Genome-Wide Identification of Long Noncoding RNA and Their Potential Interactors in ISWI Mutants
Abstract
1. Introduction
2. Results
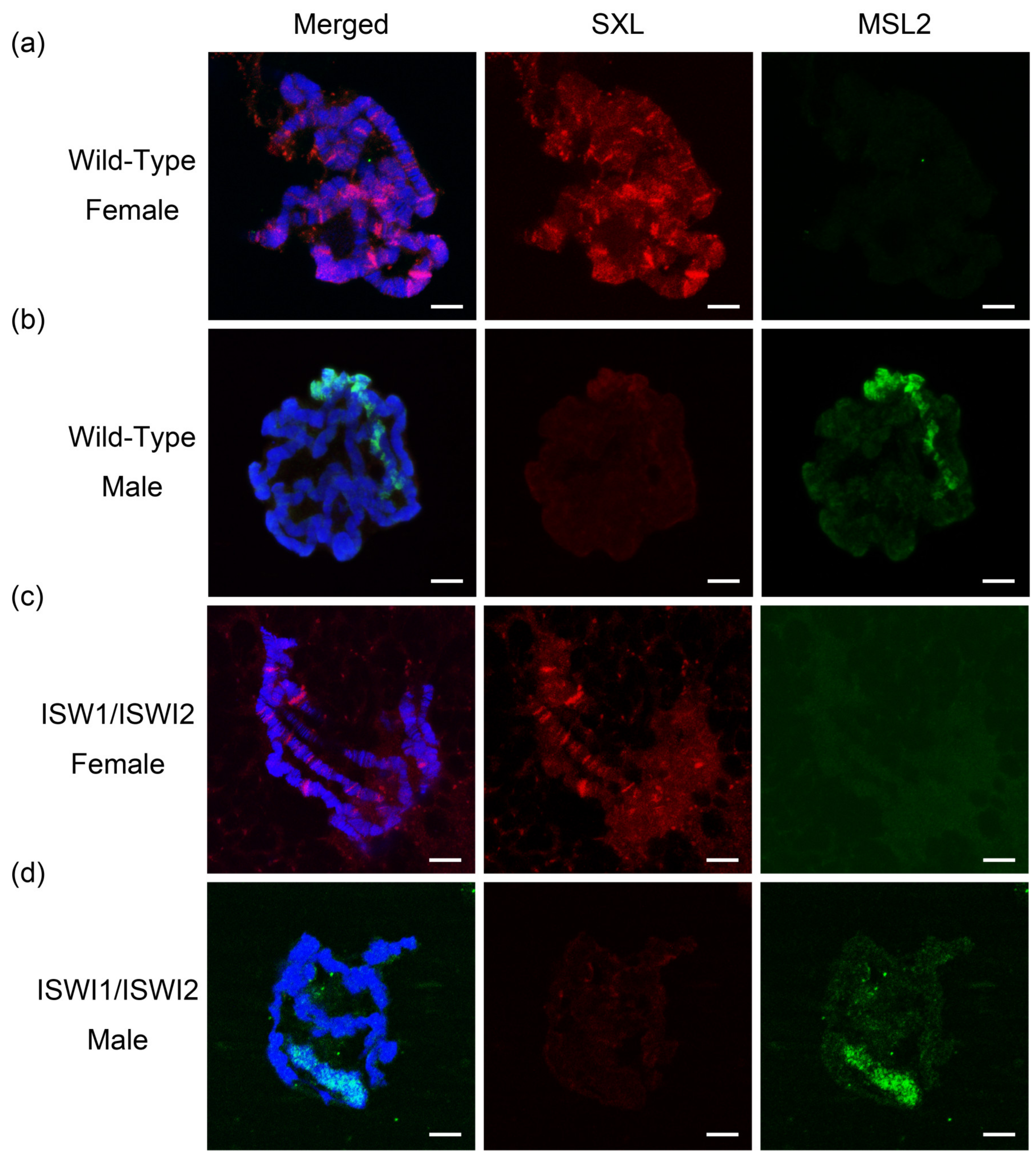
2.1. Hybridization and Identification of ISWI Mutation Lines
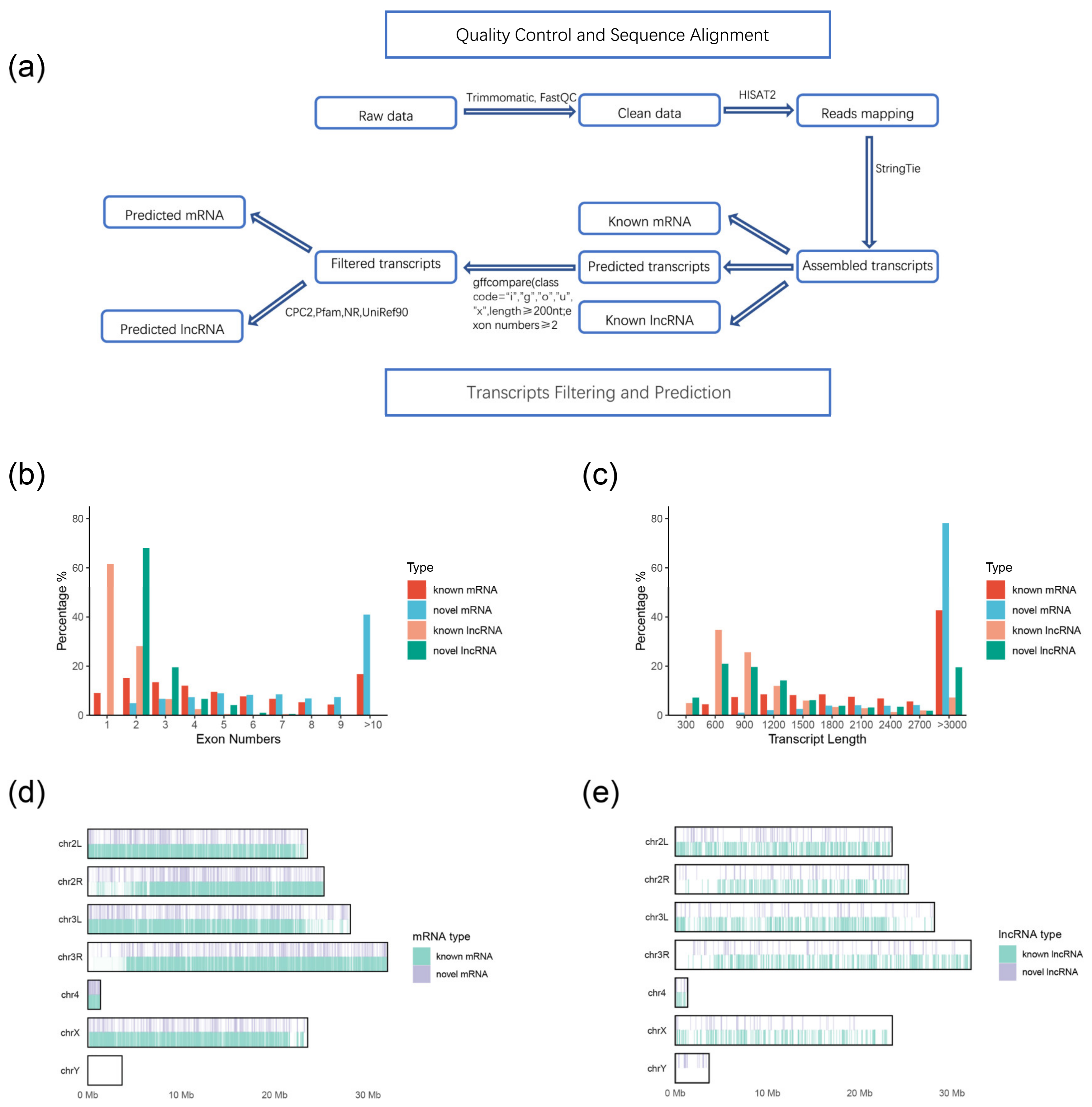
2.2. Identification and Genomic Characteristics of lncRNAs and mRNAs
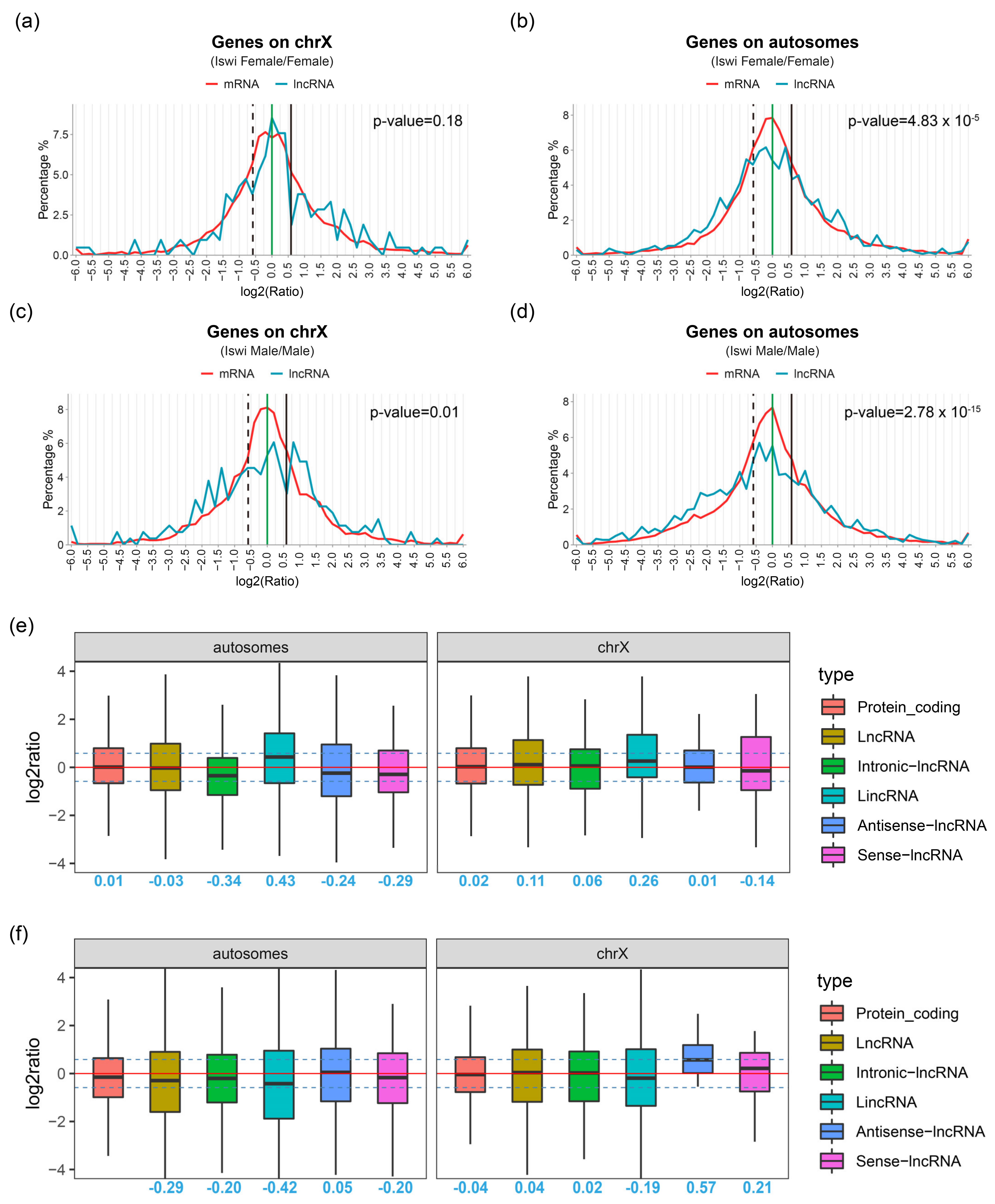
2.3. LncRNAs Expression Changes Significantly in ISWI-Mutated Background
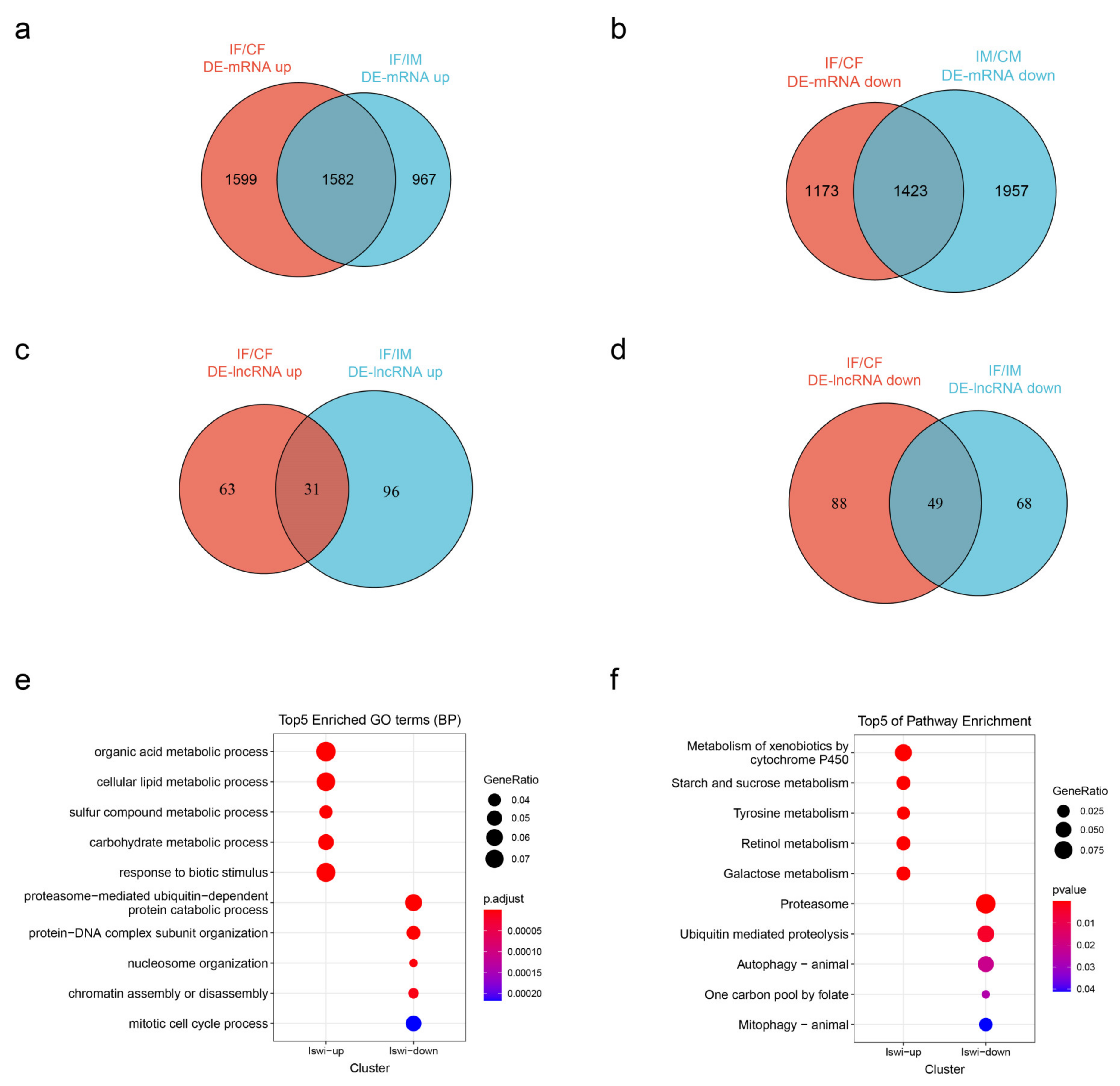
2.4. Differentially Expressed mRNAs and lncRNAs
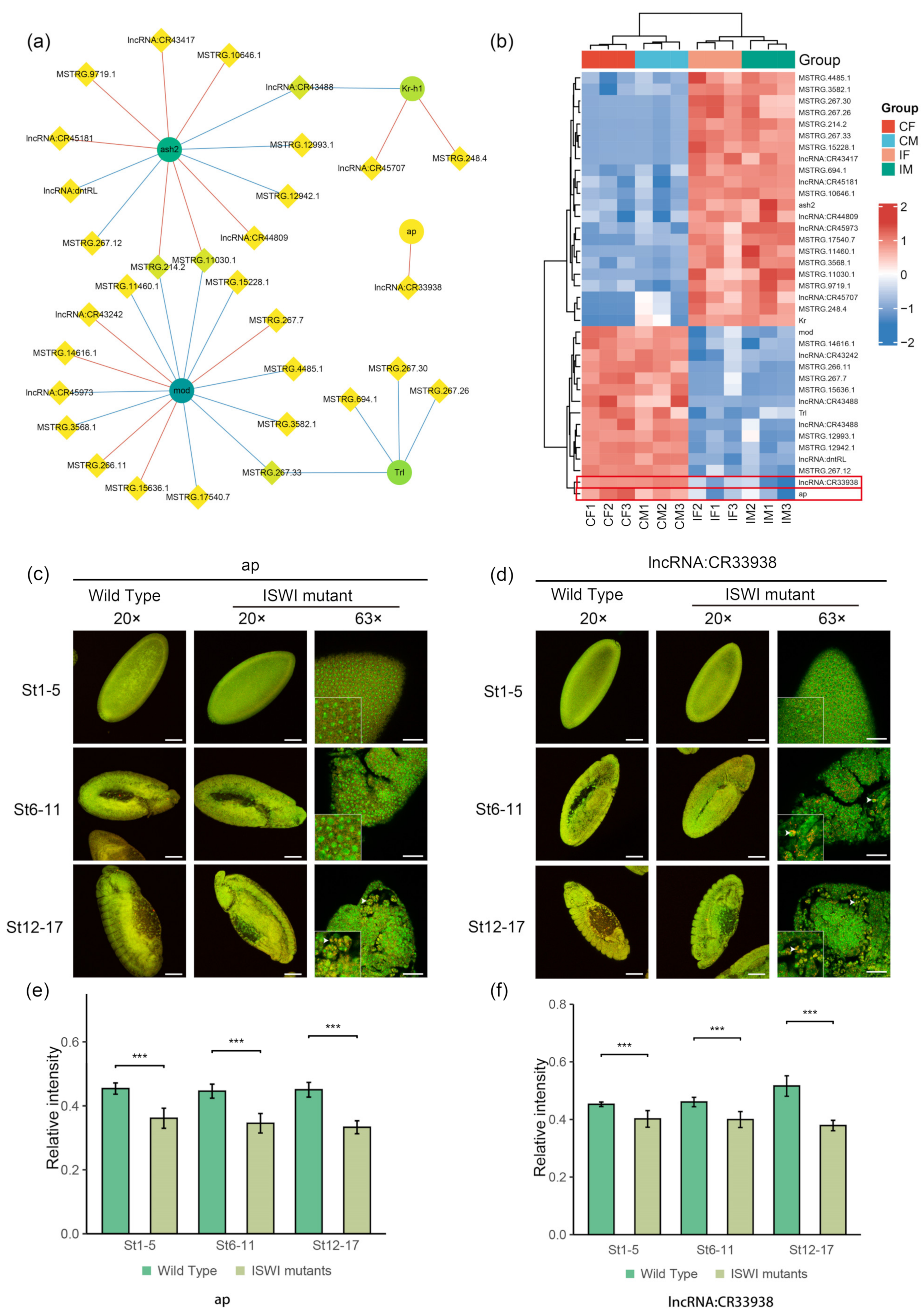
2.5. Potential Interactors of DE-lncRNAs
2.6. Differentially Expressed Transposons
3. Discussion
4. Materials and Methods
4.1. Drosophila Stocks and Crosses
4.2. Immunostaining of Chromosomes
4.3. Embryo TSA-FISH
4.4. RNA Extraction and Sequencing
4.5. RNA-Seq Reads Mapping and Transcriptome Assembly
4.6. Transcripts Filtering and Prediction
4.7. Comparisons between lncRNAs and mRNA Transcripts
4.8. Ratio Distribution
4.9. Differential Expression Analysis
4.10. Potential Reactors of DE-lncRNA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsukiyama, T.; Danial, C.; Tamkun, J.; Wu, C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 1995, 83, 1021–1026. [Google Scholar] [CrossRef]
- Li, Y.; Gong, H.; Wang, P.; Zhu, Y.; Peng, H.; Cui, Y.; Li, H.; Liu, J.; Wang, Z. The emerging role of ISWI chromatin remodeling complexes in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 346. [Google Scholar] [CrossRef] [PubMed]
