Abstract
Healthy limb joints are important for maintaining health and attaining longevity. Endochondral ossification (the replacement of cartilage with bone, occurring during skeletal development) is essential for bone formation, especially in long-axis bones. In contrast to endochondral ossification, chondrocyte populations in articular cartilage persist and maintain joint tissue into adulthood. Articular cartilage, a connective tissue consisting of chondrocytes and their surrounding extracellular matrices, plays an essential role in the mechanical cushioning of joints in postnatal locomotion. Osteoarthritis (OA) pathology relates to disruptions in the balance between anabolic and catabolic signals, that is, the loss of chondrocyte homeostasis due to aging or overuse of cartilages. The onset of OA increases with age, shortening a person’s healthy life expectancy. Although many people with OA experience pain, the mainstay of treatment is symptomatic therapy, and no fundamental treatment has yet been established. To establish regenerative or preventative therapies for cartilage diseases, further understanding of the mechanisms of cartilage development, morphosis, and homeostasis is required. In this review, we describe the general development of cartilage and OA pathology, followed by a discussion on anabolic and catabolic signals in cartilage homeostasis, mainly microRNAs.
1. Introduction
Osteoarthritis (OA) is the most common form of arthritis. It is characterized by gradual loss of articular cartilage, synovial membrane inflammation, osteophyte formation, and subchondral bone sclerosis. OA is associated with age-related loss of homeostatic balance between degradation and repair mechanisms in the articular cartilage [1,2,3,4,5]. This dysregulation induces senescence, differentiation, proliferation, and death in joint cells through gene and/or protein expression networks that switch from anabolic to catabolic outcomes. Cartilage-degrading enzymes, such as disintegrin and metalloproteinases with thrombospondin motifs (ADAMTS)-4 and ADAMTS-5, and matrix metalloproteinase (MMP)-13, are critical enzymes in OA pathogenesis [1,2,3]. Cartilage is composed of chondrocytes and an extracellular matrix whose major components include type II collagens and proteoglycans such as aggrecan. Chondrocytes in articular cartilage regulate cartilage homeostasis partly by synthesizing an extracellular matrix (ECM) rich in type II collagen, proteoglycans, and related macromolecules [6]. The advent of novel high throughput technologies has opened new perspectives in osteoarthritis research by mass spectrometry-based proteomic approaches [7]. In recent years, global proteomics studies using mass spectrometry have been widely conducted to elucidate the pathogenesis of articular cartilage [8]. Most studies have focused on proteins identified directly in the secretome of cartilage cell cultures [9,10,11,12,13,14,15,16,17,18,19], but proteomic analyses using cartilage tissue, cartilage explants [8], OA synovial fluid [20,21,22,23,24,25,26], and synovial cells have also been performed [8,27].
Once adult articular cartilage is damaged, its regeneration and repair are limited because of its hypovascularity. The catabolic and abnormal differentiation of chondrocytes due to aging or overuse of cartilage leads to loss of cartilage ECM, causing OA [28,29,30,31]. Articular cartilage maintains homeostasis by responding at the molecular level to various physiological stresses, including mechanical stress. When this balance is disrupted, osteoarthritis develops and progresses [32]. Aging is a major risk factor for OA, and aged chondrocytes exhibit multiple senescent phenotypes [33,34,35]. In particular, aging chondrocytes have reduced resistance to oxidative stress and impaired cellular homeostasis owing to autophagy dysfunction [36]. Synovitis, a common pathological change in osteoarthritic joints, is also a risk factor for OA, and severe synovitis exacerbates cartilage erosion [37,38]. Proinflammatory factors, such as interleukin-6 and tumor necrosis factor, are released from damaged joint tissue and induce synovial proliferation and inflammation, both known to contribute to synovitis [39]. OA can affect any joint but mainly affects the knee, hand, hip, and spine. The progressive and irreversible destruction of the cartilage matrix can cause joint pain and disability, which affects the quality of life [40,41]. Although many people with OA suffer from pain, available treatments in the early stage are limited to exercise therapy and symptomatic therapy, such as pharmacologic therapy, and joint replacement surgery is often indicated in the late phase of the disease. Fundamental treatments, such as pharmacologic interventions that could alter the progressive loss of articular cartilage and regenerate articular chondrocytes, are not available. To establish regenerative or preventative therapies for cartilage diseases, further understanding of the mechanisms of cartilage development, morphosis, and homeostasis is required. In this review article, we describe the general development of cartilage and OA pathology, followed by a discussion of anabolic and catabolic signals involved in cartilage homeostasis, focusing on noncoding RNAs (ncRNAs), especially microRNAs (miRNAs).
2. The Role of Sox9 on Chondrogenesis
SRY-Box Transcription Factor 9 (Sox9) is a member of the Sox family of transcription factors that are characterized by a high-mobility group (HMG)-box DNA-binding domain. Sox9 plays a pivotal role in male sex development because its sequence is at least 50% identical to that of the sex-determining factor SRY [42]. Sox9 is expressed early in mesenchymal condensations throughout the embryo and is an essential cartilage-promoting factor during cartilage development and skeletal formation. To identify the role of Sox9 in cartilage development, Bi et al. generated Sox−/− embryonic stem (ES) cells. The teratomas derived from Sox9−/− ES cells did not form any cartilage or expressed the chondrocyte-specific markers collagen type II alpha 1 (Col2a1), collagen type IX alpha 2 (Col9a2), collagen type XI alpha 2 (Col11a2), or Aggrecan (Acan). In mouse chimeras, Sox9−/− cells were unable to express any chondrocyte-specific ECM genes, such as Col2a1 [43]. In subsequent studies, Sox9 heterozygous (Sox9+/−) mice showed cartilage-related defects and died shortly after birth [44]. These studies demonstrate that Sox9 plays a key role in chondrocyte differentiation and cartilage formation.
The chondrocyte differentiation marker Col2a1, which is expressed abundantly in the early stages of embryo development, colocalizes with Sox9 in all chondroprogenitor cells. Col2a1 encodes type II collagen, which is a major structural component of the cartilage. It is expressed in chondrogenic tissues before chondrocyte differentiation [45]. Sox9 binds directly to the Col2a1 enhancer element to guide the transcription of the gene in chondrocytes. When chondrocytes hypertrophy in the growth plate, collagen type X alpha 1 (Col10a1) expression is activated. The Sox trio then disappears simultaneously, and Col2a1 slowly disappears. Col1a1 expression is activated as cartilage is replaced by bone [46]. When Sox9 activity is reduced, the production of cartilage matrix proteins such as type II collagen is inhibited, leading to major skeletal abnormalities [47].
Several studies show that SRY-Box Transcription Factor 5 (Sox5) and SRY-Box Transcription Factor 6 (Sox6) can activate Sox9 in developing cartilage cells [48,49,50]. Researchers also found that L-Sox5 (a new form of Sox5) and Sox6 are coexpressed with Sox9 during chondrogenesis and that these three Sox transcription factors cooperate with each other in the activation of the chondrocyte differentiation marker Col2a1 [46,51,52].
3. The Effect of Sox9 on Cartilage Homeostasis and OA
Cartilage is vital throughout vertebrate life, and Sox9 is essential for cartilage development. In addition, Acan is a major ECM protein of both the growth plate and articular cartilage, and its expression has been detected in all articular cartilages throughout development and beyond [53]. To examine the function of Sox9 postnatally, Haseeb et al. generated a cartilage-specific Sox9 conditional knockout Sox9fl/fl;AcanCreERT2/+ mice line, which, when given a tamoxifen shot, deletes Sox9. In 3-month-old Sox9-deleted mutant mice, the loss of proteoglycans and hypertrophic zones was observed. Additionally, upon performing destabilization of the medial meniscus (DMM) surgery on experimental mice, the Osteoarthritis Research Society International (OARSI) scale was found to be higher in Sox9-deleted mutant mice than in control mice. In conclusion, Sox9 is required to keep growth plates open and articular cartilage resistant to OA [54,55]. These results confirmed that Sox9 plays a vital role postnatally.
Oh et al. performed chromatin immunoprecipitation sequencing (ChIP-Seq) to identify genes that harbor Sox9-interaction sites and RNA sequencing to identify genes affected by Sox9. Their results show that Sox9 regulates a specific set of cartilage ECM genes, including Acan and Col2a1, and controls the differentiation of cartilage ECM cells [21,56]. Additionally, a recent ChIP-seq analysis using the CRISPR/Cas system revealed the existence of a rib cage-specific enhancer (RCSE) located approximately 1 Mb upstream of Sox9 [57]. Multiple additional analyses with CRISPR-ChIP-mass spectrometry (CRISPR-ChIP-MS) demonstrated that the transcription factor STAT3 regulates the expression of Sox9 via this RCSE region [57]. As chondrocytes are the only cell type in the cartilage ECM, cartilage repair is highly dependent on correct chondrogenic differentiation of resident progenitor cells and ECM anabolism by differentiated chondrocytes [22,58]. Studies have demonstrated that upregulation of SOX9 could inhibit IL-1β-induced inflammation in human chondrocytes, and SOX9 transduction can renew the capacity of late passage human OA articular chondrocytes to form cartilage ECM [59]. Tankyrase-mediated poly(ADP-ribosyl)ation (PARylation) of Sox9 plays an essential role in the regulation of Sox9 ubiquitination and degradation. Sox9 binds to Tnks and Tnks2, which encode tankyrase, a regulator of cartilage matrix anabolism. Inhibition of tankyrase increases Sox9 expression, promotes cartilage ECM synthesis, and enhances chondrogenic differentiation of mesenchymal stem cells. Delivery of tankyrase inhibitors can prevent OA in mouse knee joints [60]. Thus, these findings could be applied to future studies on OA therapeutic potential.
4. ncRNAs Involved in Cartilage Homeostasis and OA
More than 90% of the human DNA is actively transcribed, however, only 2% encodes proteins. The majority of the transcripts are ncRNAs. They are classified according to their biosynthesis, length, and mechanisms of action. After transcription, ncRNAs may form short, long, and circular ncRNAs with unique secondary and tertiary structures. ncRNAs are transcribed but not translated into proteins and perform their biological functions at the RNA level [61]. Short ncRNAs are less than 200 nucleotides in length, and include miRNAs, small nucleolar RNAs (snoRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), transition RNAs (tRNAs), tRNA-derived fragments (tRFs), and Y RNA fragments. Among short ncRNAs, miRNAs are the most frequently studied, but there have also been reports on other short ncRNAs associated with OA.
