Abstract
Clubroot is one of the most economically significant diseases worldwide. As a result, many investigations focus on both curing the disease and in-depth molecular studies. Although the first transcriptome dataset for the clubroot disease describing the clubroot disease was published in 2006, many different pathogen–host plant combinations have only recently been investigated and published. Articles presenting -omics data and the clubroot pathogen Plasmodiophora brassicae as well as different host plants were analyzed to summarize the findings in the richness of these datasets. Although genome data for the protist have only recently become available, many effector candidates have been identified, but their functional characterization is incomplete. A better understanding of the life cycle is clearly required to comprehend its function. While only a few proteome studies and metabolome analyses were performed, the majority of studies used microarrays and RNAseq approaches to study transcriptomes. Metabolites, comprising chemical groups like hormones were generally studied in a more targeted manner. Furthermore, functional approaches based on such datasets have been carried out employing mutants, transgenic lines, or ecotypes/cultivars of either Arabidopsis thaliana or other economically important host plants of the Brassica family. This has led to new discoveries of potential genes involved in disease development or in (partial) resistance or tolerance to P. brassicae. The overall contribution of individual experimental setups to a larger picture will be discussed in this review.
Keywords:
brassica host; clubroot; effector; genome; metabolome; microbiome; Plasmodiophora brassicae; proteome; transcriptome 1. Introduction
Understanding the interaction of host plants with pathogens is crucial for determining their life cycle but also for developing control strategies [1]. Furthermore, the host response in terms of metabolism and/or development is critical. While traditional phytopathology approaches and microscopy can be used to monitor the interaction, molecular analyses are needed for a more in-depth view. Specific transcripts, proteins, and metabolites are important for a certain stage of disease development or s combination of host and pathogen properties. When mutants are available, e.g., for the model plant Arabidopsis thaliana, functional investigations can also be performed [2,3]. Additionally, the use of susceptible and resistant Brassica cultivars for transcriptome and proteome analyses has been included (e.g., [4,5,6]). Clubroot disease is one of the most ubiquitous diseases today, having recorded breakouts on all continents [1,7], and it can be found in a plethora of countries worldwide (Figure 1). The red dots indicate the geographical origin of spore isolates/pathotypes that have been sequenced up to date. Many studies employing large datasets to understand the biology of this soilborne phytopathogenic protist have been reported. However, when looking into large sets of data, it can be difficult to extract the key message at a glance, which is what this review aims to do, and in some cases, this might lead to an oversimplified view. The time point of analysis is critical for the expected results since the life cycle of this biotrophic pathogen is so complex. Therefore, the specific and important stages will be described in Section 2 and related to information from potential effectors of P. brassicae that have been identified so far (Figure 2). The aim of this review is to evaluate how much knowledge can be obtained from published genomes and other -omics datasets. Additionally, the data should be made available in a way that allows for comparisons across different datasets utilizing, for example, the same host plant.
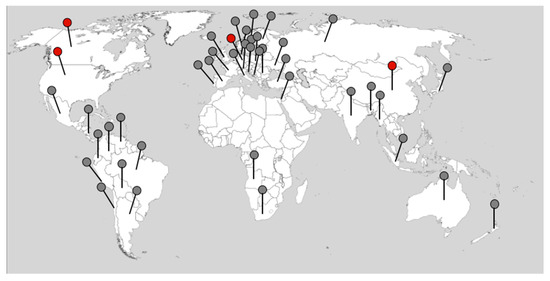
Figure 1.
The worldwide distribution of P. brassicae (based on references until the beginning of 2022). The dots show all countries where the occurrence of P. brassicae was reported. The red dots indicate the geographical origin of those where sequence information is available (based on information beginning in 2022). The map is from https://d-maps.com/carte.php?num_car=13180&lang=de (accessed on 25 February 2022).
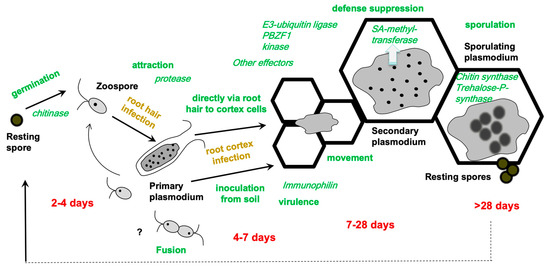
Figure 2.
The most important life stages are given together with the approximate time points of their occurrence based on different host plants and/or isolates or experimental conditions. Typical time intervals are indicated below in red, in green the various processes that are related to P. brassicae colonization are shown, and in brown, the two different phases of the life cycle are indicated. In italics, some important P. brassicae genes/proteins are given for which a function has been experimentally shown. The identification and characterization of the effectors are described in Section 4.1.
2. How to Integrate -Omics Data into the Life Cycle of Plasmodiophora brassicae
Although the life cycle has been detailed in many publications over the past two centuries (e.g., [8,9]), some details are still enigmatic (Figure 2). Recently, the combination of staining methods and microscopy has led to an improved life cycle with the addition of some more, previously questioned, phases [10]. The development of P. brassicae in its host is important to understand since it cannot be cultivated outside its host. In addition, only a few stages, i.e., resting spores and the biflagellated zoospores, exist outside of the host plant in the soil. In susceptible interactions, the stages are the same for all combinations of pathogen isolates and host plant, while they can differ in resistant interactions [11,12]. Aside from the stages, the timing of a particular developmental step within host plants can also differ depending on the virulence of the pathotype of P. brassicae in question. As a result, the time periods in Figure 2 are only estimates for the various associated stages. The life cycle starts with the germination of the resting spore, which transforms into a mobile zoospore. The latter attaches to a root hair [9] or—as recent evidence has also suggested—directly to the epidermis of a root [10]. Within the host cells, the zoospores encyst and grow into a multinucleate plasmodium. During the primary infection, the root hairs are colonized, while during the secondary infection the colonization of the complete cortex occurs. Finally, the multinucleate plasmodia mature into resting spores that are released into the soil when the tissue decomposes.
As previously stated, some stages remain unknown, such as karyogamy and meiosis, colonization of the cortex, and the movement of P. brassicae in the tissue and/or into the vasculature. The latter raises the question of how P. brassicae is restricted to the root and hypocotyl of the host plant. Although this is not the focus of this review, a few points will be added since such questions might be addressed using a combination of -omics and microscopy. Liu et al. [10] described a pair of zoospores that appear to have fused. However, the production of single spore isolates (SSI) [13,14] implies that two mating types are not required, but they do not rule out the possibility of a fusion of the same mating type. Microscopically, dikaryotic myxamoeba-like structures have also been observed in young colonized root tissue, supporting the latter idea [9]. The invasion of the root cortex is the next enigmatic step. Either the zoospores enter via the passage of a root hair to a cortex cell, or via an unknown route from the soil. Some microscopic evidence has been provided for the former [11], but as zoospore multiplication during primary infection is also a hallmark, the second route must be considered as well. While a mechanical mechanism after zoospore attachment for its protoplast injection into the root hair via a so-called Rohr and Stachel apparatus has been established [15], anything analogous to cortex invasion is still missing. The small plasmodia seem to move through the cortex after penetration [16,17,18,19] since in the beginning they are only located at the periphery, but they are rapidly found in inner cells adjacent and even within the vasculature [18]. The division of the plasmodia is accompanied by the division of host cells, resulting in clusters of infected cells, and finally colonization of the entire cortex [20,21]. The plasmodia-harboring cells elongate to provide additional space for the plasmodia to develop into resting spores [21].
3. What Has Been Compared?
The first report on a gene from P. brassicae was published in 1999, while molecular biology using individual genes and/or mutants had begun earlier. However, the gene sequence of this gene was incomplete and had no annotated function [22]. More genes were later discovered utilizing differential expression techniques such as cDNA libraries [23,24,25,26]. In the early years before the reference genome of P. brassicae had been published [27], there were reports on -omics approaches for the host–pathogen interaction, in the beginning only concentrating on the plant side [2,3]. Such datasets are of limited value if no functional analyses, such as using different ecotypes, cultivars, or mutants can be included. Since microarrays only produce plant sequences, information on P. brassicae sequences was not required for these datasets, but it would have been useful for subtractive libraries. Bulman et al. [23,28] and later Sundelin et al. [26] used a suppression subtractive hybridization strategy to identify a larger number of new P. brassicae genes. Dot-blot analyses of over 100 individual P. brassicae sequences were combined with qPCR to investigate their expression levels during the development of primary and secondary zoospores of P. brassicae [29] (Table 1).

Table 1.
The approaches historically and currently used to analyze transcripts on a more global level. The references are selected for a given technique and combination, but they are not exhaustive.
A compilation of the comparisons from several -omics datasets has been created (Figure 3). The term transcriptome covers all methods for the analysis of differential gene expressions such as ESTs, (subtractive) cDNA libraries, microarrays, and RNAseq. Furthermore, miRNAs analysis was carried out [36]. Single cell analyses were performed using laser microdissection coupled to microarray analysis [32]. Only a few experiments reported transcriptomes [37,38] or (targeted) metabolomes [39] of root and leaf tissues. Comparisons have been made for: 1. control vs. infection on different plants or on the same plant, 2. different time points, 3. leaves and roots, 4. resistant vs. susceptible plants, 5. mutant plants vs. wild type, and 6. different cell types [2,5,20,32,35]. For more references on -omics approaches see Section 5, 6 and 7. On the plant side, there are nutrients, transporters, metabolites, energy, cell cycle, defense compounds, and hormones, as well as enzymes and effectors for the pathogen. Epigenetic modifications were also investigated [40]. The soil microbiota also contribute to the outcome of the clubroot disease and were therefore studied in more detail. Analyses were done in either one condition [41], by comparing symptomatic vs. asymptomatic roots in the same field [42], resistant and susceptible cultivars [43], rotation patterns [44], or treatments with fungicides or biocontrol agents [45,46,47], and also virulent vs. avirulent pathotypes of P. brassicae on one host [48]. Only a few “multi-omics” approaches have been carried out, among them hormones and proteomes [3]. For the analysis of lipids, genome, transcriptome, proteome, and metabolome data were combined [49]. The genomic information for P. brassicae has also been translated into knowledge for the life cycle [50]. In addition to exploring individual datasets, analyzing already deposited data for a different research subject, such as done by [51], would be possible.
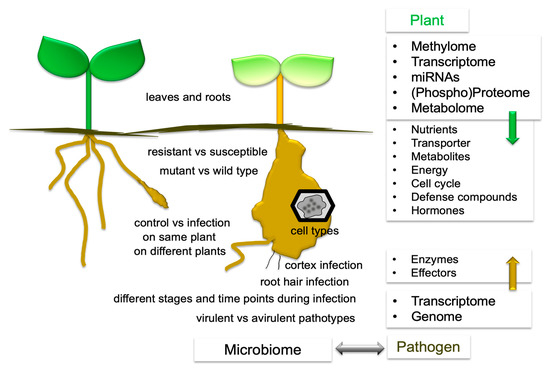
Figure 3.
The experimental setups used for -omics studies with P. brassicae, as well as a summary of the results of the respective datasets. For either plant or pathogen, the -omics techniques are compiled on the right side. The results are partially integrated into a plant model together with the different genetic levels of regulation (see Figure 4).
4. Plasmodiophora brassicae Genomes across the Globe
Since there was insufficient plasmodiophorid genomic information prior to 2015, scientists naturally focused on the plant side of the interaction [52]. The sequencing effort granted to the rhizaria supergroup remains insignificant by comparison to other eukaryotic groups [53,54]. In addition to plasmodiophorids, only three genomes from other rhizaria protists are available: the Chlorarachnea Bigelowiella natans [55], the Foraminifera Reticulomyxa filosa [56], and the Imbricatea Paulinella micropora [57]. An update on the present status of other plant pathogenic plasmodiophorids is given in Section 9.
Before the first reference genome sequence was available, analyses of genome structures were performed via PCR amplification of larger genomic DNA fragments and sequencing. For example, such investigations led to a conclusion concerning the number and size of introns in P. brassicae [28]. Pulse field gel electrophoresis combined with hybridization to marker sequences were employed to characterize spore isolates separated by the host, followed by single spore isolates. They also revealed an approximate number of chromosomes for the ca 25 MB size of the P. brassicae genome [13,58,59,60].
The first genome sequence of the German SSI e3 was released as the “reference genome” in 2015 [27], then re-sequenced in 2019 [61], followed by Canadian [62] and Chinese [63] single spore isolate (SSI) sequences, so that different parts of the world are up to now already covered (Figure 1). The e3 genome was re-sequenced using PacBio sequencing, to provide more structural information [61]. Additionally, another sequence from China was added [64]. The number of genomes in the NCBI database has increased to 50 since then (accessed 2 February 2022) and is in the majority from field isolates [65,66]. Furthermore, the mitochondrial sequence has been described in detail by now [61,67]. Differences in the size and numbers of the mitochondrial genomes seem to be influenced by the sequence technologies utilized as well as the use of two different isolates [61,67]. The possibility of using genome sequences to identify putative effectors has been reviewed by Schwelm and Ludwig-Müller [66] and Perez-Lopez et al. [68].