- Borner, K.; Jain, D.; Vazquez-Pianzola, P.; Vengadasalam, S.; Steffen, N.; Fyodorov, D.V.; Tomancak, P.; Konev, A.; Suter, B.; Becker, P.B. A role for tuned levels of nucleosome remodeler subunit ACF1 during Drosophila oogenesis. Dev. Biol. 2016, 411, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Klinker, H.; Mueller-Planitz, F.; Yang, R.L.; Forne, I.; Liu, C.F.; Nordenskiold, L.; Becker, P.B. ISWI Remodelling of Physiological Chromatin Fibres Acetylated at Lysine 16 of Histone H4. PLoS ONE 2014, 9, e88411. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Tsukiyama, T.; Wisniewski, J.; Wu, C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell 1997, 1, 141–150. [Google Scholar] [CrossRef]
- Mizutani, R.; Wakamatsu, A.; Tanaka, N.; Yoshida, H.; Tochigi, N.; Suzuki, Y.; Oonishi, T.; Tani, H.; Tano, K.; Ijiri, K.; et al. Identification and characterization of novel genotoxic stress-inducible nuclear long noncoding RNAs in mammalian cells. PLoS ONE 2012, 7, e34949. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Mo, X.; Fu, L.; Xiao, B.; Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016, 7, 8601–8612. [Google Scholar] [CrossRef]
- Scheuermann, J.C.; Boyer, L.A. Getting to the heart of the matter: Long non-coding RNAs in cardiac development and disease. EMBO J. 2013, 32, 1805–1816. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Camilleri-Robles, C.; Amador, R.; Klein, C.C.; Guigo, R.; Corominas, M.; Ruiz-Romero, M. Genomic and functional conservation of lncRNAs: Lessons from flies. Mamm. Genome 2022, 33, 328–342. [Google Scholar] [CrossRef]
- Yang, F.; Deng, X.; Ma, W.; Berletch, J.B.; Rabaia, N.; Wei, G.; Moore, J.M.; Filippova, G.N.; Xu, J.; Liu, Y.; et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015, 16, 52. [Google Scholar] [CrossRef] [PubMed]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef] [PubMed]