After the first profiling study on snoRNAs altered in OA was performed, several snoRNAs were studied through loss-of-function and gain-of-function experiments [62]. Differentially expressed snoRNAs have been identified in aging and OA, and their knockdown or overexpression have been shown to alter the expression of chondrogenesis, cartilage hyperplasia, and OA-related genes [62].
tRFs are novel regulators of post-transcriptional gene expression. However, their expression profiles and role in post-transcriptional gene regulation in chondrocytes are unknown. In 2020, a tRF in the cartilage was reported for the first time. The expression profile of tRFs is altered in OA cartilage and chondrocytes stimulated with IL-1b, whereas tRF-3003a represses Janus kinase 3 (JAK3) gene expression in chondrocytes [63]. In addition, the expression of specific tRFs was shown to be different in old chondrocytes compared to young chondrocytes [64,65].
Recently, there has been a growing interest in competing endogenous RNAs (ceRNAs), such as circular RNAs (circRNAs) and long ncRNAs (lncRNAs), which act as miRNA sponges, although the physiological significance of ceRNAs is not well understood. Several ncRNAs have been reported to be involved in cartilage development and homeostasis [66].
4.1. MiRNAs
MiRNAs are short ncRNAs that regulate gene expression by altering target mRNA stability and inhibiting protein synthesis. In most cases, miRNAs are transcribed into primary miRNAs (pri-miRNAs) by RNA polymerase II. Pri-miRNAs have a cap structure, a poly-A tail, and a loop structure. Both the cap structure and poly-A tail are cleaved by the ribonuclease enzyme III Drosha to form a precursor miRNA (pre-miRNA) that is transported to the cytoplasm by exportin 5. When the loop structure is cleaved by Dicer, a ribonuclease enzyme III, mature miRNA is generated.
MiRNAs have been found to be important for mammalian development. In 2005, Dicer was found to be essential for vertebrate limb morphogenesis. Targeted removal of Dicer in mouse limbs results in the formation of much smaller limbs [67]. The importance of Dicer in skeletal development was confirmed in 2008 when mice with their Dicer gene deleted in cartilage showed a reduction in the proliferation of chondrocytes in Dicer-null growth plates. Severe skeletal developmental defects were also observed [68]. Iliopoulos et al. screened 365 miRNA genes and found 16 miRNA gene signatures that were differentially expressed in OA [69]. Using bioinformatics analysis, Cong et al. identified 46 differentially expressed miRNAs involved in chondrocyte apoptosis, autophagy, differentiation, and ECM degradation [70].
4.1.1. MiRNA-140
Several miRNAs play important roles in cartilage development and homeostasis. MiRNA-140 (miR-140) is known to be a cartilage-specific miRNA in embryos and zebrafish and a critical regulator of cartilage development and homeostasis [71,72,73,74]. It was also found that miR-140 expression was reduced in human OA cartilage [69,73]. MiR-140 is located in intron 16 of the WW domain-containing protein 2 (Wwp2), which is a member of the C2-WW-HECT family (NEDD4 family) of E3 ubiquitin ligases [75]. It acts as an acceptor of ubiquitin from E2 enzymes and then transfers ubiquitin to a specific lysine residue on the substrate [76], which is an intronic miRNA [50]. The expression level of Wwp2, the host gene of miR-140, is reduced in OA.
The deletion of Sox9 diminishes the expression of miR-140 [74]. The miR-140 primary transcript is an intron-retained RNA coexpressed with Wwp2, which is directly induced by Sox9 through binding to intron 10 of the Wwp2 gene during chondrogenesis [77]. Nakamura et al. identified several Sox9 binding sites upstream of that for miR-140 within the Wwp2 gene in humans, mice, and zebrafish, and showed that miR-140 is downstream of the transcription factor Sox9 in developing zebrafish and mammalian cells [78]. Yamashita et al. demonstrated that the proximal upstream region of pri-miR-140, located in intron 10 of Wwp2, has in vivo promoter activity. These results suggest that miR-140 may be derived from its own specific transcript via Sox9 binding during chondrogenesis (Figure 1) [50]. Sox5 and Sox6 also function as regulators of cartilage development by boosting Sox9 activation of Col2a1 and Agc1 [46,79]. Yamashita et al. showed that the DNA-binding and/or transactivation ability of Sox9 as a homodimer is boosted by Sox5 and Sox6 in the promoter region of pri-miR-140 [50].
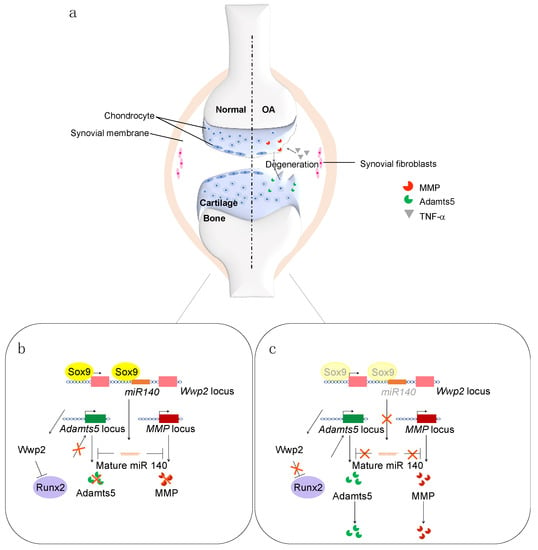
Figure 1.
(a) Schematic representation of normal (left) and OA (right) cartilages. (b) Cartilage homeostasis is maintained by the Sox9 and miR-140 pathways. (c) In OA, the expression levels of Sox9 and miR-140 are decreased.
To further understand the function of chondrocyte-specific miR-140, Miyaki et al. and Nakamura et al. generated miR-140 deletions in mice. These mice exhibited craniofacial truncation due to impaired chondrocyte differentiation [74,80]. Interestingly, Wwp2 knockout (KO) mice have been reported to exhibit a similar craniofacial truncation phenotype [81]. These reports suggest that miR-140 and its host gene Wwp2 have overlapping or coherent functions in craniofacial morphogenesis. However, this phenotype might be due to the loss of miR-140 because a gene-trap technology was used in the corresponding studies to generate Wwp2-null mice, preventing the expression of exons and introns downstream of the insertion site of the gene-trap cassette. To explore this question, Inui et al. generated Wwp2 and/or mir-140 KO mice using the CRISPR/Cas9 system. They confirmed that the skulls of miR-140 KO mice were truncated, as previously reported [74,80]. However, the skulls of Wwp2 KO mice were indistinguishable from those of wild-type (WT) mice. These results suggest that miR-140 is required for proper craniofacial development but that the Wwp2 protein is not [82]. Although we did not find a cooperative function between miR-140 and Wwp2 in craniofacial development in mice, it remains possible that these two factors cooperate in other contexts in mammals. Wwp2 protects articular cartilage by regulating Adamts5 via the Wwp2-Runx2 pathway. The regulation of a common target by both Wwp2 and miR-140 might cooperatively enhance their function in maintaining cartilage homeostasis at both the pre- and post-transcriptional stages [83].
4.1.2. MiR-455
MiR-455, located in intron 10 of collagen type XXVII alpha 1 (Col27a1) [84] has been expressed in cell culture models of chondrogenesis (along with miR-140). It regulates transforming growth factor (TGF)-β signaling by suppressing the Smad2/3 pathway [85]. MiR-455-3p has an important role in the regulation of chondrogenic differentiation of human adipose-derived stem cells (hADSCs) [86]. In addition, miR-455-3p regulates OA and the chondrogenic differentiation of human mesenchymal stem cells (hMSCs) [87,88]. Moreover, miR-455-3p functions as an activator of early chondrogenic differentiation by promoting the expression of the cartilage-specific genes Col2a1 and Comp and directly targeting and inhibiting Runt-related transcription factor 2 (Runx2) [87]. MiR-455-3p promotes chondrogenic differentiation by suppressing the expression of histone deacetylase (HDAC)2 and HDAC8, thereby maintaining an appropriate level of histone H3 acetylation at the COL2A1 promoter to promote the production of type II collagen [88]. Furthermore, it has been shown that miR-455-3p can regulate hMSC chondrogenic differentiation by directly targeting the DNA methyltransferase (DNMT) 3A 3′-UTR [88]. Studies have also shown that miR-455-3p promotes TGF-β signaling and inhibits cartilage degeneration by directly targeting P21-activated kinase (PAK2) [89]. In miR-455-3p global KO mice, obtained by using a transcription activator-like effector nuclease system, thinner cartilage thickness was observed compared to that in WT mice at six months of age [90]. Moreover, these mice showed an OA-like phenotype at five months of age, indicating that miR-455-3p is a critical regulator of cartilage homeostasis [89].
Intronic miRNAs are believed to be processed from the introns of their host transcription units and hence share common regulatory mechanisms and expression patterns with the host gene [91,92,93]. We have previously reported that miR-140 may be derived from its own specific transcript via Sox9 binding during chondrogenesis [50]. To investigate other miRNAs regulated by Sox9 in chondrocytes, a comprehensive microarray analysis was performed. Among several candidates, miRNA-455 showed enhanced expression from both 5p and 3p strands in a Sox9 concentration-dependent manner [94] and was expressed in chondrocytes in approximately equal amounts from both strands [94]. To investigate whether this was directly regulated by Sox9, we used ChIp analysis and found a binding site for Sox9 within intron 3 of Col27a1 [94]. Usually, only one strand of miRNA is incorporated into the RNA-induced silencing complex (RISC) to form a functional and mature miRNA complex [95,96,97]. However, recent reports noted that, in exceptional cases, two distinct miRNAs can be generated, although their functional relevance is not fully understood [98,99,100,101]. In order to investigate the in vivo function of miR-455, we generated miR-455 KO mice using the CRISPR/Cas9 method. Although miR-455−/− mice were born with a normal appearance and showed a normal skeletal development, their knee joints showed cartilage disruption at six months of age. This was consistent with observations from a previous study [89].