4.1. Effector Candidates
Bioinformatic approaches based on accessible genomes were used to identify putative effector candidates [27,68] that predict either signal peptides or properties known from other organisms. Several hundred effector genes were predicted by this approach in the genomes currently available, mostly from the European SSI e3 or the Canadian SSI Williams pathotype P3 [27,62,68]. The model (Figure 2) incorporates what is known about P. brassicae proteins at different stages of the life cycle, and Table 2 summarizes the individual characterized P. brassicae proteins.
Effectors are important mediators for the pathogens to regulate the defense response of the respective host [69], but what criteria must a protein meet to be classified as an effector? For example, a protease, Pro1, from P. brassicae was found to be involved in colonization [70]. It came from a screen of differentially expressed genes done by Bulman et al. [23,24], and this dataset was also the source for the SABATH-type methyltransferase PbBSMT [71]. If knocking out this protease results in loss of colonization, it is unquestionable a pathogenicity factor, but strictu sensu it is not an effector that would be involved in defense suppression. Another way to learn more about the roles of P. brassicae proteins is to express them heterologously in other plant pathogenic fungi. A putative cyclophilin gene (PbCyp3), a member of a larger family of 20 genes in the e3 genome, was expressed in Magnaporthe oryzae [72] for such a functional analysis. The strain of M. oryzae lacked the homologous gene, and the PbCyp3 protein could complement the mutant. This resulted in higher virulence on rice plants compared to the mutant strain lacking immunophilin [72]. The protein may not be characterized as an effector that suppresses plant defense because it restored the virulence in its heterologous host.
The first effector for which an in vitro function was shown, converts the defense compound salicylic acid to its methylester when expressed in E. coli [71], and Me-SA is better transported in infected A. thaliana inflorescences. The protein was coined PbBSMT and has homology to plant SABATH-type methyltransferases [71]. The gene sequence came from the dataset of differentially expressed genes that also revealed Pro1 [23,24]. It was highly expressed in the first P. brassicae genome/large-scale transcriptome of the single spore isolate e3 [27]. The high expression of PbBSMT was found throughout the life cycle in all stages [17,71]. PbBSMT mRNA was strongly associated with plasmodia as shown by RNA-FISH [17]. In addition, PbSBMT was among the highest expressed genes in a single root transcriptome assay [33]. The predicted signal peptide was later demonstrated to work in a heterologous yeast system [73]. The subcellular localization of PbBSMT in tobacco revealed its presence within the host cell [73]. Furthermore, transgenic A. thaliana plants overexpressing PbBSMT exhibited a higher Me-SA vs. SA content and were more susceptible to other pathogens [73,74]. The PbBSMT transcript was downregulated in roots of transgenic plants showing tolerance to clubroot infection by overexpression of an A. thaliana mitochondrial protein that provides constitutive SA response [51,75].
The P. brassicae e3 genome contains 139 putative E3 ubiquitin ligases, of which 115 show the conserved RING domain [76]. The authors employed heterologous expression systems approach similar as for PbBSMT to verify (a) the signal sequence in yeast and (b) in E. coli the E3-ubiquitin ligase activity. However, while for the in vitro activity an enzymatic function could be assigned, the in planta target remains unknown.
Since effectors play a role in defense suppression, Chen et al. [77] demonstrated that the P. brassicae effector PBZF1 interacts experimentally with a kinase from A. thaliana that was reported to affect the response to the clubroot pathogen. Overexpression of SnRK1.1 in A. thaliana resulted in more resistant plants. Consequently, PBZF overexpression misregulated known target genes of SnRK1.1 and also resulted in higher susceptibility to the clubroot pathogen itself [77]. Remarkably, it was discovered that key KIN10 (=SnRK1.1) pathway genetic markers [78] are significantly similarly up- and downregulated in clubroot tolerant AtOXR2-overexpressing plants [51]. A significant number of these genes were likewise misregulated in plants overexpressing the PBZF1 gene. The AtOXR2 protein was hypothesized to target the SnKR1.1. pathway to mediate the clubroot tolerance [51], although this needs to be confirmed experimentally.
Evidence for the involvement of proto-oncogenes in clubroot disease came from Bi et al. [63]. The comparison of genomes led to the identification of the so-called GPCR pathway, which stands for G-coupled protein receptor [64]. Treatment with an inhibitor of the GPCR pathway resulted in a reduction of disease symptoms.
Other P. brassicae proteins, such as the PbGH3 protein [27] and a putative homolog of indole-3-acetic acid dehydrogenase [79], have been implicated in the regulation of host hormone metabolism. For the former, an in vitro activity has been shown with the growth hormone IAA, whereas for the latter only a sequence prediction is available. The authors discuss that the potential IAA dehydrogenase could be a virulence factor since IAA is required for gall formation. These gene sequences came from the interaction of B. napus with a Canadian isolate of P. brassicae that can overcome the resistance of local cultivars. In this study, the NUDIX homolog identified by Daval et al. [43], as well as the PbBSMT sequence were also found [79]. Other hormone-related sequences that could be involved in the biosynthesis of CKs were identified in the genomes of P. brassicae (isopentenyltransferase genes) [27,62,80], but functional analyses were not performed on the encoded P. brassicae proteins. The NUDIX effector candidate was discovered during research into the effect of the microbiome on the transcriptome of P. brassicae and a host plant, B. napus, although functional analysis for this protein is lacking [43]. The gene for this protein was highly expressed under conditions where the condition of the microbiome led to high clubroot formation. A NUDIX protein of this type was already reported in a proteome dataset [3]. NUDIX effectors have been identified in other plant pathogens as well [81] and were therefore intriguing candidates in the mediation of the P. brassicae–host plant interaction.
Some effector candidates were identified via transcriptome studies rather than genomes, but the information from these datasets will be given in this context for completeness (Table 2). In the interaction between P. brassicae with A. thaliana a kinase called SSPb22 was discovered with a confirmed function and localization pattern [82]. In this report, several other candidates with putative effector motifs were reported, among them RxLR and Pexel motifs, and again, PbBSMT was identified [82].
A cysteine protease inhibitor could be another promising effector candidate [83]. Its activity has been confirmed indirectly since the resulting protein SSPbP53 was able to interact and inhibit cruciferous papain-like cysteine proteases [83]. Further, the A. thaliana mutant in CYSTEINE PEPTIDASE 1 was more resistant to clubroot than the wild type [83] indicating an important role for this effector in the development of the disease.
Putative effectors with chitin-binding domains were also discovered [84], of which the secretion peptide was confirmed in yeast assays. Co-precipitation in vitro showed binding of these domains to chitin and to P. brassicae resting spores. Furthermore, the participation of these two proteins, PbCHi2 and PbChi4, in chitin-dependent activation of the immune response in B. napus was shown [84].
In tobacco, a collection of putative effectors has been analyzed for their localization and possible function [85]. Several were able to elicit PAMP-triggered immunity in this system, and some were found in the endomembrane system, which indicated their possible translocation into the host plant. Similarly, Chen et al. [86] identified several effector proteins that could induce cell death in tobacco leaves.

Table 2.
Summary of Plasmodiophora brassicae proteins, including putative effectors, identified so far and their possible function.
Table 2.
Summary of Plasmodiophora brassicae proteins, including putative effectors, identified so far and their possible function.
| Effector | Putative Annotated Function | Pathotype | Experimental Verification | Reference |
|---|---|---|---|---|
| Pro 1 (?) 1 | Protease | SSI 2 Williams 3 P3 | In vitro protease activity shown Treatment of plants resulted in better colonization | [70] |
| PbBSMT | SABATH-type methyltransferase | SSI e3 NZ field isolate SSI Williams P3 | In vitro conversion of SA, BA, and AA to their methylester Transgenic A. thaliana plants are more susceptible to pathogens including P. brassicae and have higher Me-SA vs. SA content | [71,73,74] |
| PbCyp3 (?) | Immunophilin | SSI e3 | Heterologous expression in Magnaporthe oryzae mutant restored virulence on rice | [72] |
| PbRING1 | E3-ubiquitin ligase | SSI e3 | E3-ubiquitin ligase activity confirmed in vitro Heterologous expression in yeast confirmed signal peptide function | [76] |
| PBZF1 | RxLR effector | Chinese field isolate Present in SSI e3 and other isolates from databases | Physical interaction with kinase SnRK1.1 Heterologous expression in A. thaliana caused plants to be more susceptible to P. brassicae | [77] |
| SSPbP22 | Kinase | SSI Williams P3 | Kinase activity determined in vitro and protein modeling | [81] |
| SSPbP53 | Cysteine protease inhibitor | SSI Williams P3 | Interaction with and inhibition of cruciferous papain-like cysteine protease A. thaliana mutant in CYSTEINE PEPTIDASE 1 more resistant to clubroot | [82] |
| PbChiB2 PbChiB4 | Chitin-binding domain carbohydrate-binding module family 18 | SSI Williams P3 | Co-precipitation showed in vitro binding to spores of P. brassicae and chitin | [83] |
| GPCR pathway | G-protein coupled receptor pathway | SSI ZJ-1 | Treatment with GPCR inhibitor resulted in reduced symptoms | [64] |
| NUDIX_hydrolase | NUDIX effector | eH, Somé et al. based P1 | No experimental confirmation | [43] |
| PbGH3 | IAA conjugating enzyme | SSI e3 | In vitro conjugation of IAA to various amino acids | [27] |
| Indole-3-acetic acid dehydrogenase | Indole-3-acetic acid dehydrogenase | CCD based P5X | No experimental confirmation; Predicted function in IAA synthesis | [79] |
| Chitin synthase | Chitin synthase | SSI e3 | Sequence prediction | [27] |
| PbTPS 4 | Trehalose-P-synthase | SSI e3 | Indirect by identification of trehalose in resting spores | [87] |
1 (?) no classic effector in the sense of altering plant defense; rather factor needed for colonization; 2 abbreviations: AA: anthranilic acid; BA: benzoic acid; CCD: Canadian Clubroot Differential; IAA: indole-3-acetic acid; Me-SA: methylester of SA; NZ: New Zealand; SA: salicylic acid; SSI: single spore isolate; 3 classification system used; 4 not from an -omics study, but was the first gene fragment with an annotated function.
Some of the effector candidates identified have only been functionally evaluated in transient expression systems. Others have been predicted solely on bioinformatic sequence motifs. The latter group will presumably also contain “false positives”. Further identification of effector candidates therefore needs a different strategy not only based on effector motifs known from other plant pathogens. To summarize the numerous questions remaining: Where are the effectors localized in planta? Some data stem from heterologous transient expression in tobacco [38,85,86], which is not a transformation system close to a host of P. brassicae. Others have employed the host plant A. thaliana [73,74] to address the question of whether they are functional not only in vitro but also in planta. Since P. brassicae can so far not been routinely transformed, this is a central question that will remain. Some in vitro experiments may not be able to tell the exact substrate in planta which must be determined in order to properly comprehend the biological role of an effector. Which more closely related systems for heterologous expression can be used instead of tobacco? Some ideas have been discussed on possible cultured surrogate systems to study the effectome of P. brassicae in the future [88]. Among these are probably more closely related systems using fungal pathogens such as Verticillium dahliae or transient leaf transformation, but not using tobacco, instead using the host A. thaliana together with a hemibiotrophic bacterial pathogen.
4.2. Genomes from Host Plants Can Be Used for Functional Analyses
While genomic information for P. brassicae is gained from a small genome, this is not the case for the host plants. The genomic information is especially of interest for crop plants and not so much for the model A. thaliana, since such information can be used to obtain candidates to breed clubroot-resistant cultivars [89]. The authors review the feasibility of different types of genomic but also transcriptomic information for obtaining information that can be useful to breeders. They cover “…the impact of traditional marker-assisted selection-based breeding for clubroot resistance, and then discuss how -omics approaches have contributed to the (1) detection and genotyping of genome-wide SNP markers linked with clubroot resistance genes or quantitative trait loci, (2) understanding of host resistance mechanisms upon P. brassicae infection, and (3) acceleration of resistance breeding by identifying and characterizing candidate genes, especially those with differential efficacy against new pathotypes of P. brassicae”. Methods that have been used encompass genotyping by sequencing, high-density single nucleotide polymorphism arrays, genome-wide association studies, but also transcriptomic approaches such as (bulk-segregant) RNAseq analysis and microarrays when available for the host plant type.
4.3. Methylome and Epigenetic Regulations
Transcriptional regulation is among the well-accepted possibilities for differential gene expression (see Section 5). However, it is more and more recognized that epigenetic changes also play a role in the regulation of complete genomes. Consequently, the methylation pattern of DNA is involved in many regulatory processes. Specific sequencing techniques, such as bisulfite sequencing [90], can be used to identify the methylome.
A first link between an epigenetic factor and clubroot disease was published by Ludwig-Müller et al. [91], showing that a mutant in LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), at that time isolated as a mutant defective in indole glucosinolate levels, was more tolerant to the disease.