- Hartford CCR, L.A. When Long Noncoding Becomes Protein Coding. Mol. Cell Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef] [PubMed]
- Meller, V.H.; Rattner, B.P. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002, 21, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Hu, F.; Zhou, Y.; Wu, F.; Gaut, B.S. Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response. BMC Genom. 2019, 20, 864. [Google Scholar] [CrossRef]
- Marsano, R.M.; Dimitri, P. Constitutive Heterochromatin in Eukaryotic Genomes: A Mine of Transposable Elements. Cells 2022, 11, 761. [Google Scholar] [CrossRef]
- Percharde, M.; Sultana, T.; Ramalho-Santos, M. What Doesn’t Kill You Makes You Stronger: Transposons as Dual Players in Chromatin Regulation and Genomic Variation. Bioessays 2020, 42, e1900232. [Google Scholar] [CrossRef]
- McCullers, T.J.; Steiniger, M. Transposable elements in Drosophila. Mob. Genet. Elem. 2017, 7, 1–18. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Tenaillon, M.I.; Hufford, M.B.; Gaut, B.S.; Ross-Ibarra, J. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol. Evol. 2011, 3, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Barron, M.G.; Fiston-Lavier, A.S.; Petrov, D.A.; Gonzalez, J. Population genomics of transposable elements in Drosophila. Annu. Rev. Genet. 2014, 48, 561–581. [Google Scholar] [CrossRef] [PubMed]
- Lannoy, N.; Hermans, C. Principles of genetic variations and molecular diseases: Applications in hemophilia A. Crit. Rev. Oncol. Hematol. 2016, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pedro, D.L.F.; Lorenzetti, A.P.R.; Domingues, D.S.; Paschoal, A.R. PlaNC-TE: A comprehensive knowledgebase of non-coding RNAs and transposable elements in plants. Database 2018, 2018, bay078. [Google Scholar] [CrossRef]