We then screened for miR-455 targets using a reporter library system and identified several previously unreported miR-455 target candidates. We focused on endothelial PAS domain protein 1 (EPAS1), which encodes hypoxia inducible factor (HIF)-2α and has a seed sequence in the 3′-UTR for both miR-455s. HIF-2α is known as a catabolic transcription factor for cartilage homeostasis [102,103]. We revealed that both miR-455-5p and -3p directly regulate EPAS1 expression, suggesting that both miR-455s have anti-inflammatory functions and protect against cartilage destruction in OA. To investigate the potential therapeutic effects of miR-455s, we used the well-established surgical DMM model of OA injected with miR-455s mimics. Injection of both miR-455-5p and -3p mimics into DMM-treated knee joints significantly inhibited cartilage destruction compared to the results from the injection of control mimics. These results reveal a therapeutic effect of miR-455-5p and -3p in treating cartilage degeneration in OA, possibly by repressing Hif-2α expression [94]. Hif-2α is a potential therapeutic target for OA since it is encoded by EPAS1, an important developmental gene. Epas1−/− mice are embryonically lethal, whereas Epas1+/− mice show dwarfism [103]. Therefore, indirect suppression of EPAS1 by miR-455s may be a safe treatment for OA.
4.1.3. MiRNAs Regulating Hif-2α
Other miRNAs regulate Hif-2α expression. Zhou et al. revealed that the inhibition of SDC-4 affects cartilage homeostasis and improves the chondrocyte hypertrophy phenotype by inducing miR-96-5p expression. miR-95-5p targets HIF-2α 3′-UTR sequences and thus inhibits Hif-2α translation in murine cartilage tissue and chondrocytes [104]. MiR-365 downregulates HDAC4 and decreases chondrocyte hypertrophy; therefore, miR-365 is an important regulator of chondrocyte hypertrophy and differentiation [105]. Hwang et al. demonstrated that miR-365 levels were significantly suppressed in OA cartilage and that IL-1β decreased miR-365 levels in articular chondrocytes through the activation of the MAPK and NF-κB signaling pathways. MiR-365 suppresses IL-1β-mediated catabolic responses in monolayer and 3D cultures of articular chondrocytes, with concurrent regulation of HIF-2α expression, suggesting that miR-365 could be a useful target for OA therapy [106].
The Sp-1 and Hif-2α protein concentration reduction that occurs after overexpressing miR-138 leads to a marked reduction in the expression of major matrix collagen, COL2A1, which is critical for normal cartilage structure and function [107].
4.1.4. Other miRNAs
Several other miRNAs have been associated with growth plate maintenance and OA development. MiRNA-322 is strongly expressed in prehypertrophic to hypertrophic zones [108] and regulates the RAF/MEK/ERK pathway [109]. A disruption in this pathway in cartilage tissues causes cartilage dysplasia [110]. An analysis of cartilage tissues from miRNA-322-deficient mice (generated with the CRE-loxP system) revealed that hemizygous mutants died neonatally due to respiratory failure resulting from tracheal cartilage damage [108]. In the growth plate, miRNA-322-deficient mice exhibit a slightly reduced hypertrophic zone phenotype [108].
Growth-arrest-specific 5 (GAS5), an lncRNA, plays an important role in mammalian growth and differentiation [111]. GAS5 acts as a negative regulator of miR-21 and causes OA [112]. In chondrosarcoma-derived chondrocytes (HCS-2/8 cells) overexpressing GAS5, miR-21 expression is downregulated. Conversely, knockdown of miR-21 upregulates GAS5 [113]. In a rat model overexpressing GAS5, miR-21 expression is suppressed in growth plate chondrocytes. As a result, cell proliferation is suppressed, and apoptosis is promoted [113], suggesting that miR-21 may play an important role in the maintenance and differentiation of chondrocytes in the growth plate.
MiR-17 belongs to the miR-17-92 cluster [114]. The dysregulation of this miRNA cluster has been associated with skeletal malformations and related growth defects in humans [115]. However, the function of the miR-17-92 cluster, especially that of miR-17, in adult cartilage maintenance and OA progression has not been fully elucidated. Recently, it was reported that decreased expression of miR-17, which targets pathological catabolic factors, including MMP-3, MMP-13, ADAMTS5, and NOS2, in osteoarthritic chondrocytes, contributes to OA progression. Furthermore, miR-17 is highly expressed in both superficial and middle chondrocytes under physiological conditions and maintains the physiological catabolic and anabolic balance, potentially by restricting HIF-1α signaling. Therefore, miR-17 has dual functions: It maintains cartilage homeostasis and prevents OA [116].
Mir-379-5p, located on chromosome 14q32.31 [117], is downregulated in human osteoarthritic tissue and negatively correlated with YBX1 expression. Treating chondrocytes with IL-1β resulted in high expression of mir-379-5p, increased cell viability, increased levels of proliferation-related proteins, and overexpression of ECM-related proteins, such as collagen II and aggrecan. It also results in decreased expression of inflammatory factors and ECM-related proteins, such as MMP-1 and MMP-13. Luciferase reporter assays validated the relationship between miR-379-5p and YBX1. This function was demonstrated via the PI3K/Akt pathway and inhibited by a PI3K/Akt pathway inhibitor. These results indicate that miR-379-5p promotes the proliferation of articular chondrocytes in OA by interacting with YBX1 and regulating the PI3K/Akt pathway [118].
4.2. CircRNAs
CircRNAs are generated by back splicing and discriminated by a covalently closed-loop structure without either a 5′-3′ polyadenylated or polar tail [119,120,121]. CircRNAs derive from known protein-coding genes that comprise one or more exons. Notably, they are exceptionally stable due to their loop structures [122]. In 1976, Sanger et al. identified the first circRNAs, viroids from RNA viruses, using an electron microscope [123]. For a long time, circRNAs were regarded as transcriptional noise produced during abnormal splicing. Advances in biological research using next-generation sequencing have identified thousands of new circRNAs functionally annotated in multiple physiological and pathological processes in eukaryotes, including cancer progression [124,125], inflammation [126], aging [127], and infection [128]. Associations between circRNAs and cartilage metabolism and OA have also been reported.
4.2.1. CircRNAs and Idiopathic Short Stature (ISS)
In patients with ISS, 83 and 62 circRNAs were up and downregulated, respectively, compared with those in healthy controls. One of the circRNAs that was highly expressed in ISS, circRNA_0079201, functions as an miR-140-3p sponge. Furthermore, the proliferation, hypertrophy, and endochondral ossification of chondrocytes in ISS are regulated by the hsa_circRNA_0079201/miR-140-3p/SMAD2 pathway [129].
4.2.2. CircRNAs and OA
Differences in circRNA expression in the healthy cartilage of patients with OA have been reported. Whole transcriptome sequencing revealed that the expression of 42 circRNAs was altered in OA cartilage tissues compared with that in normal cartilage tissues [130]. It was also observed that the expression of 1380 circRNAs differed between OA and control chondrocytes [131]. Subsequently, Xiao et al. identified 197 differentially expressed circRNAs in OA knee joints [132]. Furthermore, 119 upregulated and 136 downregulated circRNAs were identified by RNA-seq in an OA mouse model induced by IL-1β [133]. A total of 11 downregulated and 101 upregulated circRNAs were identified in OA cartilage [134]. These changes in circRNA expression patterns indicate a potential function in OA. The relationship between OA and circRNAs has been gradually elucidated through mechanisms such as circRNA interference with chondrocyte proliferation and apoptosis [135], regulation of ECM degradation [136], and inflammation [137]. In the last few years, an increasing number of associations between circRNAs and OA have been reported. Here, we briefly review the latest literature. For example, circSEC24A is upregulated in OA cartilage tissues and chondrocytes. This upregulation aggravates IL-1β-induced injury by downregulating IL-1β and reducing miR-142-5p in IL-1β-stimulated chondrocytes [138]. CircSCAPER promotes IL-1β-induced ECM degradation, proliferation arrest, and apoptosis enhancement in human chondrocytes by regulating the miR-140-3p/EZH2 axis [139]. The circRNA derived from vacuolar ATPase assembly factor (VMA21) suppresses LPS-induced chondrocyte apoptosis in OA by decreasing the production of mature miR-103 [140]. Circ_0020014 acts as an miR-613 sponge to regulate ADAMTS5 expression, thereby protecting chondrocytes from IL-1β-induced inflammatory damage [141]. In addition, circ-LRP1B regulates proliferation, apoptosis, and oxidative stress in LPS-stimulated human C28/I2 chondrocytes via the miR-34a-5p/NRF1 network [142]. CircRHOT1 enhances CCND1 expression by sponging miR-142-5p to inhibit chondrocyte autophagy and promote chondrocyte proliferation in OA [143]. Circ_0005526 promotes IL-1β-induced chondrocyte injury in OA by suppressing miR-142-5p binding to transcription factor 4 [144]. Circ_0043947 contributes to interleukin 1β-induced chondrocyte injury by sponging miR-671-5p to upregulate reticulon 3 expression [145]. CircADAMTS6/miR-324-5p/PIK3R3 axis participate in IL-1β-induced human chondrocyte dysfunction via the PI3K/AKT/mTOR signaling pathway [146]. Besides, the circ_0000423/miR-27b-3p/MMP-13 axis can affect the pathogenesis of OA [134]. Nevertheless, further research on the relationship between circRNAs and OA is required.
4.3. MiRNAs and Diseases
Dysregulation of miRNAs is observed in a variety of diseases, including cancer. However, only a few congenital diseases associated with mutations in miRNA genes or their target regions have been reported.
4.3.1. Disease-Related miRNAs
The first report of a genetic disease involving miRNAs showed that point mutations in the seed region of miR-96, an miRNA expressed in the hair cells of the inner ear, resulting in autosomal dominant, progressive hearing loss [147].
4.3.2. MiRNAs and Skeletal Dysplasia
The Nosology and Classification of Genetic Skeletal Disorders published by the Nosology Committee of the International Skeletal Dysplasia Society comprises 461 different diseases classified into 42 groups based on their clinical, radiographic, and/or molecular phenotypes. Remarkably, pathogenic variants affecting 437 different genes have been found in 425/461 (92%) of these disorders [148]. However, the underlying molecular mechanisms have not been elucidated for many of them. Noteworthy, miRNAs have been associated with skeletal dysplasia.