Epigenetic variation in A. thaliana conferred a quantitative disease resistance to clubroot via transgenerational effects [40]. The involvement of DNA methylation in the development of the clubroot disease was supported by the reduction of the disease index in the A. thaliana mutant ddm1 which has an altered methylation pattern. Other variations between ecotypes could perhaps be explained by different methylation patterns, for example in regulatory promoter elements.
Long non-coding RNAs (lncRAs) can also cause epigenetic regulations [92]. Such lncRNAs have been investigated in the response of B. napus to P. brassicae. Overall, 24 differentially expressed lncRNAs were identified in the interaction of resistant and susceptible B. napus lines and P. brassicae [93]. The link between these and chromosome A08, which is known to harbor a quantitative trait locus conferring resistance to different P. brassicae pathotypes, could be used for future studies including resistance responses. However, certain lncRNAs have been discovered to be miRNA targets and thereby constituting posttranscriptional regulations (see Section 5.1) [93]. Zhu et al. [94] integrated sequences of lncRNAs and mRNAs during their investigation of B. campestris and P. brassicae. They provided evidence for a regulatory network between mRNAs and lncRNAs.
5. Transcriptome and Posttranscriptional Regulations
The transcriptome comprises, theoretically, all transcripts that are expressed at a defined time point. However, low-abundant transcripts will be ignored in many experiments, and they must be addressed in more targeted approaches. Furthermore, the choice of meaningful experimental parameters is very important. These can be differential treatments and/or different time points during growth and development. This appears to be rather simple to determine in studies with a single organism, but when two or more organisms interact, the search for such experimental conditions such as the specific time points becomes critical. For example, the early interaction is important for determining the type of interaction, susceptible or resistant/partially resistant, but transcripts connected to later infection pathways are not expressed or at low rates [30]. Genome analyses were also complemented with transcriptomes from various developmental stages of the disease and also stages of the pathogen itself, such as isolated plasmodia or resting spores [27,49,62,64]. The results of the different metabolic and regulatory processes are summarized in Figure 4.
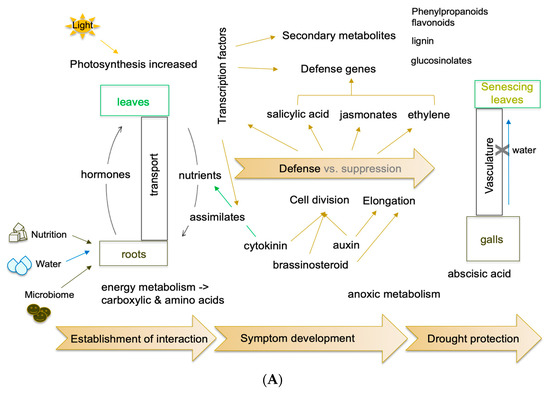
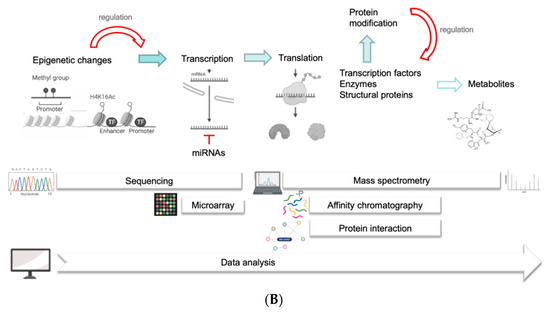
Figure 4.
(A). Model for molecular alterations in clubroots between the leaves and roots of host plants. These are generalized for different hosts (A. thaliana, various Brassica species) and compiled for different approaches to -omics data. (B). The regulatory processes, methods, and techniques are shown in the lower part of the figure. Some cartoons were taken from the free version of Biorender (last accessed 23 April 2022).
Over the years, approaches such as ESTs, (subtractive) cDNA libraries [23,24,25,26], microarray [2,30,31] and RNAseq techniques (e.g., [4,5,20,33,34,35]) have been used. Microarrays are the only method limited to detecting differences between plant tissues and were among the experiments that yielded the first information on global gene expression changes (e.g., [2,30,31]). All other techniques detected mostly the more abundant P. brassicae transcripts. With the reduction of costs, RNAseq is now the method of choice, but it requires competent bioinformatics for evaluation.
Comparing different biological functions with large datasets could help to understand interactions of pathways that are not obvious. For example, the transcriptomes of AtOXR2 and AtKin10 overexpressing plants have been compared to the transcripts of A. thaliana plants overexpressing the P. brassicae effector PbZF1, revealing a possible link between the role of AtOXR2 in tolerance induction and the AtKIN10 pathway targeted by the P. brassicae effector [51]. To back this claim, a statistically significant overlap was identified. The plethora of datasets is used through platforms such as Genevestigator or EFP browser [95,96] which give access to plant experiments. For the P. brassicae–plant interaction such experiments are not included in these databases as yet. Therefore, it is more complicated to compare different datasets.
Gene families were investigated in such large data approaches such as UDP-glucosyltransferases [97], sugar transporter families, including STP and SWEET [98,99], invertase [100], chitinase [101], auxin conjugating GH3 genes [102], pectin-related genes [103], as well as functional pathways, e.g., flavonoids [38,104] or glucosinolates [105]. Other defense-related pathways including hormones will be discussed below (see Section 5.4).
Another challenge is that many different developmental stages co-exist in clubroots with even non-infected cells present. While data evaluating an entire organ can indicate trends when genes are strongly expressed, others might be diluted simply because the host tissue contains infected cells next to uninfected cells and also different developmental stages of P. brassicae at one developmental time point (examples can be found e.g., in [21]). Such developmental stages include small plasmodia in small host cells, dividing plasmodia, and growing plasmodia in already large host cells. Finally, one can find sporulating plasmodia that are about to form the resting spores, which are the stage where, at least in the host cell, possibly not much metabolism is taking place. Therefore, Schuller et al. [32] performed a microarray analysis on cells containing distinct stages of P. brassicae with laser dissection microscopy. As an outcome, several pathways that have been previously overlooked in such studies were identified, among them a role for brassinosteroids in clubroot development. Ludwig-Müller [106] evaluated bioinformatically co-expressed genes after this investigation and identified some promising candidates for further research.
5.1. miRNAs
microRNAs are small non-coding RNAs encoded in the genome and enzymatically cleaved to yield regulatory miRNAs of various sizes. These regulate their target mRNAs by hybridization and targeted degradation [107]. The expression pattern of miRNAs in response to P. brassicae was studied in oilseed rape (or canola) at two time points after inoculation, one early and one later time point, with different developmental stages of P. brassicae present [36]. For the differential regulation of miRNAs, it needs to be anticipated that the corresponding mRNA will be adversely regulated since high expression of a miRNA will result in the degradation of the target and vice versa. Differentially expressed miRNAs were confirmed by qPCR and their RNA targets through degradome analysis and through the analysis of the potential cleavage sites for the miRNAs by RLM-RACE. Targets for the miRNAs include transcription factors (TFs), hormone-related genes (for example auxin response factors, ARFs), as well as genes associated with plant growth or stress response regulation such as cytokinin and auxin/ethylene response elements. Novel miRNAs were identified in the differentially regulated set of another important crop plant, Chinese cabbage (B. rapa) using similar experimental approaches [108]. Extending these findings to resistant and susceptible cultivars of B. napus to P. brassicae identified several specific miRNA–target RNA pairs for the resistant reaction [109]. Consequently, specific miRNAs were identified that regulate disease tolerance genes of the TIR-NBS family in B. napus [110].
5.2. Nutrition
The formation of galls is also influenced by the plant’s nutritional status. Differential regulation of various metabolic pathways can be discovered in many transcriptome datasets. These include all aspects of energy metabolism since the formation of a strong metabolic sink is the essential feature that occurs throughout gall development (Figure 4). Photosynthesis products from the leaves accumulate here together with other nutrients. Among these are amino acids which are also precursors for secondary metabolites. From such datasets, functional categories of proteins involved in nutrient transport or availability have been extracted [98,99,100].
The developing gall is a site of extensive carbohydrate consumption and the redirection of sugars is dependent on many genes including modifiers of cell development, as reviewed by Malinowski et al. [111]. Invertases that catalyze the cleavage of saccharose were upregulated in the clubroot interaction. When tissue-specific expression of an invertase inhibitor protein was utilized, this resulted in a reduction of gall size [100]. Since the sugars need to be translocated into the plasmodia the upregulation of two different classes of sugar transporters was reported [98,99]. Due to the presence of gene families, and that mutants would affect the plant’s overall sugar metabolism, transgenic or mutant approaches are more difficult here. Siemens et al. [100] therefore proposed tissue-specific reductions as a solution. Genes encoding for sucrose degrading enzymes were also detected in the comparison of a susceptible and partial resistant interaction [31].
Another interesting aspect is the energy status of such clubroots. The switch from aerobic to anaerobic metabolism was already noted in [31] and thought to occur due to the accumulation of excessive carbohydrates at the infection site. These modifications occurred in both a partial resistant and a susceptible interaction [31]. There was also an enrichment of genes implicated in hypoxia metabolism in transcriptomes of enriched single cell populations [32]. This observation was corroborated in later work by Gravot et al. [112] demonstrating that the hypoxia response in A. thaliana supported the development of clubroots. Specifically, pyruvate decarboxylase gene mutants were altered in the response to the protist. They discovered an elevation of a group of hypoxia-regulated core genes in infected vs. control roots using the dataset from [80]. Another connection between hypoxia-regulated genes and plants with an altered response to clubroot was reported in [51], where the authors found overlaps between hypoxia-regulated transcriptional datasets and A. thaliana mitochondrial gene OXR2 overexpressing plants.
The effect of nutrient (nitrogen) supply on the transcriptome of resistant and susceptible B. napus was analyzed [113]. The salicylic acid (SA) response was induced irrespective of the nutrient treatment and genotype, whereas the low nitrogen-driven resistance was dependent on low expression of nitrate uptake and nitrate reductase. Genes involved in auxin transport and signaling showed a similar response [113].
5.3. Cell biology and Growth Promoting Hormones
To establish the above-mentioned sink in clubroots (Section 5.2), nutrients need to be transported from the shoots to the infected roots, and a signal is required in the clubroots for this. Since cytokinins (CK) regulate such processes in healthy plants during development [114], they are also thought to play a role in disease development. In addition to their function in sink control, they are also important players in the establishment of the growing tissue by cell division [114]. Plant hormones regulate many features that are important for the growth of the clubroots in A. thaliana and other Brassica species. The cell cycle is important, and once the pathogen has established itself in the root cortex, cell enlargement occurs. Plant hormones orchestrate both responses. Cell cycle control is mediated mainly by CKs, brassinosteroids (BRs), and auxin [115]. The latter is also a major regulator of cell enlargement [115]. Manipulation of these hormones can change gall size, albeit it is usually critical to assess due to its effects on the entire plant growth [2,32,101,105].
Cell cycle genes were identified as critical mediators for the reprogramming of growth of root galls in datasets described for A. thaliana [20]. Further, Malinowski et al. [80] investigated cytokinin biosynthesis-related genes in developing galls based on the transcriptome from [20]. Their findings did not completely support a role for cytokinins in pathogen–plant interactions and growth conditions as strongly as those found in A. thaliana plants overexpressing cytokinin oxidase/dehydrogenase genes [2]. The outcome of experiments might not always be comparable since they strongly depend on the growth/light conditions, the pathotype, and the host genotype.
In a growing plant, auxin and brassinosteroids are responsible for cell division, but also for cell elongation [116]. Many genes encoding proteins in BR synthesis, metabolism, and perception were found to be differentially regulated in the transcriptome dataset from individual cell populations with different plasmodial stages. BRs are involved in club growth since plants mutated in the receptor BRI1 as well as plants treated with a BR biosynthesis inhibitor showed reduced gall size [32].
Auxin homeostasis genes are differentially regulated in various datasets (e.g., [2,31,32,43]), and this has been exploited in some follow-up studies. Among these are the so-called GH3 genes based on the dataset of [2], encoding for proteins involved in the conjugation of IAA to amino acids [117]. GH3 double mutant plants are marginally more susceptible to clubroot, according to functional approaches [102]. Auxin perception and transport are also important since the treatment of infected plants with the respective inhibitors caused the growth of galls to be reduced [102]. Taken together, this research found that two types of auxin receptors, as well as potassium channels, play a role in the elongation response.
Auxin has also been linked to cell wall remodeling [116,118]. Potassium transport has been indicative of acid growth-dependent cell elongation, also involving expansins [102]. Cell wall elongation connected to expansins has already been postulated for the interaction of A. thaliana with P. brassicae on the transcriptome level [2] and was later confirmed with high upregulation of expansin genes in a susceptible interaction of B. napus with P. brassicae [119]. Interestingly, an expansin was also on the gene list that was altered after the overexpression of the P. brassicae effector PBZF1 in A. thaliana [77]. Other cell-wall modifying proteins are encoded by pectin methylesterases, which were analyzed for their role during clubroot development [103] based on the dataset from [80]. Functional analyses confirmed that the club development in a pme18 mutant was altered, namely cell wall enlargement was reduced. Furthermore, the transcriptome data were supplemented by cell wall component analysis, which revealed a change in the content of methylated pectin [104].