- Moschall, R.; Gaik, M.; Medenbach, J. Promiscuity in post-transcriptional control of gene expression: Drosophila sex-lethal and its regulatory partnerships. FEBS Lett. 2017, 591, 1471–1488. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef]
- Corona, D.F.; Clapier, C.R.; Becker, P.B.; Tamkun, J.W. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002, 3, 242–247. [Google Scholar] [CrossRef]
- Judd, J.; Duarte, F.M.; Lis, J.T. Pioneer-like factor GAF cooperates with PBAP (SWI/SNF) and NURF (ISWI) to regulate transcription. Genes Dev. 2021, 35, 147–156. [Google Scholar] [CrossRef]
- Prabhakaran, M.; Kelley, R.L. Mutations in the transcription elongation factor SPT5 disrupt a reporter for dosage compensation in Drosophila. PLoS Genet. 2012, 8, e1003073. [Google Scholar] [CrossRef]
- Espinas, M.L.; Canudas, S.; Fanti, L.; Pimpinelli, S.; Casanova, J.; Azorin, F. The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep. 2000, 1, 253–259. [Google Scholar] [CrossRef]
- Shaffer, C.D.; Stephens, G.E.; Thompson, B.A.; Funches, L.; Bernat, J.A.; Craig, C.A.; Elgin, S.C. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc. Natl. Acad. Sci. USA 2002, 99, 14332–14337. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.A.; Khoury, M.J.; Wirtz-Peitz, F.; Bilder, D. Evidence for a nuclear role for Drosophila Dlg as a regulator of the NURF complex. Mol. Biol. Cell 2021, 32, ar23. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.W. Primary Sex Determination in Drosophila melanogaster Does Not Rely on the Male-Specific Lethal Complex. Genetics 2016, 202, 541–549. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Slack, C.; Alic, N.; Foley, A.; Cabecinha, M.; Hoddinott, M.P.; Partridge, L. The Ras-Erk-ETS-Signaling Pathway Is a Drug Target for Longevity. Cell 2015, 162, 72–83. [Google Scholar] [CrossRef]