The miR-17-92 cluster gene Mir17HG was the first miRNA-encoding gene whose mutation was found to cause abnormal skeletal development in humans. Its deletion causes the arm-length syndrome (Feingold syndrome type 2) [115]. The skeletal phenotype varies from case to case, one family shows skeletal overgrowth with polydactyly [149], while another study reported a case of a nine-year-old boy with developmental delay, short stature, mild macrocephaly, hypertelorism, brachydactyly, and clinodactyly [150]. The reason for these phenotypic inconsistencies is unclear. It might be explained by differences in genetic and non-genetic factors specific to each case, including the miR-17-92 expression level.
4.3.3. Gain-of-Function Mutation of miR-140
It is well known that a deficiency in miR-140 causes skeletal dysplasia. Recently, Gliogieniene et al. reported new findings on skeletal dysplasia caused by a point mutation in the miR-140 gene and on the underlying mechanism. They discovered that, in a novel autosomal dominant human skeletal dysplasia, a single nucleotide substitution occurs in miR-140, resulting in a neomorphic (gain-of-function) mutation. A single nucleotide substitution in miR-140-5p was identified by whole-genome sequencing of an ultra-rare congenital skeletal disorder [151]. Since miR-140 is completely conserved in vertebrates, a knock-in mouse model was generated with the same base substitution to examine its effect on mice. Mutant mice showed abnormalities similar to those in the patient’s skeleton, proving the causative role of this variant. Interestingly, the cartilage phenotype of miR-140G/G mice differed from that of miR-140-null (miR140−/−) mice [74,80,151]. MiR-140 mutated mice displayed a decrease in Col10a1 expression, a delay in secondary ossification of the carpal and tubular bones, a severe decline in epiphyseal mineralization, and a mild flat vertebral body. These phenotypes match the features of patients with skeletal dysplasia [151].
Furthermore, this mutant miRNA gene resulted in abundant mutant miR-140-5p expression without defects in miRNA processing. In chondrocytes, the mutation extensively derepressed the targets of WT miR-140-5p and represses those of mutant miR-140-5p, suggesting that the mutation has both loss-of-function and gain-of-function effects. Furthermore, mutant miR-140-5p competes with Ybx1, a conserved RNA-binding protein, for an overlapping binding site. This may explain why this mutant miRNA strongly represses its targets and exerts robust effects in vivo, even in the absence of evolutionarily selected miRNA-target RNA interactions [152,153]. This is the first reported case of a pathogenic gain-of-function miRNA mutation. It provides molecular insights into the novel actions of emerging or mutant miRNAs [151].
5. Conclusions
This review focuses on Sox9, the master transcription factor of cartilage, and ncRNAs, mainly miRNAs, and provides an overview of previous reports. To date, many factors involved in the maintenance of cartilage homeostasis and OA have been identified, but our understanding of the interactions and networks among these factors remains incomplete. It is hoped that the powerful tools that have emerged in recent years will lead to greater knowledge of the mechanisms of musculoskeletal congenital diseases and, ultimately, to therapeutic approaches.
Author Contributions
Y.F., L.L., L.Y. and M.I. wrote the first draft of the manuscript. Y.F., T.C. and H.A. edited the manuscript and have approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This review was supported by AMED-CREST from the Japan Agency for Medical Research and Development (AMED) (JP21gm0810008 to H.A.) and the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant numbers: 20H05696 to H.A.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank all members of the Asahara Laboratory for many productive discussions.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Acan | Aggrecan |
| ADAMTS | a disintegrin and metalloproteinases with thrombospondin motif |
| ceRNA | competing endogenous RNA |
| ChIP-Seq | chromatin immunoprecipitation sequencing |
| circRNA | circular RNA |
| Col2a1 | collagen type II alpha 1 |
| Col9a2 | collagen type IX alpha 2 |
| Col11a2 | collagen type XI alpha 2 |
| Col10a1 | collagen type X alpha 1 |
| Col27a1 | collagen type XXVII alpha 1 |
| ceRNA | competing endogenous RNA |
| CRISPR-ChIP-MS | CRISPR-ChIP-mass spectrometry |
| DMM | destabilization of the medial meniscus |
| DNMT | DNA methyltransferase |
| ECM | extracellular matrix |
| EPAS1 | endothelial PAS domain protein 1 |
| ES | embryonic stem |
| GAS5 | Growth-arrest-specific 5 |
| hADSC | human adipose-derived stem cell |
| HDAC | histone deacetylase |
| HMG | high-mobility group |
| HIF | hypoxia inducible factor |
| hMSC | human mesenchymal stem cell |
| ISS | idiopathic short stature |
| JAK3 | Janus kinase 3 |
| KO | knockout |
| lncRNA | long ncRNA |
| miR-140 | MiRNA-140 |
| miRNA | microRNA |
| MMP | matrix metalloproteinase |
| ncRNAs | noncoding RNAs |
| OA | Osteoarthritis |
| OARSI | Osteoarthritis Research Society International |
| PAK2 | P21-activated kinase |
| PARylation | poly(ADP-ribosyl)ation |
| piRNAs | Piwi-interacting RNAs |
| pre-miRNA | precursor miRNA |
| pri-miRNA | primary miRNA |
| RCSE | rib cage-specific enhancer |
| RISC | RNA-induced silencing complex |
| Runx2 | Runt-related transcription factor 2 |
| siRNA | small interfering RNA |
| snoRNA | small nucleolar RNA |
| Sox5 | SRY-box transcription factor 5 |
| Sox6 | SRY-box transcription factor 6 |
| Sox9 | SRY-box transcription factor 9 |
| tRF | tRNA-derived fragment |
| tRNA | transition RNA |
| TGFβ | transforming growth factor-β |
| VMA21 | vacuolar ATPase assembly factor 21 |
| WT | wild-type |
| Wwp2 | WW domain-containing protein 2 |
References
- Lotz, M.K. New Developments in Osteoarthritis. Posttraumatic Osteoarthritis: Pathogenesis and Pharmacological Treatment Options. Arthritis Res. Ther. 2010, 12, 211. [Google Scholar] [CrossRef]
- Hashimoto, M.; Nakasa, T.; Hikata, T.; Asahara, H. Molecular Network of Cartilage Homeostasis and Osteoarthritis. Med. Res. Rev. 2008, 28, 464–481. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.B.; Marcu, K.B. Cartilage Homeostasis in Health and Rheumatic Diseases. Arthritis Res. Ther. 2009, 11, 224. [Google Scholar] [CrossRef]
- Lotz, M. Osteoarthritis Year 2011 in Review: Biology. Osteoarthr. Cartil. 2012, 20, 192–196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, Y. Osteoarthritis Year in Review 2021: Biology. Osteoarthr. Cartil. 2022, 30, 207–215. [Google Scholar] [CrossRef]
- Mow, V.C.; Ratcliffe, A.; Robin Poole, A. Cartilage and Diarthrodial Joints as Paradigms for Hierarchical Materials and Structures. Biomaterials 1992, 13, 67–97. [Google Scholar] [CrossRef]
- Ratneswaran, A.; Rockel, J.S.; Kapoor, M. Understanding Osteoarthritis Pathogenesis: A Multiomics System-Based Approach. Curr. Opin. Rheumatol. 2020, 32, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.; Bay-Jensen, A.C.; Pap, T.; Dvir-Ginzberg, M.; Quasnichka, H.; Barrett-Jolley, R.; Mobasheri, A.; Henrotin, Y. Chondrocyte Secretome: A Source of Novel Insights and Exploratory Biomarkers of Osteoarthritis. Osteoarthr. Cartil. 2017, 25, 1199–1209. [Google Scholar] [CrossRef]
- Polacek, M.; Bruun, J.A.; Johansen, O.; Martinez, I. Differences in the Secretome of Cartilage Explants and Cultured Chondrocytes Unveiled by SILAC Technology. J. Orthop. Res. 2010, 28, 1040–1049. [Google Scholar] [CrossRef]
- Calamia, V.; Lourido, L.; Fernández-Puente, P.; Mateos, J.; Rocha, B.; Montell, E.; Vergés, J.; Ruiz-Romero, C.; Blanco, F.J. Secretome Analysis of Chondroitin Sulfate-Treated Chondrocytes Reveals Anti-Angiogenic, Anti-Inflammatory and Anti-Catabolic Properties. Arthritis Res. Ther. 2012, 14, R202. [Google Scholar] [CrossRef] [PubMed]
- Calamia, V.; Mateos, J.; Fernández-Puente, P.; Lourido, L.; Rocha, B.; Fernández-Costa, C.; Montell, E.; Vergés, J.; Ruiz-Romero, C.; Blanco, F.J. A Pharmacoproteomic Study Confirms the Synergistic Effect of Chondroitin Sulfate and Glucosamine. Sci. Rep. 2014, 4, 5069. [Google Scholar] [CrossRef]
- Catterall, J.B.; Rowan, A.D.; Sarsfield, S.; Saklatvala, J.; Wait, R.; Cawston, T.E. Development of a Novel 2D Proteomics Approach for the Identification of Proteins Secreted by Primary Chondrocytes after Stimulation by IL-1 and Oncostatin M. Rheumatology 2006, 45, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Riffault, M.; Moulin, D.; Grossin, L.; Mainard, D.; Magdalou, J.; Vincourt, J.B. Label-Free Relative Quantification Applied to LC-MALDI Acquisition for Rapid Analysis of Chondrocyte Secretion Modulation. J. Proteom. 2015, 114, 263–273. [Google Scholar] [CrossRef]