Abscisic acid is a stress signal frequently linked to drought stress. In older root galls a strong increase in the number of ABA-associated transcripts was found [2,120]. These old roots are destroyed and therefore, the water transport capacities are diminished. Further work based on gene expression has not been done on ABA-related processes, but ABA was discovered in clubroots during later stages of infection in susceptible interactions (see Section 7).
5.4. Defense Responses and Defense Signals
Clues on defense pathways can be obtained from comparisons of 1. (partial) resistant vs. susceptible cultivars of the same plant species; 2. infected vs. non-infected roots on the same plant; 3. infection of different pathotypes of P. brassicae on one host root; 4. mutants vs. wild-type plants (Figure 2). Since in RNAseq studies also P. brassicae transcripts can be detected, they were also reported in some studies [33], although the majority of examples from the plant side will be discussed in this section.
The defense response of plants is often regulated by plant hormones [121]. For biotrophic pathogens, primarily SA is discussed, but evidence for jasmonates (JA) and ethylene has also been reported in clubroot interactions [111,119]. An early response to SA signaling and a later response to JA were found in the comparison of a partially resistant interaction with a compatible of A. thaliana [62,122]. In roots and shoots of A. thaliana, the infection resulted in the upregulation of the SA- and JA-regulated defense pathways, as well as the shikimate pathway [37].
In B. napus interactions with resistant and susceptible cultivars, the resistance response was mediated mainly by SA pathways [119]. Already early in infection, genes associated with pathogen-associated molecular patterns and effector recognition, SA signaling, pathogenesis-related genes, and cell wall modification were upregulated in resistant vs. susceptible roots [4]. The authors concluded that the recognition in the resistant line was already high during early infection, while recognition pathways were suppressed in the susceptible line [4]. A strong SA-dependent response in a root gall on a single root of B. oleracea var. gongylodes compared to a non-infected one from the same plant revealed strong induction primarily of SA-dependent defense responses and the authors concluded that a non-infected root reacted similarly to a resistant one under their field conditions [33]. They allocated these findings partially to the high expression of PbBSMT in the galls (4.1).
In the resistant B. rapa ssp. rapeifera (line ECD4 from the European Clubroot Differential), carrying resistance to clubroot, many typical resistance genes were annotated in the genome, and evidence for SA- and JA-related defense genes was revealed in the transcriptome [123]. Analyzing the genetic architecture by comparing the transcriptome of different resistant Brassica species with respect to their response to P. brassicae resulted in the identification of a major group of resistance genes, but also in transcripts encoding secondary metabolite pathways and lipid transport [124]. Obviously, the SA response is not responsible for the outcome of the disease in all interactions. A weighted genetic network was constructed that could lead to the identification of more target genes for subsequent analysis. The analysis of differentially expressed genes in B. napus during infection with virulent and avirulent P. brassicae pathotypes revealed no apparent pattern that would be induced by a more or less virulent pathotype [48].
Other authors identified, next to the activation of the SA-dependent defense response, ethylene as a signal in a comparative transcriptome analysis of rutabaga cultivars, another B. napus species, in response to P. brassicae [125]. The respective defense pathways were supported by the differential regulation of WRKY genes that are known as SA response transcription factors [126], and SA- as well as ethylene-responsive genes. In another B. rapa comparison of susceptible and resistant cultivars and P. brassicae, the general induction of defense pathways was confirmed but the authors found additional evidence for upregulation of ethylene and brassinosteroids in the resistance response, whereas the SA response was downregulated [127]. In addition, the role of SA and JA as defense signals was investigated in the comparison of susceptible and resistant B. oleracea var. capitata [5]. SA, JA, ethylene, and brassinosteroids were identified in Chinese cabbage (B. rapa) transcriptomes as defense signals [128]. In conclusion, there is a lot of evidence for SA signaling during many early defense responses, but also for the participation of JA at later time points, whereas a role for ethylene has only been suggested in some experiments. However, A. thaliana mutants in ethylene perception were more susceptible to clubroot [129]. Finally, brassinosteroids have been linked to the development of galls (see Section 5.3), as well as to defense responses.
Evidence for the involvement of secondary metabolite pathways in defense is most consistent for the phenylpropanoid pathway that leads to cell wall components, flavonoids, and other phenolics, among them SA. A strong regulation of the phenylpropanoid metabolism was found after inoculation of susceptible and resistant B. napus cultivars with the same P. brassicae isolate, where, in the latter, more time points showed an elevation of defense associated genes [38]. The pathways linked to lignin and flavonoids/anthocyanidin synthesis were similarly upregulated at early time points, i.e., in the root hair of the A. thaliana–P. brassicae interaction and were regarded as being of major importance [35]. A potential role for flavonoids based on the dataset of [2] was investigated, and PCR was used to confirm the transcriptome data [104]. In situ staining of root galls with a reagent that specifically stains flavonoids and sinapates showed differential accumulation patterns for both compound classes. In addition to the phenylpropanoid pathway, genes encoding for enzymes involved in terpene and glucosinolate pathways were upregulated in an early A. thaliana–P. brassicae interaction [35,105].
6. Proteomes of Plasmodiophora brassicae Inoculated Tissues
While transcriptome studies are easier to conduct, as seen by the amount of publications on the various methodologies, some proteome data on A. thaliana and Brassica species were published early on. However, like with transcriptome data, approaches for analyzing proteomes became more sensitive, resulting in a rise in the number of proteins discovered in such datasets. Furthermore, the quantity and quality of sequences in databases are rising, making peptide sequence annotation easier. Proteins reported in datasets such as differentially expressed confirm transcriptome data (see Section 5) and complement metabolomes [59] (see Section 7). Therefore, the results will be discussed only briefly here since the pairs used for comparisons were often similar [6]. In addition, proteomes were acquired from single conditions [3].
Pioneering work was done in A. thaliana [3] and B. napus [130] and P. brassicae infections where only a small number of proteins were identified, including cell cycle and cell wall modifications [3] and hormone metabolism, calcium signaling and ROS detoxification [130]. Proteome analyses at initial infection stages of Chinese cabbage confirm a role for SA and JA in defense [131]. Other studies utilizing B. napus revealed a resistance response connected with proteins involved in secondary metabolite pathways [132] or, for B. oleracea, an involvement of ABA in the resistant interaction based on the proteins discovered [6]. A proteomics experimental strategy to elucidating Rcr1-mediated resistance in B. rapa found association with proteolysis and a unique signaling mechanism that needs further investigation [133].
Two studies [134,135] describe the protein profiles of B. rapa in response to P. brassicae utilizing a more targeted high-throughput functional proteomics technique combining iTRAQ affinity chromatography with mass spectrometry for large molecules. This results in the extraction of phosphorylated proteins, which can provide information about the post-transcriptional changes in plant proteins caused by P. brassicae interaction. During the secondary phase of the infection cycle, proteins involved in energy and lipid metabolism, plant defense, cell wall modification, and hormone production and signaling were discovered to be differently expressed [135]. The role of brassinosteroid biosynthesis was verified in this study. The other experiment revealed evidence for glutathione in the resistant and cytokinin production in the susceptible B. rapa interaction.
7. Metabolome Analyses of Plasmodiophora brassicae Inoculated Tissues
Metabolites are the end products of enzymatic reactions, which are catalyzed by proteins, and these are encoded by the transcripts. It is therefore evident that the metabolites are the end results of biochemical reactions, whereas the proteins can also encompass regulatory ones such as transcription factors or kinases.
“A hormone and proteome approach…” was the title of the first publication in this methodological aspect of clubroot research, but Devos et al. [3] reported only on a few compounds and proteins. Further work on hormones is still restricted to a more targeted approach since untargeted metabolome analyses usually do not find plant hormones that are present in lower amounts. Furthermore, such metabolite patterns have often been supported by transcriptome datasets. The presence of metabolites involved in oxidative stress [112] was also confirmed in the microarray experiment by Schuller et al. [32] who found a significant number of differentially regulated genes involved in anoxic metabolism. Another example is lipid metabolism, where several -omics techniques were used [49] which are discussed in greater detail here.
Scanning electron microscopy of P. brassicae infected roots in early publications showed the presence of lipids as putative storage vesicles in secondary plasmodia [136]. However, the analysis of lipid compounds is more complicated due to their chemical nature. While transcriptome approaches had detected differentially expressed genes in lipid pathways, their presence in the plant and/or pathogen was only demonstrated in the “multi-omics” approach using genomic, transcriptome, proteome, and (targeted) metabolome data to characterize the patterns of lipid biosynthesis associated pathways in P. brassicae [49]. The authors confirmed the presence of lipid-containing structures in resting spores, germinating spores, and primary and secondary plasmodia by Nile red staining. Physiological evidence for alterations in lipid patterns in infected vs. control roots of A. thaliana was previously reported using a TLC analysis approach with staining for triacylglycerol derivatives [137] and on a histological basis using Nile red [138]. Bi et al. [49] performed proteome and lipid analysis on isolated lipid droplets of P. brassicae. They identified over 200 proteins associated with these lipid droplets and their transcriptome. Further, lipid analysis was done to identify the free fatty acid composition. These analyses revealed that P. brassicae lipid droplets are rich in arachidonic acids [49]. Fatty acid biosynthesis and metabolism were discovered by KEGG pathway analysis for putative functions. This is probably the most comprehensive analysis for one group of metabolites, deriving from the pathogen in clubroots. Schwelm et al. [27] speculated that part of the lipid biosynthesis occurs in P. brassicae, but that the pathway is not complete in the protist, and that precursors of lipids would be needed for a complete pathway. Such a feature was discussed as a hallmark of biotrophic pathogens that may not require a complete pathway if the host can supplement missing parts.
Secondary metabolites have been studied more in depth using targeted approaches, such as indole-derived defense compounds, and these reported the production of at least 45 different metabolites in B. napus infected by P. brassicae [139]. A comparative mapping of quantitative trait loci associated with resistance was carried out using metabolome profiling and pathogen quantification in a segregating progeny of B. napus. Distinct metabolic modules were discovered, and the glucosinolate gluconasturtiin was identified in addition to two as yet unidentified compounds within them [140]. This is an intriguing example of metabolic profiling (=metabolomics) and genetics working together. The results for an untargeted analysis on B. rapa ssp. pekinensis roots were not so clear with respect to the metabolites detected in the different interactions, but glucosinolates appeared to be typically higher in infected roots compared to controls [141]. It is clear that more analyses are needed to complement the transcriptomes.
A more global hormone analysis was done by Prerostova et al. [39] comparing control and infected roots and leaves of resistant and susceptible Brassica napus plants. Two groups of growth promoting hormones, auxin, and CKs, as well as defense compounds jasmonates (JA) and SA and the abiotic stress hormone ABA were determined. The patterns reported were quite complicated and accompanied by transcriptional data for hormone-related genes based on qPCR, not RNAseq, so they are not reported in the above transcriptome section. It is noteworthy that this work is one of the few also including leaf materials of infected and control plants. Altogether, the work showed a positive correlation of gall growth with auxin and CKs, as well as upregulation of defense hormones in the resistant cultivar at earlier time points of the study. In leaves and roots, but not galls, ABA was induced during the late response [39] which corresponds to the ABA-related genes that were reported in transcriptomes [2,120]. Furthermore, a comparison of a resistant with a susceptible line and subsequent analysis of their hormonal profiles in addition to the transcriptional response linked ABA to a function in the late susceptible response in roots of B. rapa [127]. In addition to growth-promoting hormones and abiotic stress signals, the defense compounds JA, SA, and the ethylene precursor ACC were determined. A suppression of JA and SA in the resistant line at late stages was found, but their upregulation in the susceptible line [127]. In another study comparing resistant and susceptible B. rapa ssp. pekinensis the SA-dependent resistance pathway was induced in the resistant interaction based on phytohormone analyses, while the susceptible genotypes were higher with growth promoting hormones auxin and CK [141], confirming previous reports.
8. Potential Role for the Microbiome
Microbiome studies have demonstrated that the former can have a significant impact on the latter in other plant microbe interactions [142]. Since these microbiome studies result in a plethora of organisms belonging to pro- and eukaryotes (e.g., [43]), these findings will not be described in detail. Rather, the various conditions that were used to examine the microbiomes associated with clubroot disease will be reviewed. Individual soil microbes can influence the outcome of the clubroot disease, of which most are isolated from soils or the rhizosphere, and such effects of disease reduction are interesting for biocontrol mechanisms (e.g., [143,144]). Such microbes can also be present in more complex soils, so it is of interest to understand this specific aspect better in more detail.