- Lee, J.; Yoo, E.; Lee, H.; Park, K.; Hur, J.H.; Lim, C. LSM12 and ME31B/DDX6 Define Distinct Modes of Posttranscriptional Regulation by ATAXIN-2 Protein Complex in Drosophila Circadian Pacemaker Neurons. Mol. Cell 2017, 66, 129–140.e7. [Google Scholar] [CrossRef]
- Nakamura, A.; Amikura, R.; Hanyu, K.; Kobayashi, S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 2001, 128, 3233–3242. [Google Scholar] [CrossRef]
- Ray, M.; Acharya, S.; Shambhavi, S.; Lakhotia, S.C. Over-expression of Hsp83 in grossly depleted hsromge lncRNA background causes synthetic lethality and l(2)gl phenocopy in Drosophila. J. Biosci. 2019, 44, 36. [Google Scholar] [CrossRef]
- Lécuyer, E.; Yoshida, H.; Parthasarathy, N.; Alm, C.; Babak, T.; Cerovina, T.; Hughes, T.R.; Tomancak, P.; Krause, H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007, 131, 174–187. [Google Scholar] [CrossRef]
- Wilk, R.; Hu, J.; Blotsky, D.; Krause, H.M. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016, 30, 594–609. [Google Scholar] [CrossRef]
- Meyer, W.J.; Schreiber, S.; Guo, Y.; Volkmann, T.; Welte, M.A.; Muller, H.A. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet. 2006, 2, e134. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, M.P.; Bhadra, U.; Kundu, J.; Birchler, J.A. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics 2005, 169, 2061–2074. