; Yip, W. UDP-glucose pyrophosphorylase2 (OsUgp2), a pollen-preferential gene in rice, plays a critical role in starch accumulation during pollen maturation. Sci. Bull. 2009, 54, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Huang, Y.; Loka, D.A.; Bai, H.; Liu, Y.; Wang, S.; Zhou, Z. Drought-induced disturbance of carbohydrate metabolism in anthers and male abortion of two Gossypium hirsutum cultivars differing in drought tolerance. Plant Cell Rep. 2020, 39, 195–206. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid Metabolism: Critical Roles in Male Fertility and Other Aspects of Reproductive Development in Plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Moller, B.L.; Preuss, D. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef] [Green Version]
- Selinski, J.; Scheibe, R. Pollen tube growth: Where does the energy come from? Plant Signal. Behav. 2014, 9, e977200. [Google Scholar] [CrossRef] [Green Version]
- Bennett, N.K.; Mai, K.N.; Darch, M.A.; Nakaoka, H.J.; Nakamura, K. Defining the ATPome reveals cross-optimization of metabolic pathways. Nat. Commun. 2020, 11, 4319. [Google Scholar] [CrossRef]
- Zancani, M.; Braidot, E.; Filippi, A.; Lippe, G. Structural and functional properties of plant mitochondrial F-ATP synthase. Mitochondrion 2020, 53, 178–193. [Google Scholar] [CrossRef]
- Zhang, W.; Wan, B.; Zhou, F.; Chen, H.; Li, X.; Lin, Y. Up- and Down-regulated Expression of OsCPK25/26 Results in Increased Number of Stamens in Rice. Plant Mol. Biol. Rep. 2014, 32, 1114–1128. [Google Scholar] [CrossRef]
- Kwon, C.T.; Kim, S.H.; Kim, D.; Paek, N.C. The Rice Floral Repressor Early flowering1 Affects Spikelet Fertility By Modulating Gibberellin Signaling. Rice 2015, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Pu, C.X.; Han, Y.F.; Zhu, S.; Song, F.Y.; Zhao, Y.; Wang, C.Y.; Zhang, Y.C.; Yang, Q.; Wang, J.; Bu, S.L.; et al. The Rice Receptor-Like Kinases dwarf and runtish spikelet1 and 2 Repress Cell Death and Affect Sugar Utilization during Reproductive Development. Plant Cell 2017, 29, 70–89. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Hua, L.; Dong, S.; Chen, H.; Zhu, X.; Jiang, J.; Zhang, F.; Li, Y.; Fang, X.; Chen, F. OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 2015, 84, 672–681. [Google Scholar] [CrossRef]
- Gomez, J.F.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Wang, F.; Geng, S.; Guan, J.; Tao, S.; Jia, M.; Sun, G.; Wang, Z.; Wang, K.; Ye, X.; et al. The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 2021, 20, 75–88. [Google Scholar] [CrossRef]
- Ma, W.; Wu, F.; Sheng, P.; Wang, X.; Zhang, Z.; Zhou, K.; Zhang, H.; Hu, J.; Lin, Q.; Cheng, Z.; et al. The LBD12-1 Transcription Factor Suppresses Apical Meristem Size by Repressing Argonaute 10 Expression. Plant Physiol. 2017, 173, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Xiang, X.J.; Sun, L.P.; Yu, P.; Yang, Z.F.; Zhang, P.P.; Zhang, Y.X.; Wu, W.X.; Chen, D.B.; Zhan, X.D.; Khan, R.M.; et al. The MYB transcription factor Baymax1 plays a critical role in rice male fertility. Theor. Appl. Genet. 2021, 134, 453–471. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, L.; Sun, L.; Yu, P.; Zhang, P.; Abbas, A.; Xiang, X.; Wu, W.; Zhang, Y.; Cao, L.; et al. OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 2019, 99, 175–191. [Google Scholar] [CrossRef]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 451. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zhang, Q.; Zhou, H.; Ni, F.; Wu, X.; Qi, Y. Rice MicroRNA effector complexes and targets. Plant Cell 2009, 21, 3421–3435. [Google Scholar] [CrossRef] [Green Version]
