Tissue-Based Markers as a Tool to Assess Response to Neoadjuvant Radiotherapy in Rectal Cancer—Systematic Review
Abstract
1. Introduction
2. Results
2.1. Biopolymers of Cancer Cells
2.1.1. Proteins
2.1.2. Genetic Markers—Mutations and Expression of Protein Coding Genes
2.1.3. Micro-RNA
2.2. Immunological Markers—Blood-Based and Tissue-Based
2.2.1. Blood-Based Immunological Markers
2.2.2. Tissue-Based Immunological Markers
2.2.3. Immunological Biomarkers within Tumors
2.3. Other
3. Discussion
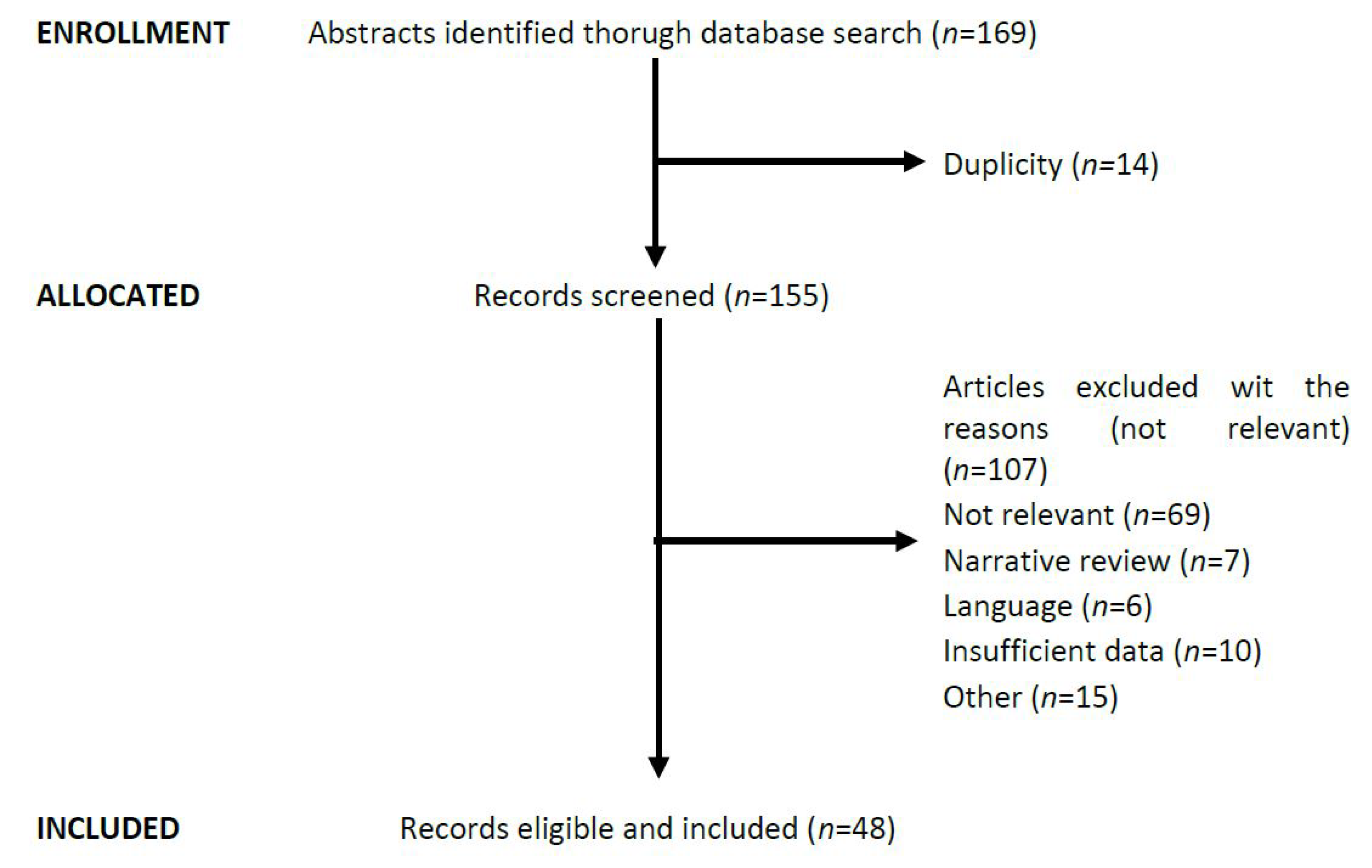
4. Materials and Methods
Literature Search and Inclusion Criteria
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Trans. Oncol. 2021, 14, 101–174. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Garrido-Laguna, I.; et al. Rectal cancer, version 6.2020: Featured updates to the NCCN guidelines. JNCCN J. Natl. Compr. Cancer Netw. 2020, 18, 807–815. [Google Scholar]
- Dou, R.; He, S.; Deng, Y.; Wang, J. Comparison of guidelines on rectal cancer: Exception proves the rule? Gastroenterol. Rep. 2021, 9, 290–298. [Google Scholar] [CrossRef] [PubMed]
- You, Y.N.; Hardiman, K.M.; Bafford, A.; Poylin, V.; Francone, T.D.; Davis, K.; Paquette, I.M.; Steele, S.R.; Feingold, D.L. The American Society of Colon and Rectal Surgeons Clinical Practice Guidelines for the Management of Rectal Cancer. Dis. Colon Rectum 2020, 63, 1191–1222. [Google Scholar] [CrossRef]
- Hiotis, S.P.; Weber, S.M.; Cohen, A.M.; Minsky, B.D.; Paty, P.B.; Guillem, J.G.; Wagman, R.; Saltz, L.B.; Wong, D.W. Assessing the predictive value of clinical complete response to neoadjuvant therapy for rectal cancer: An analysis of 488 patients. J. Am. Coll. Surg. 2002, 194, 131–135. [Google Scholar] [CrossRef]
- Biondo, S.; Navarro, M.; Marti-Rague, J.; Arriola, E.; Pares, D.; Del Rio, C.; Cambray, M.; Novell, V. Response to neoadjuvant therapy for rectal cancer: Influence on long-term results. Colorectal Dis. 2005, 7, 472–479. [Google Scholar] [CrossRef]
- Van der Valk, M.J.M.; Hilling, D.E.; Bastiaannet, E.; Kranenbarg, E.M.; Beets, G.L.; Figueiredo, N.L. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): An international multicentre registry study. Lancet 2018, 391, 2537–2545. [Google Scholar]
- Dworak, O.; Keilholz, L.; Hoffmann, A. Pathological features of rectal cancer after preoperative radiochemotherapy. Int. J. Colorectal Dis. 1997, 12, 19–23. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hansen, A.R.; Ratain, M.J.; Siu, L.L. Tumour heterogeneity in the clinic. Nature 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Patel, S.V.; Roxburgh, C.S.; Vakiani, E.; Shia, J.; Smith, J.J.; Temple, L.K.; Paty, P.; Garcia-Aguilar, J.; Nash, G.; Guillem, J.; et al. Distance to the anal verge is associated with pathologic complete response to neoadjuvant therapy in locally advanced rectal cancer. J. Surg. Oncol. 2016, 114, 637–641. [Google Scholar] [CrossRef]