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, W.; Birchler, J.A. Identification of Inverse Regulator-a (Inr-a) as Synonymous with Pre-mRNA Cleavage Complex II Protein (Pcf11) in Drosophila. G3 2012, 2, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.; Kim, H.Y.; Chen, X.; Shen, W.; Jang, J.S.; Stein, S.N.; Cormier, O.; Pereira, L.; Shih, C.R.Y.; Krieger, C.; et al. 20-hydroxyecdysone (20E) signaling regulates amnioserosa morphogenesis during Drosophila dorsal closure: EcR modulates gene expression in a complex with the AP-1 subunit, Jun. Biol. Open 2021, 10, bio058605. [Google Scholar] [CrossRef] [PubMed]
- Keegan, S.E.; Hughes, S.C. Role of nuclear-cytoplasmic protein localization during Drosophila neuroblast development. Genome 2021, 64, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Arama, E.; Dickman, D.; Kimchie, Z.; Shearn, A.; Lev, Z. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 2000, 19, 3706–3716. [Google Scholar] [CrossRef]
- Perkins, A.D.; Tanentzapf, G. An ongoing role for structural sarcomeric components in maintaining Drosophila melanogaster muscle function and structure. PLoS ONE 2014, 9, e99362. [Google Scholar] [CrossRef]
- Li, D.; Liu, J.; Liu, W.; Li, G.; Yang, Z.; Qin, P.; Xu, L. The ISWI remodeler in plants: Protein complexes, biochemical functions, and developmental roles. Chromosoma 2017, 126, 365–373. [Google Scholar] [CrossRef]
- Yaniv, M. Chromatin remodeling: From transcription to cancer. Cancer Genet. 2014, 207, 352–357. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Misteli, T. Beyond the sequence: Cellular organization of genome function. Cell 2007, 128, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R. Sophisticated Conversations between Chromatin and Chromatin Remodelers, and Dissonances in Cancer. Int. J. Mol. Sci. 2021, 22, 5578. [Google Scholar] [CrossRef] [PubMed]