A study employing symptomatic and asymptomatic roots of B. napus from the same field gave evidence for a strong alteration in the microbiome between such roots [42]. The number of microbes, both bacteria and fungi, associated with the roots showing disease symptoms was lower than those associated with healthy roots. A more complex setup was investigated by Daval et al. [43]. They compared two different B. napus cultivars with different resistances to P. brassicae and three soils harboring high, medium, or low microbiota diversities and levels of richness. Clearly, the soil microbiota had an impact on the outcome of the interaction in the genotypes as well as over time. Other treatments and experimental conditions that resulted in microbiome modifications included different rotation patterns for fields to reduce the clubroot incidence [44], and various biocontrol trials such as rice straw, which stabilizes the microbiome and reduces clubroot [45], and the effect of fungicides [47]. For the latter condition, one would, of course, assume that their application will alter the microbes’ composition, but it is nevertheless necessary to analyze such predictions.
When such experiments are taken to the field alterations in the microbiome were seen when the plants were treated with Streptomyces alfalfae to reduce clubroot symptoms [46]. Such considerations are critical for long-term treatments in nature and their effect on the natural microbial populations. Under natural field conditions, the comparison between infected and asymptomatic roots of B. napus also showed differences in the endobiome of roots [42]. The microbial population and its relative abundance in the asymptomatic roots in this study was far higher than that in the symptomatic roots, and many microorganisms in asymptomatic roots have biological control and plant growth promotion functions that could alter the clubroot symptoms. It should be possible to identify even novel microbes originating from natural conditions that are suitable for biocontrol, using such methods.
As already mentioned, not only the root-associated microbes are altered, but also the endophytic community has been analyzed for changes in clubroot infected and control tissues of B. juncea [145]. There were distinct differences in the comparison, such as the dominance of Pseudomonas at the genus level in clubroots, whereas Rhodanobacter was the dominant genus in healthy roots, but there were also some parallels [145].
9. Status of Other Plasmodiophorid Genomes
The systematics for plasmodiophorids, including the pathogenic groups, has been recently again updated [146]. In addition to P. brassicae, two other plasmodiophorid genera are known that infect important crop plants (Figure 5). Polymyxa betae and P. graminis confer the disease symptoms to their hosts by transmitting viral diseases as vectors [147]. Spongospora subterraneana is also a transmitter for a viral disease, but also causes root galls on their host, albeit much smaller than P. brassicae [147,148]. Interestingly, for P. brassicae there is no evidence that it is a vector for the transmission of viral diseases since only the clubroot symptoms have been described. Other pathogenic genera e.g., Maullinia braseltonii invade host plants with low economic importance, such as brown algae [149,150].
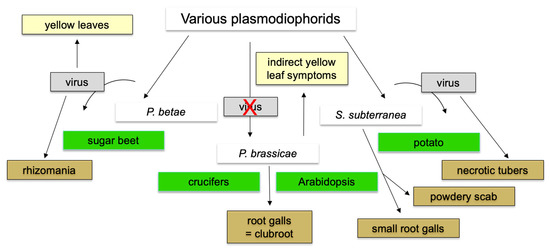
Figure 5.
Comparing three plant pathogenic plasmodiophorids, P. brassicae, P. betae, and S. subterranea for their disease symptoms or virus transmitter capability. Only those with currently available genome information are shown. The red cross indicates no virus transmission.
In a first approach with another plasmodiophorid organism, Mutasa-Göttgens et al. [151] generated 11 Polymyxa betae Expressed Sequence Tags (EST) using a phagemid library of cDNA clones from infected sugar beet roots. The same strategy was applied for Polymyxa graminis, generating four EST [152] and for P. betae, generating 76 P. betae EST as well as 120 sugar beet ESTs upregulated during the different stages of the infection process [153]. Following that, cDNA from five rhizarian species including P. brassicae and Spongospora subterranea ESTs was sequenced using the 454-pyrosequencing technology [154]. Bulman et al. [24] employed dual culture systems to add some more sequences from S. subterranea and P. brassicae that were differentially expressed. With the genomes of S. subterranea in 2018 [155] and P. betae in 2019 [156] available, the molecular biology of plasmodiophorids can now move further. Key genes identified in the P. brassicae interaction could also be important for other plasmodiophorid–host interactions. While the S. subterranea genome is still in a draft status [155], the genome of P. betae has been further improved and the mitochondrial sequence was also annotated and published [156]. The latter revealed that there are differences between P. brassicae and P. betae in genome size and thus also in the number of proteins that are encoded [27,156].
More protist genomes are needed for comparative analyses that fully encompass their diversity. Indeed, there are other pathogenic plasmodiophorids than the ones mentioned above, among them organisms able to colonize algae or oomycota, and therefore being adapted to a different lifestyle, for example in water [157]. Genome data can provide insight into such adaptations.
10. Conclusions
The period of -omics research in clubroot started in the early 2000s. There is a large amount of data that were gathered over the years that followed, and the new data are increasing rapidly. Further, the genomes for P. brassicae pathotypes have added to the complexity. Previously, subtractive cDNA libraries could only discover a few genes from the pathogen, but since this was not exhaustive, one must recognize that the large number of P. brassicae genes identified was attributable to the genomes. It would be ideal if such datasets are readily available to the community. The plant side is better covered, but also here the availability of the datasets does not necessarily guarantee their widespread use, such as for comparisons.
While effectors have been predicted using bioinformatics on genomes, more methods for their functional analysis are required. Other gene predictions need to be carried out as well. Since the sequence of P. brassicae differs significantly from other organisms, the functional prediction should be complemented by other methods. These can include the modeling of protein structures based on web-based tools such as SWISS MODEL or others (e.g., [158]). Several of the above-mentioned effectors, for example, have been modeled and the functional prediction could be supported by other experimental approaches [71,76].
Heterologous transformation will remain a method of choice since the clubroot pathogen up to now cannot be routinely transformed. Expression studies in microorganisms, such as E. coli, have led to the identification of a putative protease function during early infection [70], a SABATH-type methyltransferase (PbBSMT) that can methylate the defense compounds salicylic acid (SA), benzoic acid and anthranilic acid [71] and a GH3-type auxin conjugate synthetase [27]. The expression of a gene encoding a RING domain protein has also given some clues to its function [76]. The function of the secretory peptide of the PbBSMT protein was validated in vitro using a heterologous artificial yeast system [73]. In addition, the heterologous expression of P. brassicae candidate genes in respective mutants of fungi has been done and confirmed the role of another P. brassicae protein in vitro [72] (Table 2).
Transient transformation of tobacco leaves is another potential strategy for heterologous transformation; however, this is possibly an even more artificial system, although results can be generated relatively quickly, similar to microbes. The gene encoding PbBSMT was transiently expressed in tobacco and the authors showed a higher susceptibility to a leaf pathogen, Pseudomonas [62,73]. Finally, the genes can be heterologously transferred into host plants like A. thaliana. This was done to show that overexpression of PbBSMT caused more susceptible plants [74] as well as higher levels of the SA methylester (Me-SA) vs. SA [73]. Other examples used a larger-scale experimental approach with tobacco leaves and transient transformation of numerous effectors [84,85]. These also led to some information on their localization but were all rather artificial involving a no-host organism and also the “wrong” organs, namely leaves. As discussed in [88], other model systems for functional analyses are therefore required.
Funding
This review article received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Apologies to those who were not cited due to oversight of recent work, “hidden” datasets, or the need to concentrate here.
Conflicts of Interest
The author declare no conflict of interest.
References
- Dixon, G.R. The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Siemens, J.; Keller, I.; Sarx, J.; Kunz, S.; Schuller, A.; Nagel, W.; Schmülling, T.; Parniske, M.; Ludwig-Müller, J. Transcriptome Analysis of Arabidopsis Clubroots Indicate a Key Role for Cytokinins in Disease Development. Mol. Plant Microbe Interact. 2006, 19, 480–494. [Google Scholar] [CrossRef] [Green Version]
- Devos, S.; Laukens, K.; Deckers, P.; Van Der Straeten, D.; Beeckman, T.; Inzé, D.; Van Onckelen, H.; Witters, E.; Prinsen, E. A Hormone and Proteome Approach to Picturing the Initial Metabolic Events during Plasmodiophora brassicae Infection on Arabidopsis. Mol. Plant Microbe Interact. 2006, 19, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome Analysis of Brassica rapa Near-Isogenic Lines Carrying Clubroot-Resistant and –Susceptible Alleles in Response to Plasmodiophora brassicae during Early Infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef] [Green Version]
- Ning, Y.; Wang, Y.; Fang, Z.; Zhuang, M. Comparative Transcriptome Analysis of Cabbage (Brassica oleracea var. capitata) Infected by Plasmodiophora brassicae Reveals Drastic Defense Response at Secondary Infection Stage. Plant Soil 2019, 443, 167–183. [Google Scholar] [CrossRef]
- Moon, J.Y.; Tae, S.; Gyung, K.; Choi, J.; Yoon, S.; Cho, H.S.; Kim, H.S. Comparative Proteomic Analysis of Host Responses to Plasmodiophora brassicae Infection in Susceptible and Resistant Brassica oleracea. Plant Biotechnol. Rep. 2020, 14, 263–274. [Google Scholar] [CrossRef]
- Gossen, B.D.; Strelkov, S.E.; Todd, C.D. Clubroot Disease in Latin America: Distribution and Management Strategies. Plant Pathol. 2019, 68, 827–833. [Google Scholar] [CrossRef] [Green Version]
- Tommerup, I.; Ingram, D. The Life-Cycle of Plasmodiophora brassicae Woron. In Brassica Tissue Cultures and in Intact Roots. New Phytol. 1971, 70, 327–332. [Google Scholar] [CrossRef]
- Kageyama, K.; Asano, T. Life Cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar] [CrossRef]
- Liu, L.; Qin, L.; Zhou, Z.; Hendriks, W.G.H.M.; Liu, S.; Wei, Y. Refining the Life Cycle of Plasmodiophora brassicae. Phytopathology 2020, 110, 1704–1712. [Google Scholar] [CrossRef]
- Donald, E.C.; Jaudzems, G.; Porter, I.J. Pathology of Cortical Invasion by Plasmodiophora brassicae in Clubroot Resistant and Susceptible Brassica oleracea Hosts. Plant Pathol. 2008, 57, 201–209. [Google Scholar] [CrossRef]