- Lin, A.Y.; Wong, W.D.; Shia, J.; Minsky, B.D.; Temple, L.K.; Guillem, J.G.; Paty, P.B.; Weiser, M.R. Predictive clinicopathologic factors for limited response of T3 rectal cancer to combined modality therapy. Int. J. Colorectal Dis. 2007, 23, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Al-Sukhni, E.; Attwood, K.; Mattson, D.M.; Gabriel, E.; Nurkin, S.J. Predictors of Pathologic Complete Response Following Neoadjuvant Chemoradiotherapy for Rectal Cancer. Ann. Surg. Oncol. 2016, 23, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Janssen, M.H.; Öllers, M.C.; van Stiphout, R.G.; Riedl, R.G.; Bogaard, J.V.D.; Buijsen, J.; Lambin, P.; Lammering, G. PET-Based Treatment Response Evaluation in Rectal Cancer: Prediction and Validation. Int. J. Radiat. Oncol. 2012, 82, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.K.T.; Tait, D.; Chau, I.; Brown, G. MRI predictive factors for tumor response in rectal cancer following neoadjuvant chemoradiation therapy—Implications for induction chemotherapy? Int. J. Radiat. Oncol. Biol. Phys. 2013, 87, 505–511. [Google Scholar] [CrossRef]
- Wu, L.-M.; Zhu, J.; Hu, J.; Yin, Y.; Gu, H.-Y.; Hua, J.; Chen, J.; Xu, J.-R. Is there a benefit in using magnetic resonance imaging in the prediction of preoperative neoadjuvant therapy response in locally advanced rectal cancer? Int. J. Colorectal Dis. 2013, 28, 1225–1238. [Google Scholar] [CrossRef]
- Zhou, X.; Yi, Y.; Liu, Z.; Cao, W.; Lai, B.; Sun, K.; Li, L.; Zhou, Z.; Feng, Y.; Tian, J. Radiomics-Based Pretherapeutic Prediction of Non-response to Neoadjuvant Therapy in Locally Advanced Rectal Cancer. Ann. Surg. Oncol. 2019, 26, 1676–1684. [Google Scholar] [CrossRef] [PubMed]
- Buijsen, J.; Van Stiphout, R.G.; Menheere, P.P.C.A.; Lammering, G.; Lambin, P. Blood biomarkers are helpful in the prediction of response to chemoradiation in rectal cancer: A prospective, hypothesis driven study on patients with locally advanced rectal cancer. Radiother. Oncol. 2014, 111, 237–242. [Google Scholar] [CrossRef]
- Zeestraten, E.C.M.; Kuppen, P.J.K.; Van de Velde, C.J.H.; Marijnen, C.A.M. Prediction in Rectal Cancer. Semin. Radiat. Oncol. 2012, 22, 175–183. [Google Scholar] [CrossRef]
- Lu, X.; Cheng, C.; Zhu, S.; Yang, Y.; Zheng, L.; Wang, G.; Shu, X.; Wu, K.; Liu, K.; Tong, Q. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol. Rep. 2010, 24, 981–987. [Google Scholar]
- Meng, W.-J.; Yan, H.; Zhou, B.; Zhang, W.; Kong, X.-H.; Wang, R.; Zhan, L.; Li, Y.; Zhou, Z.-G.; Sun, X.-F. Correlation of SATB1 overexpression with the progression of human rectal cancer. Int. J. Colorectal Dis. 2011, 27, 143–150. [Google Scholar] [CrossRef]