- Nonomura, K.; Morohoshi, A.; Nakano, M.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. A germ cell specific gene of the Argonaute family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 2007, 19, 2583–2594. [Google Scholar] [CrossRef] [Green Version]
- Qin, F.J.; Sun, Q.W.; Huang, L.M.; Chen, X.S.; Zhou, D.X. Rice SUVH histone methyltransferase genes display specific functions in chromatin modification and retrotransposon repression. Mol. Plant 2010, 3, 773–782. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, D.; Zhong, X.; Zhang, C.; Zhang, Q.; Zhou, D.X. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA 2012, 109, 5773–5778. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Shen, J.; Mao, H.; Xie, W.; Li, X.; Zhang, Q. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 2012, 5, 1210–1216. [Google Scholar] [CrossRef] [Green Version]
- Tütüncü Konyar, S.; Dane, F. Cytochemistry of pollen development in Campsis radicans (L.) Seem. (Bignoniaceae). Plant Syst. Evol. 2012, 299, 87–95. [Google Scholar] [CrossRef]
- Pinheiro, H.A.; DaMatta, F.M.; Chaves, A.R.M.; Fontes, E.P.B.; Loureiro, M.E. Drought tolerance in relation to protection against oxidative stress in clones of Coffea canephora subjected to long-term drought. Plant Sci. 2004, 167, 1307–1314. [Google Scholar] [CrossRef]
- Pompelli, M.F.; Martins, S.C.; Antunes, W.C.; Chaves, A.R.; DaMatta, F.M. Photosynthesis and photoprotection in coffee leaves is affected by nitrogen and light availabilities in winter conditions. J. Plant Physiol. 2010, 167, 1052–1060. [Google Scholar] [CrossRef]
- Han, Y.; Gao, Y.; Zhou, H.; Zhai, X.; Ding, Q.; Ma, L. Mitochondrial genes are involved in the fertility transformation of the thermosensitive male-sterile line YS3038 in wheat. Mol. Breed. 2021, 41, 61. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, L.; Hao, Y.; Liu, X.; Shao, L.; Wang, H.; Zhou, H.; Zhang, D.; Zhu, T.; Ding, Q.; Ma, L. Proteomic and Phosphoproteomic Analyses Reveal a Complex Network Regulating Pollen Abortion and Potential Candidate Proteins in TCMS Wheat. Int. J. Mol. Sci. 2022, 23, 6428. https://doi.org/10.3390/ijms23126428
Ma L, Hao Y, Liu X, Shao L, Wang H, Zhou H, Zhang D, Zhu T, Ding Q, Ma L. Proteomic and Phosphoproteomic Analyses Reveal a Complex Network Regulating Pollen Abortion and Potential Candidate Proteins in TCMS Wheat. International Journal of Molecular Sciences. 2022; 23(12):6428. https://doi.org/10.3390/ijms23126428
Chicago/Turabian StyleMa, Liting, Yuran Hao, Xiaorong Liu, Leilei Shao, Hairong Wang, Hao Zhou, Dazhong Zhang, Ting Zhu, Qin Ding, and Lingjian Ma. 2022. "Proteomic and Phosphoproteomic Analyses Reveal a Complex Network Regulating Pollen Abortion and Potential Candidate Proteins in TCMS Wheat" International Journal of Molecular Sciences 23, no. 12: 6428. https://doi.org/10.3390/ijms23126428
APA StyleMa, L., Hao, Y., Liu, X., Shao, L., Wang, H., Zhou, H., Zhang, D., Zhu, T., Ding, Q., & Ma, L. (2022). Proteomic and Phosphoproteomic Analyses Reveal a Complex Network Regulating Pollen Abortion and Potential Candidate Proteins in TCMS Wheat. International Journal of Molecular Sciences, 23(12), 6428. https://doi.org/10.3390/ijms23126428