- Jandura, A.; Krause, H.M. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. 2017, 33, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Constanty, F.; Shkumatava, A. lncRNAs in development and differentiation: From sequence motifs to functional characterization. Development 2021, 148, dev182741. [Google Scholar] [CrossRef]
- Li, K.; Tian, Y.; Yuan, Y.; Fan, X.; Yang, M.; He, Z.; Yang, D. Insights into the Functions of LncRNAs in Drosophila. Int. J. Mol. Sci. 2019, 20, 4646. [Google Scholar] [CrossRef]
- Zhao, W.; Geng, D.; Li, S.; Chen, Z.; Sun, M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018, 7, 842–855. [Google Scholar] [CrossRef]
- Drummond-Barbosa, D. Local and Physiological Control of Germline Stem Cell Lineages in Drosophila melanogaster. Genetics 2019, 213, 9–26. [Google Scholar] [CrossRef]
- Birchler, J.A. Parallel Universes for Models of X Chromosome Dosage Compensation in Drosophila: A Review. Cytogenet. Genome Res. 2016, 148, 52–67. [Google Scholar] [CrossRef]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Pradhan, R.K.; Ramakrishna, W. Transposons: Unexpected players in cancer. Gene 2022, 808, 145975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Qi, H.; Huang, C.; Yuan, L.; Zhang, L.; Wang, R.; Tian, Y.; Sun, L. Interaction of Male Specific Lethal complex and genomic imbalance on global gene expression in Drosophila. Sci. Rep. 2021, 11, 19679. [Google Scholar] [CrossRef] [PubMed]