- Polacek, M.; Bruun, J.A.; Elvenes, J.; Figenschau, Y.; Martinez, I. The Secretory Profiles of Cultured Human Articular Chondrocytes and Mesenchymal Stem Cells: Implications for Autologous Cell Transplantation Strategies. Cell Transplant. 2011, 20, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Polacek, M.; Bruun, J.A.; Johansen, O.; Martinez, I. Comparative Analyses of the Secretome from Dedifferentiated and Redifferentiated Adult Articular Chondrocytes. Cartilage 2011, 2, 186–196. [Google Scholar] [CrossRef]
- Haglund, L.; Bernier, S.M.; Önnerfjord, P.; Recklies, A.D. Proteomic Analysis of the LPS-Induced Stress Response in Rat Chondrocytes Reveals Induction of Innate Immune Response Components in Articular Cartilage. Matrix Biol. 2008, 27, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Lourido, L.; Calamia, V.; Fernández-Puente, P.; Mateos, J.; Oreiro, N.; Blanco, F.J.; Ruiz-Romero, C. Secretome Analysis of Human Articular Chondrocytes Unravels Catabolic Effects of Nicotine on the Joint. Proteom. Clin. Appl. 2016, 10, 671–680. [Google Scholar] [CrossRef]
- Stenberg, J.; Rüetschi, U.; Skiöldebrand, E.; Kärrholm, J.; Lindahl, A. Quantitative Proteomics Reveals Regulatory Differences in the Chondrocyte Secretome from Human Medial and Lateral Femoral Condyles in Osteoarthritic Patients. Proteome Sci. 2013, 11, 43. [Google Scholar] [CrossRef]
- Taylor, D.W.; Ahmed, N.; Parreno, J.; Lunstrum, G.P.; Gross, A.E.; Diamandis, E.P.; Kandel, R.A. Collagen Type XII and Versican Are Present in the Early Stages of Cartilage Tissue Formation by Both Redifferentating Passaged and Primary Chondrocytes. Tissue Eng. Part A 2015, 21, 683–693. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Bhattacharjee, M.; Ahmad, S.; Nirujogi, R.S.; Renuse, S.; Subbannayya, Y.; Marimuthu, A.; Srikanth, S.M.; Raju, R.; Dhillon, M.; et al. Differential Proteomic Analysis of Synovial Fluid from Rheumatoid Arthritis and Osteoarthritis Patients. Clin. Proteom. 2014, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Guo, J.; Luo, Y.; Zhang, W.; Cui, Y.; Wang, Q.; Zhang, Z.; Wang, T. Functional Proteomics Revealed IL-1β Amplifies TNF Downstream Protein Signals in Human Synoviocytes in a TNF-Independent Manner. Biochem. Biophys. Res. Commun. 2014, 450, 538–544. [Google Scholar] [CrossRef]
- Gobezie, R.; Kho, A.; Krastins, B.; Sarracino, D.A.; Thornhill, T.S.; Chase, M.; Millett, P.J.; Lee, D.M. High Abundance Synovial Fluid Proteome: Distinct Profiles in Health and Osteoarthritis. Arthritis Res. Ther. 2007, 9, R36. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, L.; Nirujogi, R.S.; Ahmad, S.; Bhattacharjee, M.; Manda, S.S.; Renuse, S.; Kelkar, D.S.; Subbannayya, Y.; Raju, R.; Goel, R.; et al. Poteomic Analysis of Human Osteoarthritis Synovial Fluid. Clin. Proteomics 2014, 11, 6. [Google Scholar] [CrossRef]
- Kamphorst, J.J.; Van Der Heijden, R.; DeGroot, J.; Lafeber, F.P.J.G.; Reijmers, T.H.; Van El, B.; Tjaden, U.R.; Van Der Greef, J.; Hankemeier, T. Profiling of Endogenous Peptides in Human Synovial Fluid by NanoLC-MS: Method Validation and Peptide Identification. J. Proteome Res. 2007, 6, 4388–4396. [Google Scholar] [CrossRef]
- Ritter, S.Y.; Subbaiah, R.; Bebek, G.; Crish, J.; Scanzello, C.R.; Krastins, B.; Sarracino, D.; Lopez, M.F.; Crow, M.K.; Aigner, T.; et al. Proteomic Analysis of Synovial Fluid from the Osteoarthritic Knee: Comparison with Transcriptome Analyses of Joint Tissues. Arthritis Rheum. 2013, 65, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Yamagiwa, H.; Sarkar, G.; Charlesworth, M.C.; McCormick, D.J.; Bolander, M.E. Two-Dimensional Gel Electrophoresis of Synovial Fluid: Method for Detecting Candidate Protein Markers for Osteoarthritis. J. Orthop. Sci. 2003, 8, 482–490. [Google Scholar] [CrossRef]
- Russo, R.; Vassallo, V.; Stellavato, A.; Valletta, M.; Cimini, D.; Pedone, P.V.; Schiraldi, C.; Chambery, A. Differential Secretome Profiling of Human Osteoarthritic Synoviocytes Treated with Biotechnological Unsulfated and Marine Sulfated Chondroitins. Int. J. Mol. Sci. 2020, 21, 3746. [Google Scholar] [CrossRef]
- Lotz, M. Cytokines in Cartilage Injury and Repair. Clin. Orthop. Relat. Res. 2001, 391, S108–S115. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.B. The Role of the Chondrocyte in Osteoarthritis. Arthritis Rheum. 2000, 43, 1916–1926. [Google Scholar] [CrossRef]
- Gravallese, E.M.; Goldring, S.R. Cellular mechanisms and the role of cytokines in bone erosions in rheumatoid arthritis. Arthritis Rheum. 2000, 43, 2143–2151. [Google Scholar] [CrossRef]
- Hayami, T.; Pickarski, M.; Zhuo, Y.; Wesolowski, G.A.; Rodan, G.A.; Duong, L.T. Characterization of Articular Cartilage and Subchondral Bone Changes in the Rat Anterior Cruciate Ligament Transection and Meniscectomized Models of Osteoarthritis. Bone 2006, 38, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Bleuel, J.; Zaucke, F.; Brüggemann, G.P.; Niehoff, A. Effects of Cyclic Tensile Strain on Chondrocyte Metabolism: A Systematic Review. PLoS ONE 2015, 10, e0119816. [Google Scholar] [CrossRef]
- Lotz, M.; Loeser, R.F. Effects of Aging on Articular Cartilage Homeostasis. Bone 2012, 51, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Collins, J.A.; Diekman, B.O. Ageing and the Pathogenesis of Osteoarthritis. Nat. Rev. Rheumatol. 2016, 12, 412–420. [Google Scholar] [CrossRef]
- McCulloch, K.; Litherland, G.J.; Rai, T.S. Cellular Senescence in Osteoarthritis Pathology. Aging Cell 2017, 16, 210–218. [Google Scholar] [CrossRef]
- Kurakazu, I.; Akasaki, Y.; Tsushima, H.; Sueishi, T.; Toya, M.; Kuwahara, M.; Uchida, T.; Lotz, M.K.; Nakashima, Y. TGFβ1 Signaling Protects Chondrocytes against Oxidative Stress via FOXO1–Autophagy Axis. Osteoarthr. Cartil. 2021, 29, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Roemer, F.W.; Guermazi, A.; Felson, D.T.; Niu, J.; Nevitt, M.C.; Crema, M.D.; Lynch, J.A.; Lewis, C.E.; Torner, J.; Zhang, Y. Presence of MRI-Detected Joint Effusion and Synovitis Increases the Risk of Cartilage Loss in Knees without Osteoarthritis at 30-Month Follow-up: The MOST Study. Ann. Rheum. Dis. 2011, 70, 1804–1809. [Google Scholar] [CrossRef]
- Xie, Y.; Zinkle, A.; Chen, L.; Mohammadi, M. Fibroblast Growth Factor Signalling in Osteoarthritis and Cartilage Repair. Nat. Rev. Rheumatol. 2020, 16, 547–564. [Google Scholar] [CrossRef]
- Liu-Bryan, R.; Terkeltaub, R. Emerging Regulators of the Inflammatory Process in Osteoarthritis. Nat. Rev. Rheumatol. 2015, 11, 35–44. [Google Scholar] [CrossRef]
- Peffers, M.J.; Milner, P.I.; Tew, S.R.; Clegg, P.D. Regulation of SOX9 in Normal and Osteoarthritic Equine Articular Chondrocytes by Hyperosmotic Loading. Osteoarthr. Cartil. 2010, 18, 1502–1508. [Google Scholar] [CrossRef]
- Bian, Q.; Wang, Y.-J.; Liu, S.-F.; Li, Y.-P. Osteoarthritis: Genetic Factors, Animal Models, Mechanisms, and Therapies. Front. Biosci. 2012, 4, 74–100. [Google Scholar] [CrossRef]
- de Crombrugghe, B.; Lefebvre, V.; Nakashima, K. Regulatory Mechanisms in the Pathways of Cartilage and Bone Formation. Curr. Opin. Cell Biol. 2001, 13, 721–728. [Google Scholar] [CrossRef]
- Bi, W.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; De Crombrugghe, B. Sox9 Is Required for Cartilage Formation. Nat. Genet. 1999, 22, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Huang, W.; Whitworth, D.J.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; De Crombrugghe, B. Haploinsufficiency of Sox9 Results in Defective Cartilage Primordia and Premature Skeletal Mineralization. Proc. Natl. Acad. Sci. USA 2001, 98, 6698–6703. [Google Scholar] [CrossRef]
- Cheah, K.S.E.; Lau, E.T.; Au, P.K.C.; Tam, P.P.L. Expression of the Mouse A1(II) Collagen Gene Is Not Restricted to Cartilage during Development. Development 1991, 111, 945–953. [Google Scholar] [CrossRef]
- Lefebvre, V.; Li, P.; De Crombrugghe, B. A New Long Form of Sox5 (L-Sox5), Sox6 and Sox9 Are Coexpressed in Chondrogenesis and Cooperatively Activate the Type II Collagen Gene. EMBO J. 1998, 17, 5718–5733. [Google Scholar] [CrossRef]
- Lefebvre, V.; Huang, W.; Harley, V.R.; Goodfellow, P.N.; de Crombrugghe, B. SOX9 Is a Potent Activator of the Chondrocyte-Specific Enhancer of the pro Alpha1(II) Collagen Gene. Mol. Cell. Biol. 1997, 17, 2336–2346. [Google Scholar] [CrossRef]
- Wright, E.; Hargrave, M.R.; Christiansen, J.; Cooper, L.; Kun, J.; Evans, T.; Gangadharan, U.; Greenfield, A.; Koopman, P. The Sry-Related Gene Sox9 Is Expressed during Chondrogenesis in Mouse Embryos. Nat. Genet. 1995, 9, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Kawaguchi, H.; Kamekura, S.; Ogata, N.; Mori, Y.; Nakamura, K.; Ikegawa, S.; Chung, U. Il Distinct Roles of Sox5, Sox6, and Sox9 in Different Stages of Chondrogenic Differentiation. J. Bone Miner. Metab. 2005, 23, 337–340. [Google Scholar] [CrossRef]