- Kobelt, P.; Siemens, J.; Sacristán, M.D. Histological Characterisation of the Incompatible Interaction between Arabidopsis thaliana and the Obligate Biotrophic Pathogen Plasmodiophora brassicae. Mycol. Res. 2000, 104, 220–225. [Google Scholar] [CrossRef]
- Fähling, M.; Graf, H.; Siemens, J. Characterization of a Single-Spore Isolate Population of Plasmodiophora brassicae Resulting from a Single Club. J. Phytopathol. 2004, 152, 438–444. [Google Scholar] [CrossRef]
- Klewer, A.; Luerßen, H.; Graf, H.; Siemens, J. Restriction Fragment Length Polymorphism Markers to Characterize Plasmodiophora brassicae Single-Spore Isolates with Different Virulence Patterns. J. Phytopathol. 2001, 149, 121–128. [Google Scholar] [CrossRef]
- Buczacki, S.T.; Clay, C.M. Some Observations on Secondary Zoospore Development in Plasmodiophora brassicae. Trans. Br. Mycol. Soc. 1984, 82, 339–342. [Google Scholar] [CrossRef]
- Asano, T.; Kageyama, K. Growth and Movement of Secondary Plasmodia of Plasmodiophora brassicae in Turnip Suspension-Culture Cells. Plant Pathol. 2006, 55, 145–151. [Google Scholar] [CrossRef]
- Badstöber, J.; Gachon, C.M.M.; Ludwig-Müller, J.; Sandbichler, A.M.; Neuhauser, S. Demystifying Biotrophs: FISHing for mRNAs to Decipher Plant and Algal Pathogen–Host Interaction at the Single Cell Level. Sci. Rep. 2020, 10, 14269. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Bennett, R.; Kiddle, G.; Ihmig, S.; Ruppel, M.; Hilgenberg, W. The Host Range of Plasmodiophora brassicae and Its Relationship to Endogenous Glucosinolate Content. New Phytol. 1999, 141, 443–458. [Google Scholar] [CrossRef] [Green Version]
- Mühlenberg, I.; Schuller, A.; Siemens, J.; Kobelt, P.; Ludwig-Müller, J. Plasmodiophora brassicae, the Causal Agent of Clubroot Disease, May Penetrate Plant Cell Walls via Cellulase. Plant Prot. Sci. 2003, 38, 69–72. [Google Scholar] [CrossRef] [Green Version]
- Olszak, M.; Truman, W.; Stefanowicz, K.; Sliwinska, E.; Ito, M.; Walerowski, P.; Rolfe, S.; Malinowski, R. Transcriptional Profiling Identifies Critical Steps of Cell Cycle Reprogramming Necessary for Plasmodiophora brassicae-Driven Gall Formation in Arabidopsis. Plant J. 2019, 97, 715–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuller, A.; Ludwig-Müller, J. Histological Methods to Detect the Clubroot Pathogen Plasmodiophora brassicae during Its Complex Life Cycle. Plant Pathol. 2016, 65, 1223–1237. [Google Scholar] [CrossRef]
- Ito, S.; Ichinose, H.; Yanagi, C.; Tanaka, S.; Kameya-Iwaki, M.; Kishi, F. Identification of an in Planta-Induced mRNA of Plasmodiophora brassicae. J. Phytopathol. 1999, 147, 79–82. [Google Scholar] [CrossRef]
- Bulman, S.; Siemens, J.; Ridgway, H.J.; Eady, C.; Conner, A.J. Identification of Genes from the Obligate Intracellular Plant Pathogen, Plasmodiophora brassicae. FEMS Microbiol. Lett. 2006, 264, 198–204. [Google Scholar] [CrossRef] [Green Version]
- Bulman, S.; Candy, J.M.; Fiers, M.; Lister, R.; Conner, A.J.; Eady, C.C. Genomics of Biotrophic, Plant-Infecting Plasmodiophorids Using In Vitro Dual Cultures. Protist 2011, 162, 449–461. [Google Scholar] [CrossRef]
- Siemens, J.; Graf, H.; Bulman, S.; In, O.; Ludwig-Müller, J. Monitoring Expression of Selected Plasmodiophora brassicae Genes during Clubroot Development in Arabidopsis thaliana. Plant Pathol. 2009, 58, 130–136. [Google Scholar] [CrossRef]
- Sundelin, T.; Jensen, D.F.; Lübeck, M. Identification of Expressed Genes during Infection of Chinese Cabbage (Brassica rapa subsp. pekinensis) by Plasmodiophora brassicae. J. Eukaryot. Microbiol. 2011, 58, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Fogelqvist, J.; Knaust, A.; Jülke, S.; Lilja, T.; Bonilla-Rosso, G.; Karlsson, M.; Shevchenko, A.; Dhandapani, V.; Choi, S.R.; et al. The Plasmodiophora brassicae Genome Reveals Insights in Its Life Cycle and Ancestry of Chitin Synthases. Sci. Rep. 2015, 5, 11153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulman, S.; Ridgway, H.J.; Eady, C.; Conner, A.J. Intron-Rich Gene Structure in the Intracellular Plant Parasite Plasmodiophora brassicae. Protist 2007, 158, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Hwang, S.F.; Strelkov, S.E. Assessment of Gene Expression Profiles in Primary and Secondary Zoospores of Plasmodiophora brassicae by Dot Blot and Real-Time PCR. Microbiol. Res. 2013, 168, 518–524. [Google Scholar] [CrossRef]
- Agarwal, A.; Kaul, V.; Faggian, R.; Rookes, J.E.; Ludwig-Müller, J.; Cahill, D.M. Analysis of Global Host Gene Expression during the Primary Phase of the Arabidopsis thaliana–Plasmodiophora brassicae Interaction. Funct. Plant Biol. 2011, 38, 462–478. [Google Scholar] [CrossRef] [Green Version]
- Jubault, M.; Lariagon, C.; Taconnat, L.; Renou, J.P.; Gravot, A.; Delourme, R.; Manzanares-Dauleux, M.J. Partial Resistance to Clubroot in Arabidopsis Is Based on Changes in the Host Primary Metabolism and Targeted Cell Division and Expansion Capacity. Funct. Integr. Genom. 2013, 13, 191–205. [Google Scholar] [CrossRef] [Green Version]
- Schuller, A.; Kehr, J.; Ludwig-Müller, J. Laser Microdissection Coupled to Transcriptional Profiling of Arabidopsis Roots Inoculated by Plasmodiophora brassicae Indicates a Role for Brassinosteroids in Clubroot Formation. Plant Cell Physiol. 2014, 55, 392–411. [Google Scholar] [CrossRef]
- Ciaghi, S.; Schwelm, A.; Neuhauser, S. Transcriptomic Response in Symptomless Roots of Clubroot Infected Kohlrabi (Brassica oleracea var. gongylodes) Mirrors Resistant Plants. BMC Plant Biol. 2019, 19, 288. [Google Scholar]
- Zhang, X.; Liu, Y.; Fang, Z.; Li, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. Comparative Transcriptome Analysis between Broccoli (Brassica oleracea var. italica) and Wild Cabbage (Brassica macrocarpa Guss.) in Response to Plasmodiophora brassicae during Different Infection Stages. Front. Plant Sci. 2016, 7, 1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Bi, K.; Gao, Z.; Chen, T.; Liu, H.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D. Transcriptome Analysis of Arabidopsis thaliana in Response to Plasmodiophora brassicae during Early Infection. Front. Microbiol. 2017, 8, 673. [Google Scholar] [CrossRef]
- Verma, S.S.; Rahman, M.H.; Deyholos, M.K.; Basu, U.; Kav, N.N.V. Differential Expression of miRNAs in Brassica napus Root Following Infection with Plasmodiophora brassicae. PLoS ONE 2014, 9, e86648. [Google Scholar] [CrossRef] [PubMed]
- Irani, S.; Trost, B.; Waldner, M.; Nayidu, N.; Tu, J.; Kusalik, A.J.; Todd, C.D.; Wei, Y.; Bonham-Smith, P.C. Transcriptome Analysis of Response to Plasmodiophora brassicae Infection in the Arabidopsis Shoot and Root. BMC Genom. 2018, 19, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irani, S.; Todd, C.D.; Wei, Y.; Bonham-Smith, P.C. Changes in Phenylpropanoid Pathway Gene Expression in Roots and Leaves of Susceptible and Resistant Brassica napus Lines in Response to Plasmodiophora brassicae Inoculation. Physiol. Mol. Plant Pathol. 2019, 106, 196–203. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Konradyova, V.; Knirsch, V.; Gaudinova, A.; Kramna, B.; Kazda, J.; Ludwig-Müller, J.; Vankova, R. Hormonal Responses to Plasmodiophora brassicae Infection in Brassica napus Cultivars Differing in Their Pathogen Resistance. Int. J. Mol. Sci. 2018, 19, 4024. [Google Scholar] [CrossRef] [Green Version]
- Liégard, B.; Baillet, V.; Etcheverry, M.; Joseph, E.; Lariagon, C.; Lemoine, J.; Evrard, A.; Colot, V.; Gravot, A.; Manzanares-Dauleux, M.J.; et al. Quantitative Resistance to Clubroot Infection Mediated by Transgenerational Epigenetic Variation in Arabidopsis. New Phytol. 2019, 222, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, L.; Guillerm-Erckelboudt, A.Y.; Gazengel, K.; Linglin, J.; Ourry, M.; Glory, P.; Sarniguet, A.; Daval, S.; Manzanares-Dauleux, M.J.; Mougel, C. Temporal Dynamics of Bacterial and Fungal Communities during the Infection of Brassica rapa Roots by the Protist Plasmodiophora brassicae. PLoS ONE 2019, 14, e0204195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Gao, Z.; Tian, B.; Bi, K.; Chen, T.; Liu, H.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D. Endosphere Microbiome Comparison between Symptomatic and Asymptomatic Roots of Brassica napus Infected with Plasmodiophora brassicae. PLoS ONE 2017, 12, e0185907. [Google Scholar] [CrossRef] [Green Version]
- Daval, S.; Gazengel, K.; Belcour, A.; Linglin, J.; Sarniguet, A.; Manzanares-Dauleux, M.J.; Mougel, C. Soil Microbiota Influences Clubroot Disease by Modulating Plasmodiophora brassicae and Brassica napus Transcriptomes. Microb. Biotechnol. 2020, 13, 1648–1672. [Google Scholar] [CrossRef]
- Yang, H.; Zheng, J.; Fu, Y.D.; Zhang, Y.H.; Yi, C.L. Specific Genes and Sequence Variation in Pathotype 7 of the Clubroot Pathogen Plasmodiophora brassicae. Eur. J. Plant Pathol. 2020, 157, 17–28. [Google Scholar] [CrossRef]
- Han, Z.; Di, C.; Khashi, M.; Gao, D.; Wu, F.; Pan, K. Repeated Application of Rice Straw Stabilizes Soil Bacterial Community Composition and Inhibits Clubroot Disease. Agriculture 2021, 11, 108. [Google Scholar] [CrossRef]
- Hu, Y.; Qiu, L.; Zhang, Z.; Liu, K.; Xia, X.; Xiong, S.; Zhao, S.; Zhao, Z.; Hu, Y.; Liang, Y. Control of Streptomyces Alfalfae XY25 T Over Clubroot Disease and Its Effect on Rhizosphere Microbial Community in Chinese Cabbage Field Trials. Front. Microbiol. 2021, 12, 641556. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Luo, L.; Zhang, L.; Wang, L.; Shi, X.; Yang, H.; Tan, S.; Tan, L. Comparison of the Effects of Three Fungicides on Clubroot Disease of Tumorous Stem Mustard and Soil Bacterial Community. J. Soils Sediments 2022, 22, 256–271. [Google Scholar] [CrossRef]
- Jiang, J.; Fredua-Agyeman, R.; Hwang, S.-F.; Strelkov, S.E. Differentially expressed genes in canola (Brassica napus) during infection by virulent and avirulent Plasmodiophora brassicae pathotypes. Plant Pathol. 2021, 70, 50–60. [Google Scholar] [CrossRef]
- Bi, K.; He, Z.; Gao, Z.; Zhao, Y.; Fu, Y.; Cheng, J.; Xie, J.; Jiang, D.; Chen, T. Integrated Omics Study of Lipid Droplets from Plasmodiophora brassicae. Sci. Rep. 2016, 6, 36965. [Google Scholar] [CrossRef] [Green Version]
- Schwelm, A.; Dixelius, C.; Ludwig-Müller, J. New Kid on the Block—The Clubroot Pathogen Genome Moves the Plasmodiophorids into the Genomic Era. Eur. J. Plant Pathol. 2016, 145, 531–542. [Google Scholar] [CrossRef]
- Mencia, R.; Welchen, E.; Auer, S.; Ludwig-Müller, J. A Novel Target (Oxidation Resistant 2) in Arabidopsis thaliana to Reduce Clubroot Disease Symptoms via the Salicylic Acid Pathway without Growth Penalties. Horticulturae 2022, 8, 9. [Google Scholar] [CrossRef]
- Neuhauser, S.; Bulman, S.; Kirchmair, M. Plasmodiophorids: The challenge to understand soil-borne, obligate biotrophs with a multiphasic life cycle. In Molecular Identification of Fungi; Gherbawy, Y., Voigt, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 51–78. [Google Scholar]
- Burki, F.; Keeling, P.J. Rhizaria. Curr. Biol. 2014, 24, 103–107. [Google Scholar] [CrossRef] [Green Version]
- Sibbald, S.J.; Archibald, J.M. More protist genomes needed. Nat. Ecol. Evol. 2017, 1, 145. [Google Scholar] [CrossRef]
- Curtis, B.; Tanifuji, G.; Burki, F.; Gruber, A.; Irimia, M.; Maruyama, S.; Arias, M.C.; Ball, S.G.; Gile, G.H.; Hirakawa, Y.; et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 2012, 492, 59–65. [Google Scholar] [CrossRef]