- Meng, W.-J.; Pathak, S.; Ding, Z.-Y.; Zhang, H.; Adell, G.; Holmlund, B.; Li, Y.; Zhou, Z.-G.; Sun, X.-F. Special AT-rich sequence binding protein 1 expression correlates with response to preoperative radiotherapy and clinical outcome in rectal cancer. Cancer Biol. Ther. 2015, 16, 1738–1745. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Ramos, C.M.; Habr-Gama, A.; Quevedo, B.D.S.; Felício, N.M.; Bettoni, F.; Koyama, F.C.; Asprino, P.F.; Galante, P.A.; Gama-Rodrigues, J.; Camargo, A.A.; et al. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med. Genom. 2014, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, P.R.; Hanenberg, H. XRCC2 (X-ray repair cross complementing 2). Atlas Genet. Cytogenet. Oncol. Haematol. 2019, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Song, X.; Chen, Z.; Qin, C.; He, Y. XRCC2 rs3218536 polymorphism decreases the sensitivity of colorectal cancer cells to poly(ADP-ribose) polymerase 1 inhibitor. Oncol. Lett. 2014, 8, 1222–1228. [Google Scholar] [CrossRef][Green Version]
- Qin, C.-J.; Song, X.-M.; Chen, Z.-H.; Ren, X.-Q.; Xu, K.-W.; Jing, H.; He, Y.-L. XRCC2 as a predictive biomarker for radioresistance in locally advanced rectal cancer patients undergoing preoperative radiotherapy. Oncotarget 2015, 6, 32193–32204. [Google Scholar] [CrossRef]
- Wang, X.; Li, N.; Li, H.; Liu, B.; Qiu, J.; Chen, T.; Cao, X. Silencing of Human Phosphatidylethanolamine-Binding Protein 4 Sensitizes Breast Cancer Cells to Tumor Necrosis Factor-α–Induced Apoptosis and Cell Growth Arrest. Clin. Cancer Res. 2005, 11, 7545–7553. [Google Scholar] [CrossRef]
- Yu, G.P.; Chen, G.Q.; Wu, S.; Shen, K.; Ji, Y. The expression of PEBP4 protein in lung squamous cell carcinoma. Tumour Biol. 2011, 32, 1257–1263. [Google Scholar] [CrossRef]
- Luo, Z.K.; Chen, Q.F.; Qu, X.; Zhou, X.Y. The roles and signaling pathways of phosphatidylethanolamine-binding protein 4 in tumors. Onco Targets Ther. 2019, 12, 7685–7690. [Google Scholar] [CrossRef]
- Qiu, J.; Yang, G.; Shen, Z.; Xie, Y.; Wang, L. HPEBP4 as a predictive marker for the pathological response of rectal cancer to preoperative radiotherapy. Int. J. Colorectal Dis. 2013, 28, 241–246. [Google Scholar] [CrossRef]
- Qiu, J.; Tao, Y.; Yang, G.; Xu, K.; Lin, A.L.; Li, L. Effect of a chemical inhibitor of human phosphatidylethanolamine-binding protein 4 on radiosensitivity of rectal cancer cells. World J. Surg. Oncol. 2016, 14, 16–20. [Google Scholar] [CrossRef]