- Yamashita, S.; Miyaki, S.; Kato, Y.; Yokoyama, S.; Sato, T.; Barrionuevo, F.; Akiyama, H.; Scherer, G.; Takada, S.; Asahara, H. L-Sox5 and Sox6 Proteins Enhance Chondrogenic MiR-140 MicroRNA Expression by Strengthening Dimeric Sox9 Activity. J. Biol. Chem. 2012, 287, 22206–22215. [Google Scholar] [CrossRef]
- Lefrebvre, V.; de Crombrugghe, B. Toward Understanding S0X9 Function in Chondrocyte Differentiation. Matrix Biol. 1998, 16, 529–540. [Google Scholar] [CrossRef]
- Zhao, Q.; Eberspaecher, H.; Lefebvre, V.; De Crombrugghe, B. Parallel Expression of Sox9 and Col2a1 in Cells Undergoing Chondrogenesis. Dev. Dyn. 1997, 209, 377–386. [Google Scholar] [CrossRef]
- Chambers, M.G.; Kuffner, T.; Cowan, S.K.; Cheah, K.S.E.; Mason, R.M. Expression of Collagen and Aggrecan Genes in Normal and Osteoarthritic Murine Knee Joints. Osteoarthr. Cartil. 2002, 10, 51–61. [Google Scholar] [CrossRef]
- Haseeb, A.; Kc, R.; Angelozzi, M.; de Charleroy, C.; Rux, D.; Tower, R.J.; Yao, L.; da Silva, R.P.; Pacifici, M.; Qin, L.; et al. SOX9 Keeps Growth Plates and Articular Cartilage Healthy by Inhibiting Chondrocyte Dedifferentiation/ Osteoblastic Redifferentiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2019152118. [Google Scholar] [CrossRef]
- Henry, S.P.; Jang, C.W.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; De Crombrugghe, B. Generation of Aggrecan-CreERT2 Knockin Mice for Inducible Cre Activity in Adult Cartilage. Genesis 2009, 47, 805–814. [Google Scholar] [CrossRef]
- Oh, C.D.; Lu, Y.; Liang, S.; Mori-Akiyama, Y.; Chen, D.; De Crombrugghe, B.; Yasuda, H. SOX9 Regulates Multiple Genes in Chondrocytes, Including Genes Encoding ECM Proteins, ECM Modification Enzymes, Receptors, and Transporters. PLoS ONE 2014, 9, e107577. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, Y.; Chiba, T.; Kataoka, K.; Yamashita, S.; Sato, T.; Kato, T.; Takahashi, K.; Miyamoto, T.; Kitazawa, M.; Hatta, T.; et al. Combinatorial CRISPR/Cas9 Approach to Elucidate a Far-Upstream Enhancer Complex for Tissue-Specific Sox9 Expression. Dev. Cell 2018, 46, 794–806.e6. [Google Scholar] [CrossRef]
- Johnson, K.; Zhu, S.; Tremblay, M.S.; Payette, J.N.; Wang, J.; Bouchez, L.C.; Meeusen, S.; Althage, A.; Cho, C.Y.; Wu, X.; et al. A Stem Cell-Based Approach to Cartilage Repair. Science 2012, 336, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Wang, W.; Tu, B.; Zhu, Y.; Fan, C.; Li, Y. Overexpression of SOX9 Alleviates the Progression of Human Osteoarthritis in Vitro and in Vivo. Drug Des. Dev. Ther. 2019, 13, 2833–2842. [Google Scholar] [CrossRef]
- Kim, S.; Han, S.; Kim, Y.; Kim, H.S.; Gu, Y.R.; Kang, D.; Cho, Y.; Kim, H.; Lee, J.; Seo, Y.; et al. Tankyrase Inhibition Preserves Osteoarthritic Cartilage by Coordinating Cartilage Matrix Anabolism via Effects on SOX9 PARylation. Nat. Commun. 2019, 10, 4898. [Google Scholar] [CrossRef]
- Hattori, M. Finishing the Euchromatic Sequence of the Human Genome. Tanpakushitsu Kakusan Koso 2005, 50, 162–168. (In Japanese) [Google Scholar]
- Peffers, M.J.; Chabronova, A.; Balaskas, P.; Fang, Y.; Dyer, P.; Cremers, A.; Emans, P.J.; Feczko, P.Z.; Caron, M.M.; Welting, T.J.M. SnoRNA Signatures in Cartilage Ageing and Osteoarthritis. Sci. Rep. 2020, 10, 10641. [Google Scholar] [CrossRef]
- Green, J.A.; Ansari, M.Y.; Ball, H.C.; Haqqi, T.M. TRNA-Derived Fragments (TRFs) Regulate Post-Transcriptional Gene Expression via AGO-Dependent Mechanism in IL-1β Stimulated Chondrocytes. Osteoarthr. Cartil. 2020, 28, 1102–1110. [Google Scholar] [CrossRef]
- Balaskas, P.; Green, J.A.; Haqqi, T.M.; Dyer, P.; Kharaz, Y.A.; Fang, Y.; Liu, X.; Welting, T.J.M.; Peffers, M.J. Small Non-Coding Rnaome of Ageing Chondrocytes. Int. J. Mol. Sci. 2020, 21, 5675. [Google Scholar] [CrossRef]
- Zacharjasz, J.; Mleczko, A.M.; Bakowski, P.; Piontek, T.; Bakowska-żywicka, K. Small Noncoding Rnas in Knee Osteoarthritis: The Role of Micrornas and Trna-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 5711. [Google Scholar] [CrossRef]
- Young, D.A.; Barter, M.J.; Soul, J. Osteoarthritis Year in Review: Genetics, Genomics, Epigenetics. Osteoarthr. Cartil. 2022, 30, 216–225. [Google Scholar] [CrossRef]
- Harfe, B.D.; McManus, M.T.; Mansfield, J.H.; Hornstein, E.; Tabin, C.J. The RNaseIII Enzyme Dicer Is Required for Morphogenesis but Not Patterning of the Vertebrate Limb. Proc. Natl. Acad. Sci. USA 2005, 102, 10898–10903. [Google Scholar] [CrossRef]
- Kobayashi, T.; Lu, J.; Cobb, B.S.; Rodda, S.J.; McMahon, A.P.; Schipani, E.; Merkenschlager, M.; Kronenberg, H.M. Dicer-Dependent Pathways Regulate Chondrocyte Proliferation and Differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 1949–1954. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Malizos, K.N.; Oikonomou, P.; Tsezou, A. Integrative MicroRNA and Proteomic Approaches Identify Novel Osteoarthritis Genes and Their Collaborative Metabolic and Inflammatory Networks. PLoS ONE 2008, 3, e3740. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Zhu, Y.; Tu, G. A Bioinformatic Analysis of MicroRNAs Role in Osteoarthritis. Osteoarthr. Cartil. 2017, 25, 1362–1371. [Google Scholar] [CrossRef]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H.A. MicroRNA Expression in Zebrafish Embryonic Development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef]
- Tuddenham, L.; Wheeler, G.; Ntounia-Fousara, S.; Waters, J.; Hajihosseini, M.K.; Clark, I.; Dalmay, T. The Cartilage Specific MicroRNA-140 Targets Histone Deacetylase 4 in Mouse Cells. FEBS Lett. 2006, 580, 4214–4217. [Google Scholar] [CrossRef]
- Miyaki, S.; Nakasa, T.; Otsuki, S.; Grogan, S.P.; Higashiyama, R.; Inoue, A.; Kato, Y.; Sato, T.; Lotz, M.K.; Asahara, H. MicroRNA-140 Is Expressed in Differentiated Human Articular Chondrocytes and Modulates Interleukin-1 Responses. Arthritis Rheum. 2009, 60, 2723–2730. [Google Scholar] [CrossRef]
- Miyaki, S.; Sato, T.; Inoue, A.; Otsuki, S.; Ito, Y.; Yokoyama, S.; Kato, Y.; Takemoto, F.; Nakasa, T.; Yamashita, S.; et al. MicroRNA-140 Plays Dual Roles in Both Cartilage Development and Homeostasis. Genes Dev. 2010, 24, 1173–1185. [Google Scholar] [CrossRef]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT Family of E3 Ubiquitin Ligases: Multiple Players in Cancer Development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological Functions of the HECT Family of Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Yang, J.; Qin, S.; Yi, C.; Ma, G.; Zhu, H.; Zhou, W.; Xiong, Y.; Zhu, X.; Wang, Y.; He, L.; et al. MiR-140 Is Co-Expressed with Wwp2-C Transcript and Activated by Sox9 to Target Sp1 in Maintaining the Chondrocyte Proliferation. FEBS Lett. 2011, 585, 2992–2997. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; He, X.; Kato, H.; Wakitani, S.; Kobayashi, T.; Watanabe, S.; Iida, A.; Tahara, H.; Warman, M.L.; Watanapokasin, R.; et al. Sox9 Is Upstream of MicroRNA-140 in Cartilage. Appl. Biochem. Biotechnol. 2012, 166, 64–71. [Google Scholar] [CrossRef]
- Han, Y.; Lefebvre, V. L-Sox5 and Sox6 Drive Expression of the Aggrecan Gene in Cartilage by Securing Binding of Sox9 to a Far-Upstream Enhancer. Mol. Cell. Biol. 2008, 28, 4999–5013. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Inloes, J.B.; Katagiri, T.; Kobayashi, T. Chondrocyte-Specific MicroRNA-140 Regulates Endochondral Bone Development and Targets Dnpep to Modulate Bone Morphogenetic Protein Signaling. Mol. Cell. Biol. 2011, 31, 3019–3028. [Google Scholar] [CrossRef]
- Zou, W.; Chen, X.; Shim, J.H.; Huang, Z.; Brady, N.; Hu, D.; Drapp, R.; Sigrist, K.; Glimcher, L.H.; Jones, D. The E3 Ubiquitin Ligase Wwp2 Regulates Craniofacial Development through Mono-Ubiquitylation of Goosecoid. Nat. Cell Biol. 2011, 13, 59–65. [Google Scholar] [CrossRef]
- Inui, M.; Mokuda, S.; Sato, T.; Tamano, M.; Takada, S.; Asahara, H. Dissecting the Roles of MiR-140 and Its Host Gene. Nat. Cell Biol. 2018, 20, 516–518. [Google Scholar] [CrossRef]
- Mokuda, S.; Nakamichi, R.; Matsuzaki, T.; Ito, Y.; Sato, T.; Miyata, K.; Inui, M.; Olmer, M.; Sugiyama, E.; Lotz, M.; et al. Wwp2 Maintains Cartilage Homeostasis through Regulation of Adamts5. Nat. Commun. 2019, 10, 2429. [Google Scholar] [CrossRef] [PubMed]
- Lalevée, S.; Lapaire, O.; Bühler, M. MiR455 Is Linked to Hypoxia Signaling and Is Deregulated in Preeclampsia. Cell Death Dis. 2014, 5, e1408. [Google Scholar] [CrossRef]
- Swingler, T.E.; Wheeler, G.; Carmont, V.; Elliott, H.R.; Barter, M.J.; Abu-Elmagd, M.; Donell, S.T.; Boot-Handford, R.P.; Hajihosseini, M.K.; Münsterberg, A.; et al. The Expression and Function of MicroRNAs in Chondrogenesis and Osteoarthritis. Arthritis Rheum. 2012, 64, 1909–1919. [Google Scholar] [CrossRef]