- Lecuyer, E.; Parthasarathy, N.; Krause, H.M. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol. Biol. 2008, 420, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Yuan, Z.; Guo, D.; Hou, B.; Yin, C.; Zhang, W.; Li, F. Genome-wide identification of long noncoding RNA genes and their potential association with fecundity and virulence in rice brown planthopper, Nilaparvata lugens. BMC Genom. 2015, 16, 749. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Chen, L.; Shi, G.; Chen, G.; Li, J.; Li, M.; Zou, C.; Fang, C.; Li, C. Transcriptome Analysis Suggests the Roles of Long Intergenic Non-coding RNAs in the Growth Performance of Weaned Piglets. Front. Genet. 2019, 10, 196. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- Pirooznia, M.; Perkins, E.J.; Deng, Y. Batch Blast Extractor: An automated blastx parser application. BMC Genom. 2008, 9 (Suppl. S2), S10. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, S.; Wang, R.; Sun, L. Genome-Wide Identification of Long Noncoding RNA and Their Potential Interactors in ISWI Mutants. Int. J. Mol. Sci. 2022, 23, 6247. https://doi.org/10.3390/ijms23116247
Zhang L, Zhang S, Wang R, Sun L. Genome-Wide Identification of Long Noncoding RNA and Their Potential Interactors in ISWI Mutants. International Journal of Molecular Sciences. 2022; 23(11):6247. https://doi.org/10.3390/ijms23116247
Chicago/Turabian StyleZhang, Ludan, Shuai Zhang, Ruixue Wang, and Lin Sun. 2022. "Genome-Wide Identification of Long Noncoding RNA and Their Potential Interactors in ISWI Mutants" International Journal of Molecular Sciences 23, no. 11: 6247. https://doi.org/10.3390/ijms23116247
APA StyleZhang, L., Zhang, S., Wang, R., & Sun, L. (2022). Genome-Wide Identification of Long Noncoding RNA and Their Potential Interactors in ISWI Mutants. International Journal of Molecular Sciences, 23(11), 6247. https://doi.org/10.3390/ijms23116247