- Glöckner, G.; Hülsmann, N.; Schleicher, M.; Noegel, A.A.; Eichinger, L.; Gallinger, C.; Pawlowski, J.; Sierra, R.; Euteneuer, U.; Pillet, L.; et al. The genome of the foraminiferan Reticulomyxa filosa. Curr. Biol. 2014, 24, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Lhee, D.; Lee, J.; Cho, C.H.; Ha, J.-S.; Jeong, S.E.; Jeon, C.O.; Zelzion, U.; Price, D.C.; Chan, Y.-F.; Gabr, A.; et al. Paulinella micropora KR01 holobiont genome assembly for studying primary plastid evolution. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Fähling, M.; Graf, H.; Siemens, J. Pathotype Separation of Plasmodiophora brassicae by the Host Plant. J. Phytopathol. 2003, 151, 425–430. [Google Scholar] [CrossRef]
- Graf, H.; Sokolowski, F.; Klewer, A.; Diederichsen, E.; Luerßen, H.; Siemens, J. Electrophoretic Karyotype of the Obligate Biotrophic Parasite Plasmodiophora brassicae Wor. J. Phytopathol. 2001, 149, 313–318. [Google Scholar] [CrossRef]
- Graf, H.; Fähling, M.; Siemens, J. Chromosome Polymorphism of the Obligate Biotrophic Parasite Plasmodiophora brassicae. J. Phytopathol. 2004, 152, 86–91. [Google Scholar] [CrossRef]
- Stjelja, S.; Fogelqvist, J.; Tellgren-roth, C.; Dixelius, C. The Architecture of the Plasmodiophora brassicae Nuclear and Mitochondrial Genomes. Sci. Rep. 2019, 9, 15753. [Google Scholar] [CrossRef] [Green Version]
- Rolfe, S.A.; Strelkov, S.E.; Links, M.G.; Clarke, W.E.; Robinson, S.J.; Djavaheri, M.; Malinowski, R.; Haddadi, P.; Kagale, S.; Parkin, I.A.P.; et al. The Compact Genome of the Plant Pathogen Plasmodiophora brassicae Is Adapted to Intracellular Interactions with Host Brassica spp. BMC Genom. 2016, 17, 272. [Google Scholar] [CrossRef] [Green Version]
- Bi, K.; Chen, T.; He, Z.; Gao, Z.; Zhao, Y.; Fu, Y.; Cheng, J.; Xie, J.; Jiang, D. Proto-Oncogenes in a Eukaryotic Unicellular Organism Play Essential Roles in Plasmodial Growth in Host Cells. BMC Genom. 2018, 19, 881. [Google Scholar] [CrossRef] [Green Version]
- Bi, K.; Chen, T.; He, Z.; Gao, Z.; Zhao, Y.; Liu, H.; Fu, Y.; Xie, J.; Cheng, J.; Jiang, D. Comparative Genomics Reveals the Unique Evolutionary Status of Plasmodiophora brassicae and the Essential Role of GPCR Signaling Pathways. Phytopathol. Res. 2019, 1, 12. [Google Scholar] [CrossRef]
- Sedaghatkish, A.; Gossen, B.D.; Yu, F.; Torkamaneh, D.; Mcdonald, M.R. Whole-Genome DNA Similarity and Population Structure of Plasmodiophora brassicae Strains from Canada. BMC Genom. 2019, 20, 744. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Ludwig-Müller, J. Molecular Pathotyping of Plasmodiophora brassicae—Genomes, Marker Genes, and Obstacles. Pathogens 2021, 10, 259. [Google Scholar] [CrossRef]
- Daval, S.; Belcour, A.; Gazengel, K.; Legrand, L.; Gouzy, J.; Cottret, L.; Lebreton, L.; Aigu, Y.; Mougel, C.; Manzanares-Dauleux, M.J. Computational Analysis of the Plasmodiophora brassicae Genome: Mitochondrial Sequence Description and Metabolic Pathway Database Design. Genomics 2019, 111, 1629–1640. [Google Scholar] [CrossRef]
- Pérez-López, E.; Waldner, M.; Hossain, M.; Kusalik, A.J.; Wei, Y.; Bonham-Smith, P.C.; Todd, C.D. Identification of Plasmodiophora brassicae Effectors—A Challenging Goal. Virulence 2018, 9, 1344–1353. [Google Scholar] [CrossRef]
- Toruno, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Hwang, R.; Hwang, S.-F.; Strelkov, S.E.; Gossen, B.D.; Zhou, Q.I.X.; Peng, G. Molecular Characterization of a Serine Protease Pro1 from Plasmodiophora brassicae That Stimulates Resting Spore Germination. Mol. Plant Pathol. 2010, 11, 503–512. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Jülke, S.; Geiß, K.; Richter, F.; Mithöfer, A.; Sola, I.; Rusak, G.; Keenan, S.; Bulman, S. A Novel Methyltransferase from the Intracellular Pathogen Plasmodiophora brassicae Methylates Salicylic Acid. Mol. Plant Pathol. 2015, 16, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Tzelepis, G.; Zouhar, M.; Ryšánek, P.; Dixelius, C. The Immunophilin Repertoire of Plasmodiophora brassicae and Functional Analysis of PbCYP3 Cyclophilin. Mol. Genet. Genom. 2017, 293, 381–390. [Google Scholar] [CrossRef] [Green Version]
- Djavaheri, M.; Ma, L.; Klessig, D.F.; Mithöfer, A.; Gropp, G.; Borhan, H. Mimicking the Host Regulation of Salicylic Acid: A Virulence Strategy by the Clubroot Pathogen Plasmodiophora brassicae. Mol. Plant Microbe Interact. 2019, 32, 296–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulman, S.; Richter, F.; Marschollek, S.; Benade, F.; Jülke, S.; Ludwig-Müller, J. Arabidopsis thaliana Expressing PbBSMT, a Gene Encoding a SABATH-Type Methyltransferase from the Plant Pathogenic Protist Plasmodiophora brassicae, Show Leaf Chlorosis and Altered Host Susceptibility. Plant Biol. 2019, 21, 120–130. [Google Scholar] [CrossRef]
- Mencia, R.; Céccoli, G.; Fabro, G.; Torti, P.; Colombatti, F. OXR2 Increases Plant Defense against a Hemibiotrophic Pathogen via the Salicylic Acid Pathway 1. Plant Physiol. 2020, 184, 1112–1127. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Wang, S.; Zhang, W.; Tang, J.; Wang, H.; Yu, L.; Zhang, X.; Fei, Z.; Li, J. Genome-Wide Identification of Genes Encoding Putative Secreted E3 Ubiquitin Ligases and Functional Characterization of PbRING1 in the Biotrophic Protist Plasmodiophora brassicae. Curr. Genet. 2019, 65, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, Y.; Yan, R.; Ren, L.; Liu, F.; Zeng, L.; Sun, S.; Yang, H.; Chen, K.; Xu, L.; et al. SnRK1.1-Mediated Resistance of Arabidopsis thaliana to Clubroot Disease Is Inhibited by the Novel Plasmodiophora brassicae Effector PBZF1. Mol. Plant Pathol. 2021, 22, 1057–1069. [Google Scholar] [CrossRef]
- Baena-Gonzalez, E.; Rolland, F.; Thevelein, J.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Galindo-González, L.; Hwang, S.; Strelkov, S.E. Candidate Effectors of Plasmodiophora brassicae Pathotype 5X During Infection of Two Brassica napus Genotypes. Front. Microbiol. 2021, 12, 742268. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, R.; Novák, O.; Borhan, M.H.; Spíchal, L.; Strnad, M.; Rolfe, S.A. The Role of Cytokinins in Clubroot Disease. Eur. J. Plant Pathol. 2016, 145, 543–557. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.; Wang, Y. Nudix effectors: A common weapon in the arsenal of plant pathogens. PLoS Pathog. 2016, 12, e1005704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-López, E.; Hossain, M.; Tu, J.; Waldner, M.; Todd, C.D.; Kusalik, A.J.; Wei, Y.; Bonham-Smith, P.C. Transcriptome Analysis Identifies Plasmodiophora brassicae Secondary Infection Effector Candidates. J. Eukaryot. Microbiol. 2020, 67, 337–351. [Google Scholar] [CrossRef] [Green Version]
- Pérez-López, E.; Hossain, M.; Wei, Y.; Todd, C.D.; Bonham-Smith, P.C. A Clubroot Pathogen Effector Targets Cruciferous Cysteine Proteases to Suppress Plant Immunity. Virulence 2021, 12, 2327–2340. [Google Scholar] [CrossRef]
- Muirhead, K.; Pérez-López, E. Plasmodiophora brassicae chitin-binding effectors guard and mask spores during infection. bioRxiv 2020. preprint. [Google Scholar] [CrossRef]
- Hossain, M.; Pérez-López, E.; Todd, C.D.; Wei, Y.; Bonham-Smith, P.C. Endomembrane-Targeting Plasmodiophora brassicae Effectors Modulate PAMP Triggered Immune Responses in Plants. Front. Microbiol. 2021, 12, 651279. [Google Scholar] [CrossRef]
- Chen, W.; Li, Y.; Yan, R.; Xu, L.; Ren, L.; Liu, F.; Zeng, L.; Yang, H.; Chi, P.; Wang, X.; et al. Identification and Characterization of Plasmodiophora brassicae Primary Infection Effector Candidates That Suppress or Induce Cell Death in Host and Nonhost Plants. Phytopathology 2019, 109, 1689–1697. [Google Scholar] [CrossRef]
- Brodmann, D.; Schuller, A.; Ludwig-Müller, J.; Aeschbacher, R.A.; Wiemken, A.; Boller, T.; Wingler, A. Induction of Trehalase in Arabidopsis Plants Infected with the Trehalose-Producing Pathogen Plasmodiophora brassicae. Mol. Plant Microbe Interact. 2002, 15, 693–700. [Google Scholar] [CrossRef] [Green Version]
- González-García, M.; Pérez-López, E. Looking for a Cultured Surrogate for Effectome Studies of the Clubroot Pathogen. Front. Microbiol. 2021, 12, 650307. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Galindo-González, L.; Hwang, S.; Strelkov, S.E. Application of Genomics and Transcriptomics to Accelerate Development of Clubroot Resistant Canola Can. J. Plant Pathol. 2021, 43, 189–208. [Google Scholar] [CrossRef]
- Yong, W.S.; Hsu, F.M.; Chen, P.Y. Profiling genome-wide DNA methylation. Epigenet. Chromatin 2016, 9, 26. [Google Scholar] [CrossRef] [Green Version]
- Ludwig-Müller, J.; Pieper, K.; Ruppel, M.; Cohen, J.D.; Epstein, E.; Kiddle, G.; Bennett, R. Indole Glucosinolate and Auxin Biosynthesis in Arabidopsis thaliana (L.) Heynh. Glucosinolate Mutants and the Development of Clubroot Disease. Planta 1999, 208, 409–419. [Google Scholar] [CrossRef]
- Heo, J.B.; Lee, Y.-S.; Sung, S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013, 21, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Summanwar, A.; Basu, U.; Rahman, H.; Kav, N. Identification of LncRNAs Responsive to Infection by Plasmodiophora brassicae in Clubroot-Susceptible and -Resistant Brassica napus Lines Carrying Resistance Introgressed from Rutabaga. Mol. Plant Microbe Interact. 2019, 32, 1360–1377. [Google Scholar] [CrossRef]
- Zhu, H.; Li, X.; Xi, D.; Zhai, W.; Zhang, Z. Integrating Long Noncoding RNAs and MRNAs Expression Profiles of Response to Plasmodiophora brassicae Infection in Pakchoi (Brassica campestris ssp. chinensis Makino). PLoS ONE 2019, 14, e0224927. [Google Scholar] [CrossRef] [Green Version]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A reference expression database for the metaanalysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.; Provart, N. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative Genomic and Transcriptomic Analyses of Family-1 UDP Glycosyltransferase in Three Brassica Species and Arabidopsis Indicates Stress-Responsive Regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.; Li, J. Genome-Wide Characterization and Expression Profiling of SWEET Genes in Cabbage (Brassica oleracea var. capitata L.) Reveal Their Roles in Chilling and Clubroot Disease Responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Yu, L.; Wang, H.; Li, J. Genome-Wide Identification and Expression Profiling of Sugar Transporter Protein (STP) Family Genes in Cabbage (Brassica oleracea var. capitata L.) Reveals Their Involvement in Clubroot Disease Responses. Genes 2019, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Siemens, J.; Gonzales, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-Müller, J. Extracellular Invertase Is Involved in the Regulation of Clubroot Disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011, 12, 247–262. [Google Scholar] [CrossRef]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-Wide Identification and Expression Analysis of Chitinase Gene Family in Brassica rapa Reveals Its Role in Clubroot Resistance. Plant Sci. 2018, 270, 257–267. [Google Scholar] [CrossRef]
- Jahn, L.; Mucha, S.; Bergmann, S.; Horn, C.; Staswick, P.; Steffens, B.; Siemens, J.; Ludwig-Müller, J. The Clubroot Pathogen (Plasmodiophora brassicae) Influences Auxin Signaling to Regulate Auxin Homeostasis in Arabidopsis. Plants 2013, 2, 726–749. [Google Scholar] [CrossRef] [Green Version]
- Stefanowicz, K.; Szymanska-chargot, M.; Truman, W.; Walerowski, P. Cell Enlargement and Loss of Cellular Integrity in Root Systems Are Mediated by Pectin Demethylation. Front. Plant Sci. 2021, 12, 711838. [Google Scholar] [CrossRef] [PubMed]
- Päsold, S.; Siegel, I.; Seidel, C.; Ludwig-Müller, J. Flavonoid Accumulation in Arabidopsis thaliana Root Galls Caused by the Obligate Biotrophic Pathogen Plasmodiophora brassicae. Mol. Plant Pathol. 2010, 11, 545–562. [Google Scholar] [CrossRef]