- Zhang, Z.; Kang, Y.; Zhang, Z.; Zhang, H.; Duan, X.; Liu, J.; Li, X.; Liao, W. Expression of MicroRNAs during Chondrogenesis of Human Adipose-Derived Stem Cells. Osteoarthr. Cartil. 2012, 20, 1638–1646. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, C.; Meng, F.; Zhao, X.; Zhang, Z.; Huang, G.; Chen, W.; Fu, M.; Liao, W. MiR-455-3p Regulates Early Chondrogenic Differentiation via Inhibiting Runx2. FEBS Lett. 2015, 589, 3671–3678. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, L.; Zhang, Z.; Meng, F.; Huang, G.; Sheng, P.; Zhang, Z.; Liao, W. MicroRNA-455-3p Modulates Cartilage Development and Degeneration through Modification of Histone H3 Acetylation. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2881–2891. [Google Scholar] [CrossRef]
- Hu, S.; Zhao, X.; Mao, G.; Zhang, Z.; Wen, X.; Zhang, C.; Liao, W.; Zhang, Z. MicroRNA-455-3p Promotes TGF-β Signaling and Inhibits Osteoarthritis Development by Directly Targeting PAK2. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, X.; Zhang, C.; Zhang, Z.; Lun, J.; Liao, W.; Zhang, Z. MiR-455-3p Inhibits the Degenerate Process of Chondrogenic Differentiation through Modification of DNA Methylation Article. Cell Death Dis. 2018, 9, 537. [Google Scholar] [CrossRef] [PubMed]
- Baskerville, S.; Bartel, D.P. Microarray Profiling of MicroRNAs Reveals Frequent Coexpression with Neighboring MiRNAs and Host Genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of Mammalian MicroRNA Host Genes and Transcription Units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Dudek, K.A.; Lafont, J.E.; Martinez-Sanchez, A.; Murphy, C.L. Type II Collagen Expression Is Regulated by Tissue-Specific MiR-675 in Human Articular Chondrocytes. J. Biol. Chem. 2010, 285, 24381–24387. [Google Scholar] [CrossRef]
- Ito, Y.; Matsuzaki, T.; Ayabe, F.; Mokuda, S.; Kurimoto, R.; Matsushima, T.; Tabata, Y.; Inotsume, M.; Tsutsumi, H.; Liu, L.; et al. Both MicroRNA-455-5p and -3p Repress Hypoxia-Inducible Factor-2α Expression and Coordinately Regulate Cartilage Homeostasis. Nat. Commun. 2021, 12, 4148. [Google Scholar] [CrossRef]
- Slezak-Prochazka, I.; Selvi, D.; Kroesen, B.J.; Van Den Berg, A. MicroRNAs, Macrocontrol: Regulation of MiRNA Processing. RNA 2010, 16, 1087–1095. [Google Scholar] [CrossRef]
- Trabucchi, M.; Briata, P.; Filipowicz, W.; Rosenfeld, M.G.; Ramos, A.; Gherzi, R. How to Control MiRNA Maturation? RNA Biol. 2009, 6, 536–540. [Google Scholar] [CrossRef][Green Version]
- Newman, M.A.; Hammond, S.M. Emerging Paradigms of Regulated MicroRNA Processing. Genes Dev. 2010, 24, 1086–1092. [Google Scholar] [CrossRef]
- Sakurai, K.; Furukawa, C.; Haraguchi, T.; Inada, K.I.; Shiogama, K.; Tagawa, T.; Fujita, S.; Ueno, Y.; Ogata, A.; Ito, M.; et al. MicroRNAs MiR-199a-5p and -3p Target the Brm Subunit of SWI/SNF to Generate a Double-Negative Feedback Loop in a Variety of Human Cancers. Cancer Res. 2011, 71, 1680–1689. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray Analysis Shows That Some MicroRNAs Downregulate Large Numbers Of-Target MRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Mataki, H.; Seki, N.; Mizuno, K.; Nohata, N.; Kamikawaji, K.; Kumamoto, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Dual-Strand Tumor-Suppressor MicroRNA-145 (MiR-145-5p and MiR-145-3p) Coordinately Targeted MTDH in Lung Squamous Cell Carcinoma. Oncotarget 2016, 7, 72084–72098. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Kim, J.; Ryu, J.H.; Oh, H.; Chun, C.H.; Kim, B.J.; Min, B.H.; Chun, J.S. Hypoxia-Inducible Factor-2α Is a Catabolic Regulator of Osteoarthritic Cartilage Destruction. Nat. Med. 2010, 16, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Fukai, A.; Mabuchi, A.; Ikeda, T.; Yano, F.; Ohba, S.; Nishida, N.; Akune, T.; Yoshimura, N.; Nakagawa, T.; et al. Transcriptional Regulation of Endochondral Ossification by HIF-2α during Skeletal Growth and Osteoarthritis Development. Nat. Med. 2010, 16, 678–686. [Google Scholar] [CrossRef]
- Zhou, K.; He, S.; Yu, H.; Pei, F.; Zhou, Z. Inhibition of Syndecan-4 Reduces Cartilage Degradation in Murine Models of Osteoarthritis through the Downregulation of HIF-2α by MiR-96-5p. Lab. Investig. 2021, 101, 1060–1070. [Google Scholar] [CrossRef]
- Guan, Y.-J.; Yang, X.; Wei, L.; Chen, Q. MiR-365: A Mechanosensitive MicroRNA Stimulates Chondrocyte Differentiation through Targeting Histone Deacetylase 4. FASEB J. 2011, 25, 4457–4466. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.S.; Park, S.J.; Lee, M.H.; Kim, H.A. MicroRNA-365 Regulates IL-1β-Induced Catabolic Factor Expression by Targeting HIF-2α in Primary Chondrocytes. Sci. Rep. 2017, 7, 17889. [Google Scholar] [CrossRef]
- Seidl, C.I.; Martinez-Sanchez, A.; Murphy, C.L. Derepression of MicroRNA-138 Contributes to Loss of the Human Articular Chondrocyte Phenotype. Arthritis Rheumatol. 2016, 68, 398–409. [Google Scholar] [CrossRef]
- Bluhm, B.; Ehlen, H.W.A.; Holzer, T.; Georgieva, V.S.; Heilig, J.; Pitzler, L.; Etich, J.; Bortecen, T.; Frie, C.; Probst, K.; et al. MiR-322 Stabilizes MEK1 Expression to Inhibit RAF/MEK/ERK Pathway Activation in Cartilage. Development 2017, 144, 3562–3577. [Google Scholar] [CrossRef]
- Marchand, A.; Atassi, F.; Mougenot, N.; Clergue, M.; Codoni, V.; Berthuin, J.; Proust, C.; Trégouët, D.A.; Hulot, J.S.; Lompré, A.M. MiR-322 Regulates Insulin Signaling Pathway and Protects against Metabolic Syndrome-Induced Cardiac Dysfunction in Mice. Biochim. Biophys. Acta Mol. Basis Dis. 2016, 1862, 611–621. [Google Scholar] [CrossRef]
- Boucherat, O.; Nadeau, V.; Be, F.; Charron, J.; Jeannotte, L.; Boucherat, O.; Nadeau, V.; Be, F.; Charron, J.; Jeannotte, L. Erratum to Crucial Requirement of Erk/Mapk Signaling in Respiratory Tract Development. Development 2015, 142, 3197–3211, Erratum in Development 2015, 141, 3801. [Google Scholar] [CrossRef]
- Coccia, E.M.; Cicala, C.; Charlesworth, A.; Ciccarelli, C.; Rossi, G.B.; Philipson, L.; Sorrentino, V. Regulation and Expression of a Growth Arrest-Specific Gene (Gas5) during Growth, Differentiation, and Development. Mol. Cell. Biol. 1992, 12, 3514–3521. [Google Scholar] [CrossRef]
- Song, J.; Ahn, C.; Chun, C.H.; Jin, E.J. A Long Non-Coding RNA, GAS5, Plays a Critical Role in the Regulation of MiR-21 during Osteoarthritis. J. Orthop. Res. 2014, 32, 1628–1635. [Google Scholar] [CrossRef]
- Liu, X.; She, Y.; Wu, H.; Zhong, D.; Zhang, J. Long Non-Coding RNA Gas5 Regulates Proliferation and Apoptosis in HCS-2/8 Cells and Growth Plate Chondrocytes by Controlling FGF1 Expression via MIR-21 Regulation. J. Biomed. Sci. 2018, 25, 18. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted Deletion Reveals Essential and Overlapping Functions of the MiR-17∼92 Family of MiRNA Clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef]
- De Pontual, L.; Yao, E.; Callier, P.; Faivre, L.; Drouin, V.; Cariou, S.; Van Haeringen, A.; Geneviève, D.; Goldenberg, A.; Oufadem, M.; et al. Germline Deletion of the MiR-17∼92 Cluster Causes Skeletal and Growth Defects in Humans. Nat. Genet. 2011, 43, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.Y.; Li, S.; Jin, P.; Shang, T.; Sun, R.; Kang, Y.; Zhu, W.; Wang, Q.; Zhang, X.; Yin, F.; et al. Dual Functions of MicroRNA-17 in Maintaining Cartilage Homeostasis and Protection against Osteoarthritis. Nat. Commun. 2022, 13, 2447. [Google Scholar] [CrossRef]
- Khan, S.; Brougham, C.L.; Ryan, J.; Sahrudin, A.; O’Neill, G.; Wall, D.; Curran, C.; Newell, J.; Kerin, M.J.; Dwyer, R.M. MiR-379 Regulates Cyclin B1 Expression and Is Decreased in Breast Cancer. PLoS ONE 2013, 8, e68753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, W.; Li, D.; Zheng, J. MiR-379-5p Promotes Chondrocyte Proliferation via Inhibition of PI3K/Akt Pathway by Targeting YBX1 in Osteoarthritis. Cartilage 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Yang, L. Regulation of CircRNA Biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and Characterizing Circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 426. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids Are Single Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base Paired Rod like Structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; He, J.; Qi, L.; Ren, X.; Zhang, C.; Duan, Z.; Yang, K.; Wang, W.; Lu, Q.; Li, Z. Emerging Landscape of Circular RNAs as Biomarkers and Pivotal Regulators in Osteosarcoma. J. Cell. Physiol. 2020, 235, 9037–9058. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Lin, T.; Zhang, X.; Lu, Y.; Gong, L. Identification of Circular RNA Related to Inflammation-Induced Lymphangiogenesis by Microarray Analysis. DNA Cell Biol. 2019, 38, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Li, Y.; Niringiyumukiza, J.D.; Su, P.; Xiang, W. Circular RNA Involvement in Aging: An Emerging Player with Great Potential; Elsevier Ireland Ltd: Shannon, Ireland, 2019; Volume 178. ISBN 860278369 2605.