- Halberg, N.; Sengelaub, C.A.; Navrazhina, K.; Molina, H.; Uryu, K.; Tavazoie, S.F. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell 2016, 29, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Png, K.J.; Halberg, N.; Yoshida, M.; Tavazoie, S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2011, 481, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Shao, R.; Li, J.; Huang, H.; Wang, Y.; Zhang, M.; Cao, J.; Zhang, J.; Bu, J. PITPNC1 fuels radioresistance of rectal cancer by inhibiting reactive oxygen species production. Ann. Transl. Med. 2020, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ding, W.; Ge, H.; Ponnusamy, M.; Wang, Q.; Hao, X.; Wu, W.; Zhang, Y.; Yu, W.; Ao, X.; et al. FOXK transcription factors: Regulation and critical role in cancer. Cancer Lett. 2019, 458, 1–12. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, M.; Chen, J.; Chen, K.; Zhuang, J.; Yang, Y.; Liu, X.; Guan, G. Prognostic Value of the FOXK Family Expression in Patients with Locally Advanced Rectal Cancer Following Neoadjuvant Chemoradiotherapy. Onco Targets Ther. 2020, 13, 9185–9201. [Google Scholar] [CrossRef]
- Yun, Y.-R.; Won, J.E.; Jeon, E.; Lee, S.; Kang, W.; Jo, H.; Jang, J.H.; Shin, U.S.; Kim, H.W. Fibroblast Growth Factors: Biology, Function, and Application for Tissue Regeneration. J. Tissue Eng. 2010, 2010, 218142. [Google Scholar] [CrossRef]
- Harpain, F.; Ahmed, M.A.; Hudec, X.; Timelthaler, G.; Jomrich, G.; Müllauer, L.; Selzer, E.; Dörr, W.; Bergmann, M.; Holzmann, K.; et al. FGF8 induces therapy resistance in neoadjuvantly radiated rectal cancer. J. Cancer Res. Clin. Oncol. 2018, 145, 77–86. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Donovan, N.; Brennan, D.J.; Gallagher, W.M.; Ryan, B.M. Survivin: A promising tumor biomarker. Cancer Lett. 2007, 249, 49–60. [Google Scholar] [CrossRef]
- Yu, J., II; Lee, H.; Park, H.C.; Choi, D.H.; Choi, Y.-L.; Do, I.G.; Kim, H.C.; Lee, W.Y.; Yun, S.H.; Cho, Y.B.; et al. Prognostic significance of survivin in rectal cancer patients treated with surgery and postoperative concurrent chemoradiation therapy. Oncotarget 2016, 7, 62676–62686. [Google Scholar]
- Ahmed, M.A.; Selzer, E.; Doerr, W.; Jomrich, G.; Harpain, F.; Silberhumer, G.R.; Müllauer, L.; Holzmann, K.; Grasl-Kraupp, B.; Grusch, M.; et al. Fibroblast growth factor receptor 4 induced resistance to radiation therapy in colorectal cancer. Oncotarget 2016, 7, 69976–69990. [Google Scholar] [CrossRef]
- Sulzmaier, F.J.; Jean, C.; Schlaepfer, D.D. FAK in cancer: Mechanistic findings and clinical applications. Nat. Rev. Cancer 2014, 14, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Jes, M.; Mar, J.P.; Puerto-nevado, L.; Mart, J.; Caram, C.; Vega-bravo, R. Focal adhesion kinase: Predictor of tumour response and risk factor for recurrence after neoadjuvant chemoradiation in rectal cancer. J. Cell. Mol. Med. 2016, 20, 1729–1736. [Google Scholar]
- Xie, C.; Yao, M.; Dong, Q. Proliferating cell unclear antigen-associated factor (PAF15): A novel oncogene. Int. J. Biochem. Cell Biol. 2014, 50, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhu, K.; Dang, C.; Lan, K.; Wang, H.; Yuan, D.; Chen, W.; Meltzer, S.J.; Li, K. Paf15 expression correlates with rectal cancer prognosis, cell proliferation and radiation response. Oncotarget 2016, 7, 38750–38761. [Google Scholar] [CrossRef]