- Li, L.; Long, Y.; Li, H.; Wu, X. Comparative Transcriptome Analysis Reveals Key Pathways and Hub Genes in Rapeseed During the Early Stage of Plasmodiophora brassicae Infection. Front. Genet. 2020, 10, 1275. [Google Scholar] [CrossRef] [Green Version]
- Ludwig-Müller, J. Auxin Homeostasis, Signaling and Interaction with Other Growth Hormones during the Clubroot Disease of Brassicaceae. Plant Signal. Behav. 2014, 9, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Xu, W.; Yuan, Y.; Yao, Q.; Zhao, Y.; Wang, Z.; Jiang, W.; Zhang, X. Genome-Wide Investigation of MicroRNAs and Their Targets in Brassica rapa ssp. pekinensis Root with Plasmodiophora brassicae Infection. Hortic. Plant J. 2016, 2, 209–216. [Google Scholar] [CrossRef]
- Li, Q.; Shah, N.; Zhou, X.; Wang, H.; Yu, W.; Luo, J.; Liu, Y.; Li, G.; Liu, C.; Zhang, C.; et al. Identification of Micro Ribonucleic Acids and Their Targets in Response to Plasmodiophora brassicae Infection in Brassica napus. Front. Plant Sci. 2021, 12, 734419. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Chhapekar, S.S.; Rameneni, J.J.; Oh, S.H.; Dhandapani, V.; Subburaj, S.; Shin, S.; Ramchiary, N. MiR1885 Regulates Disease Tolerance Genes in Brassica rapa during Early Infection with Plasmodiophora brassicae. Int. J. Mol. Sci. 2021, 22, 9433. [Google Scholar] [CrossRef]
- Malinowski, R.; Truman, W.; Blicharz, S. Genius Architect or Clever Thief—How Plasmodiophora brassicae Reprograms Host Development to Establish a Pathogen-Oriented Physiological Sink. Mol. Plant Microbe Interact. 2019, 32, 1259–1266. [Google Scholar] [CrossRef] [Green Version]
- Gravot, A.; Richard, G.; Lime, T.; Lemarié, S.; Jubault, M.; Lariagon, C.; Lemoine, J.; Vicente, J.; Robert-Seilaniantz, A.; Holdsworth, M.J.; et al. Hypoxia Response in Arabidopsis Roots Infected by Plasmodiophora brassicae Supports the Development of Clubroot. BMC Plant Biol. 2016, 16, 251. [Google Scholar] [CrossRef] [PubMed]
- Aigu, Y.; Daval, S.; Gazengel, K.; Marnet, N.; Lariagon, C.; Laperche, A.; Legeai, F.; Manzanares-Dauleux, M.J.; Gravot, A. Multi-Omic Investigation of Low-Nitrogen Conditional Resistance to Clubroot Reveals Brassica napus Genes Involved in Nitrate Assimilation. Front. Plant Sci. 2022, 13, 790563. [Google Scholar] [CrossRef]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Shimotohno, A.; Aki, S.S.; Takahashi, N.; Umeda, M. Regulation of the Plant Cell Cycle in Response to Hormones and the Environment. Annu. Rev. Plant Biol. 2021, 72, 273–296. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef] [Green Version]
- Galindo-Gonzalez, L.; Manolii, V.; Hwang, S.; Strelkov, S.E. Response of Brassica napus to Plasmodiophora brassicae Involves Salicylic Acid-Mediated Immunity: An RNA-Seq-Based Study. Front. Plant Sci. 2020, 11, 1025. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Plant Defence—What Can We Learn from Clubroots? Australas. Plant Pathol. 2009, 38, 318–324. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015, 56, 2158–2168. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-G.; Noh, E.; Choi, S.; Choi, B.; Shin, I.-G.; Yoo, S.-I.; Lee, D.; Ji, S.; Kim, H.-S.; Hwang, Y.-J.; et al. Draft Genome Assembly and Transcriptome Dataset for European Turnip (Brassica rapa L. ssp. rapifera), ECD4 Carrying Clubroot Resistance. Front. Genet. 2021, 12, 651298. [Google Scholar] [CrossRef]
- Hejna, O.; Havlickova, L.; He, Z.; Bancroft, I.; Curn, V. Analysing the Genetic Architecture of Clubroot Resistance Variation in Brassica napus by Associative Transcriptomics. Mol. Breed. 2019, 39, 112. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Galindo-Gonzalez, L.; Manolii, V.; Hwang, S.; Strelkov, S.E. Comparative Transcriptome Analysis of Rutabaga (Brassica napus) Cultivars Indicates Activation of Salicylic Acid and Ethylene-Mediated Defenses in Response to Plasmodiophora brassicae. Int. J. Mol. Sci. 2020, 21, 8381. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY Transcription Factors in Defense Signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.; Zhang, X.; Yuan, Y. Root Transcriptome and Metabolome Profiling Reveal Key Phytohormone-Related Genes and Pathways Involved Clubroot Resistance in Brassica rapa L. Front. Plant Sci. 2021, 12, 759623. [Google Scholar] [CrossRef]
- Fu, P.; Piao, Y.; Zhan, Z.; Zhao, Y.; Pang, W.; Li, X. Transcriptome Profile of Brassica Rapa L. Reveals the Involvement of Jasmonic Acid, Ethylene, and Brassinosteroid Signaling Pathways in Clubroot Resistance. Agronomy 2019, 9, 589. [Google Scholar] [CrossRef] [Green Version]
- Knaust, A.; Ludwig-Müller, J. The Ethylene Signaling Pathway Is Needed to Restrict Root Gall Growth in Arabidopsis after Infection with the Obligate Biotrophic Protist Plasmodiophora brassicae. J. Plant Growth Regul. 2013, 32, 9–21. [Google Scholar] [CrossRef]
- Cao, T.; Srivastava, S.; Rahman, M.; Kav, N.; Hotte, N.; Deyholos, M.; Strelkov, S. Proteome-Level Changes in the Roots of Brassica napus as a Result of Plasmodiophora brassicae Infection. Plant Sci. 2007, 174, 97–115. [Google Scholar] [CrossRef]
- Ji, R.; Wang, Y.; Wang, X.; Liu, Y.; Shen, X.; Feng, H. Proteomic Analysis of the Interaction between Plasmodiophora brassicae and Chinese Cabbage (Brassica rapa L. ssp. pekinensis) at the Initial Infection Stage. Sci. Hortic. 2018, 233, 386–393. [Google Scholar] [CrossRef]
- Adhikary, D.; Mehta, D.; Uhrig, R.G.; Rahman, H.; Kav, N.N.V. A Proteome-Level Investigation into Plasmodiophora brassicae Resistance in Brassica napus Canola. Front. Plant Sci. 2022, 13, 860393. [Google Scholar] [CrossRef]
- Song, T.; Chu, M.; Lahlali, R.; Yu, F.; Peng, G. Shotgun Label-Free Proteomic Analysis of Clubroot (Plasmodiophora brassicae) Resistance Conferred by the Gene Rcr1 in Brassica rapa. Front. Plant Sci. 2016, 7, 1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, M.; Li, G.; Hu, J.; Yang, H.; Zhang, L.; Xu, X.; Liu, J. ITRAQ-Based Quantitative Analysis Reveals Proteomic Changes in Chinese Cabbage (Brassica rapa L.) in Response to Plasmodiophora brassicae Infection. Sci. Rep. 2019, 9, 12058. [Google Scholar] [CrossRef]
- Su, T.; Yu, S.; Wang, W.; Li, P.; Zhang, F.; Yu, Y.; Zhang, D.; Zhao, X. ITRAQ Analysis of Protein Profile during the Secondary Stage of Infection of Plasmodiophora brassicae in Chinese Cabbage (Brassica rapa). J. Plant Pathol. 2018, 100, 533–542. [Google Scholar] [CrossRef]
- Ikegami, H.; Naiki, T.; Ito, T.; Imuro, Y. Ultrastructural Growth Process of Plasmodiophora brassicae in Infected Cells of Chinese Cabbage and Turnip. Res. Bull. Fac. Agric. Gifu Univ. 1982, 46, 9–19. [Google Scholar]
- Jülke, S.; Ludwig-Müller, J. Modulation of Lipid Transfer Proteins Alters Clubroot Development in Arabidopsis thaliana. Acta Hortic. 2010, 867, 165–172. [Google Scholar] [CrossRef]
- Tu, J.; Bush, J.; Bonham-Smith, P.; Wei, Y. Live Cell Imaging of Plasmodiophora brassicae—Host Plant Interactions Based on a Two-Step Axenic Culture System. Microbiologyopen 2018, 8, e00765. [Google Scholar] [CrossRef] [Green Version]
- Pedras, M.S.C.; Zheng, Q.-A.; Strelkov, S. Metabolic Changes in Roots of the Oilseed Canola Infected with the Biotroph Plasmodiophora brassicae: Phytoalexins and Phytoanticipins. J. Agric. Food Chem. 2008, 56, 9949–9961. [Google Scholar] [CrossRef]
- Wagner, G.; Laperche, A.; Lariagon, C.; Marnet, N.; Renault, D.; Guitton, Y.; Bouchereau, A.; Delourme, R.; Manzanares-dauleux, M.J.; Gravot, A. Resolution of Quantitative Resistance to Clubroot into QTL-Specific Metabolic Modules. J. Exp. Bot. 2019, 70, 5375–5390. [Google Scholar] [CrossRef] [PubMed]
- Lan, M.E.I.; Hu, J.I.; Yang, H.O.; Zhang, L.I.; Xu, X.U.; He, J.I. Phytohormonal and Metabolism Analysis of Brassica rapa L. ssp. pekinensis with Different Resistance during Plasmodiophora brassicae Infection. Biocell 2020, 44, 751–767. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef]
- Lahlali, R.; Peng, G. Suppression of Clubroot by Clonostachys rosea via Antibiosis and Induced Host Resistance. Plant Pathol. 2014, 63, 447–455. [Google Scholar] [CrossRef]
- Zhu, M.; He, Y.; Li, Y.; Ren, T.; Liu, H.; Huang, J.; Jiang, D. Two New Biocontrol Agents Against Clubroot Caused by Plasmodiophora brassicae. Front. Microbiol. 2020, 10, 3099. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sun, T.; Zhao, S.; Pan, L.; Liu, H.; Tian, X. Physiological Change Alters Endophytic Bacterial Community in Clubroot of Tumorous Stem Mustard Infected by Plasmodiophora brassicae. BMC Microbiol. 2020, 20, 244. [Google Scholar] [CrossRef]
- Hittorf, M.; Letsch-Praxmarer, S.; Windegger, A.; Bass, D.; Kirchmair, M.; Neuhauser, S. Revised Taxonomy and Expanded Biodiversity of the Phytomyxea (Rhizaria, Endomyxa). J. Eukaryot. Microbiol. 2020, 67, 648–659. [Google Scholar] [CrossRef]
- Tamada, T.; Kondo, H. Biological and Genetic Diversity of Plasmodiophorid-Transmitted Viruses and Their Vectors. J Gen. Plant Pathol. 2013, 79, 307–320. [Google Scholar] [CrossRef]
- Balendres, M.A.; Tegg, R.S.; Wilson, C.R. Key Events in Pathogenesis of Spongospora Diseases in Potato: A Review. Australas. Plant Pathol. 2016, 45, 229–240. [Google Scholar] [CrossRef]
- Murúa, P.; Goecke, F.; Westermeier, R.; van West, P.; Küpper, F.C.; Neuhauser, S. Maullinia braseltonii Sp. Nov. (Rhizaria, Phytomyxea, Phagomyxida): A Cyst-Forming Parasite of the Bull Kelp Durvillaea Spp. (Stramenopila, Phaeophyceae, Fucales). Protist 2017, 168, 468–480. [Google Scholar] [CrossRef] [Green Version]
- Maier, I.; Parodi, E.; Westermeier, R.; Müller, D.G. Maullinia ectocarpii Gen. et Sp. Nov. (Plasmodiophorea), an Intracellular Parasite in Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) and Other Filamentous Brown Algae. Protist 2000, 151, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Mutasa-Göttgens, E.S.; Chwarzczynska, D.; Halsey, K.; Asher, M.J.C. Specific Polyclonal Antibodies for the Obligate Plant Parasite Polymyxa—A Targeted Recombinant DNA Approach. Plant Pathol. 2000, 49, 276–287. [Google Scholar] [CrossRef]
- Šubr, Z.W.; Kastirr, U.; Kühne, T. Subtractive cloning of DNA from Polymyxa graminis—An obligate parasitic plasmodiophorid. J. Phytopathol. 2002, 150, 564–568. [Google Scholar] [CrossRef]
- Desoignies, N.; Carbonell, J.; Moreau, J.S.; Conesa, A.; Dopazo, J.; Legrève, A. Molecular Interactions between Sugar Beet and Polymyxa betae during Its Life Cycle. Ann. Appl. Biol. 2014, 164, 244–256. [Google Scholar] [CrossRef]
- Burki, F.; Kudryavtsev, A.; Matz, M.V.; Aglyamova, G.V.; Bulman, S.; Fiers, M.; Keeling, P.J.; Pawlowski, J. Evolution of Rhizaria: New Insights from Phylogenomic Analysis of Uncultivated Protists. BMC Evol. Biol. 2010, 10, 377. [Google Scholar] [CrossRef]
- Ciaghi, S.; Neuhauser, S.; Schwelm, A. Draft Genome Resource for the Potato Powdery Scab Pathogen Spongospora subterranea. Mol. Plant Microbe Interact. 2018, 31, 1227–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decroes, A.; Li, J.; Richardson, L.; Mutasa-Gottgens, E.; Lima-Mendez, G.; Mahillon, M.; Bragard, C.; Finn, R.D.; Legr, A. Genomics Metagenomics Approach for Polymyxa betae Genome Assembly Enables Comparative Analysis towards Deciphering the Intracellular Parasitic Lifestyle of the Plasmodiophorids. Genomics 2022, 114, 9–22. [Google Scholar] [CrossRef]
- Neuhauser, S.; Kirchmair, M.; Bulman, S.; Bass, D. Cross-Kingdom Host Shifts of Phytomyxid Parasites. BMC Evol. Biol. 2014, 14, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).