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated CircRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef]
- Liu, X.; Yan, C.; Deng, X.; Jia, J. Hsa_circularRNA_0079201 Suppresses Chondrocyte Proliferation and Endochondral Ossification by Regulating the MicroRNA-140-3p/SMAD2 Signaling Pathway in Idiopathic Short Stature. Int. J. Mol. Med. 2020, 46, 1993–2006. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, H.H.; Sun, Z.G.; Tang, H.B.; Min, J.K. Whole-Transcriptome Sequencing of Knee Joint Cartilage from Osteoarthritis Patients. Bone Jt. Res. 2019, 8, 290–303. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.; Zhang, F.; Zhang, Y.; Ren, Z.; Lammi, M.J.; Guo, X. Screening for Differentially Expressed Circular RNAs in the Cartilage of Osteoarthritis Patients for Their Diagnostic Value. Genet. Test. Mol. Biomark. 2019, 23, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Xia, Z.; Feng, B.; Bian, Y.; Fan, Y.; Li, Z.; Wu, Z.; Qiu, G.; Weng, X. Circular RNA Expression Profile of Knee Condyle in Osteoarthritis by Illumina HiSeq Platform. J. Cell. Biochem. 2019, 120, 17500–17511. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Du, D.; Chen, A.; Zhu, L. Circular RNA Expression Profile of Articular Chondrocytes in an IL-1β-Induced Mouse Model of Osteoarthritis. Gene 2018, 644, 20–26. [Google Scholar] [CrossRef]
- Li, X.; Xie, C.; Xiao, F.; Su, H.; Li, Z.; Weng, J.; Huang, Y.; He, P. Circular RNA Circ_0000423 Regulates Cartilage ECM Synthesis via Circ_0000423/MiRNA-27b-3p/MMP-13 Axis in Osteoarthritis. Aging 2022, 14, 3400–3415. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yuan, B.; Pei, Z.; Zhang, K.; Ding, Z.; Zhu, S.; Wang, Y.; Guan, Z.; Cao, Y. Circ_0136474 and MMP-13 Suppressed Cell Proliferation by Competitive Binding to MiR-127-5p in Osteoarthritis. J. Cell. Mol. Med. 2019, 23, 6554–6564. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Zhang, Y.; Wang, J.-J. CircRNA Hsa_circ_0005105 Upregulates NAMPT Expression and Promotes Chondrocyte Extracellular Matrix Degradation by Sponging MiR-26a. Cell Biol. Int. 2017, 41, 1283–1289. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, Y.; Wang, C.; Zhang, X.; He, D. CircGCN1L1 Promotes Synoviocyte Proliferation and Chondrocyte Apoptosis by Targeting MiR-330-3p and TNF-α in TMJ Osteoarthritis. Cell Death Dis. 2020, 11, 284. [Google Scholar] [CrossRef]
- Wang, Z.; Rao, Z.; Wang, X.; Jiang, C.; Yang, Y. CircPhc3 Sponging MicroRNA-93-3p Is Involved in the Regulation of Chondrocyte Function by Mechanical Instability in Osteoarthritis. Int. J. Mol. Med. 2022, 49, 6. [Google Scholar] [CrossRef]
- Luobu, Z.; Wang, L.; Jiang, D.; Liao, T.; Luobu, C.; Qunpei, L. CircSCAPER Contributes to IL-1β-Induced Osteoarthritis in Vitro via MiR-140-3p/EZH2 Axis. Bone Jt. Res. 2022, 11, 61–72. [Google Scholar] [CrossRef]
- Yang, D.; Hu, X.; Chen, Y.; Wang, C. Circular RNA Derived from Vacuolar ATPase Assembly Factor VMA21 Suppresses Lipopolysaccharide-Induced Apoptosis of Chondrocytes in Osteoarthritis (OA) by Decreasing Mature MiR-103 Production. Mol. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Yu, Z.; Cong, F.; Zhang, W.; Song, T.; Zhang, S.; Jiang, R. Circular RNA Circ_0020014 Contributes to Osteoarthritis Progression via MiR-613/ADAMTS5 Axis. Bosn. J. Basic Med. Sci. 2022, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Luo, J.; Zeng, S. Circ-LRP1B Functions as a Competing Endogenous RNA to Regulate Proliferation, Apoptosis and Oxidative Stress of LPS-Induced Human C28/I2 Chondrocytes. J. Bioenerg. Biomembr. 2022, 54, 93–108. [Google Scholar] [CrossRef]
- Man, G.; Yang, H.; Shen, K.; Zhang, D.; Zhang, J.; Wu, H.; Zhang, H.; Wang, J. Circular RNA RHOT1 Regulates MiR-142-5p/CCND1 to Participate in Chondrocyte Autophagy and Proliferation in Osteoarthritis. J. Immunol. Res. 2022, 2022, 4370873. [Google Scholar] [CrossRef] [PubMed]
- Wahafu, P.; Xu, A.; Zhao, B.; Tuo, Y.; Yang, J. Circ_0005526 Contributes to Interleukin-1β-Induced Chondrocyte Injury in Osteoarthritis via Upregulating Transcription Factor 4 by Interacting with MiR-142-5p. Bioengineered 2022, 13, 8407–8418. [Google Scholar] [CrossRef]
- He, M.; Jia, Z.; Wen, Y.; Chen, X. Circ_0043947 Contributes to Interleukin 1β-Induced Injury in Chondrocytes by Sponging MiR-671-5p to up-Regulate RTN3 Expression in Osteoarthritis Pathology. J. Orthop. Surg. Res. 2022, 17, 177. [Google Scholar] [CrossRef]
- Shen, L.; Ji, C.; Lin, J.; Yang, H. Regulation of CircADAMTS6-MiR-324-5p-PIK3R3 CeRNA Pathway May Be a Novel Mechanism of IL-1β-Induced Osteoarthritic Chondrocytes. J. Bone Miner. Metab. 2022, 40, 389–401. [Google Scholar] [CrossRef]
- Mencía, A.; Modamio-Høybjør, S.; Redshaw, N.; Morín, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; Del Castillo, I.; Steel, K.P.; Dalmay, T.; et al. Mutations in the Seed Region of Human MiR-96 Are Responsible for Nonsyndromic Progressive Hearing Loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar] [CrossRef]
- Mortier, G.R.; Cohn, D.H.; Cormier-Daire, V.; Hall, C.; Krakow, D.; Mundlos, S.; Nishimura, G.; Robertson, S.; Sangiorgi, L.; Savarirayan, R.; et al. Nosology and Classification of Genetic Skeletal Disorders: 2019 Revision. Am. J. Med. Genet. Part A 2019, 179, 2393–2419. [Google Scholar] [CrossRef] [PubMed]
- Kannu, P.; Campos-Xavier, A.B.; Hull, D.; Martinet, D.; Ballhausen, D.; Bonafé, L. Post-Axial Polydactyly Type A2, Overgrowth and Autistic Traits Associated with a Chromosome 13q31.3 Microduplication Encompassing MiR-17-92 and GPC5. Eur. J. Med. Genet. 2013, 56, 452–457. [Google Scholar] [CrossRef]
- Hemmat, M.; Rumple, M.J.; Mahon, L.W.; Strom, C.M.; Anguiano, A.; Talai, M.; Nguyen, B.; Boyar, F.Z. Short Stature, Digit Anomalies and Dysmorphic Facial Features Are Associated with the Duplication of MiR-17 ∼ 92 Cluster. Mol. Cytogenet. 2014, 7, 27. [Google Scholar] [CrossRef]
- Grigelioniene, G.; Suzuki, H.I.; Taylan, F.; Mirzamohammadi, F.; Borochowitz, Z.U.; Ayturk, U.M.; Tzur, S.; Horemuzova, E.; Lindstrand, A.; Weis, M.A.; et al. Gain-of-Function Mutation of MicroRNA-140 in Human Skeletal Dysplasia. Nat. Med. 2019, 25, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Rajewsky, N. The Evolution of Gene Regulation by Transcription Factors and MicroRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E. Evolution of MicroRNA Diversity and Regulation in Animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).