- Isaji, T.; Im, S.; Gu, W.; Wang, Y.; Hang, Q.; Lu, J.; Fukuda, T.; Hashii, N.; Takakura, D.; Kawasaki, N.; et al. An Oncogenic Protein Golgi Phosphoprotein 3 Up-regulates Cell Migration via Sialylation. J. Biol. Chem. 2014, 289, 20694–20705. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, B.; Chen, L.; Di, J.; Cui, M.; Liu, M.; Ma, Y.; Yang, H.; Xing, J.; Zhang, C.; et al. GOLPH3 predicts survival of colorectal cancer patients treated with 5-fluorouracil-based adjuvant chemotherapy. J. Transl. Med. 2014, 12, 15. [Google Scholar] [CrossRef]
- Guo, Y.-T.; Qiu, C.; Huang, Z.-X.; Yu, W.-S.; Yang, X.-F.; Wang, M.-Z. Correlational research of Golgi phosphorylation protein 3 expression in colorectal cancer. World J. Gastroenterol. 2015, 21, 13473–13479. [Google Scholar] [CrossRef]
- Zhu, K.; Zhao, Q.; Yue, J.; Shi, P.; Yan, H.; Xu, X.; Wang, R. GOLPH3 overexpression correlates with poor response to neoadjuvant therapy and prognosis in locally advanced rectal cancer. Oncotarget 2016, 7, 68328–68338. [Google Scholar] [CrossRef]
- Li, W.; Zhang, L. Regulation of ATG and Autophagy Initiation. Adv. Exp. Med. Biol. 2019, 1206, 41–65. [Google Scholar]
- Zaanan, A.; Park, J.M.; Tougeron, D.; Huang, S.; Wu, T.-T.; Foster, N.R.; Sinicrope, F.A. Association of beclin 1 expression with response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal carcinoma. Int. J. Cancer 2015, 137, 1498–1502. [Google Scholar] [CrossRef]
- Ghosh, S.; Hayden, M.S. Celebrating 25 years of NF-κB research. Immunol. Rev. 2012, 246, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Voboril, R.; Rychterova, V.; Voborilova, J.; Kubecova, M.; Fanta, J.; Dvorak, J. NF-κB / p65 expression before and after treatment in rectal cancer patients undergoing neoadjuvant (chemo) radiotherapy and surgery: Prognostic marker for disease progression and survival. Neoplasma 2016, 63, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Cholewa, B.D.; Liu, X.; Ahmad, N. The role of polo-like kinase 1 in carcinogenesis: Cause or consequence? Cancer Res. 2013, 73, 6848–6855. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, A.; del Pulgar, T.G.; Fernández-Aceñero, M.J.; Borrero-Palacios, A.; del Puerto-Nevado, L.; Martínez-Useros, J.; Marín-Arango, J.P.; Caramés, C.; Vega-Bravo, R.; Rodríguez-Remírez, M.; et al. Decreased PLK1 expression denotes therapy resistance and unfavourable disease-free survival in rectal cancer patients receiving neoadjuvant chemoradiotherapy. Pathol. Res. Pract. 2016, 212, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.; Chung, L.; Revoltar, M.; Lim, S.H.; Tut, T.-G.; Abubakar, A.; Henderson, C.J.; Chua, W.; Ng, W.; Lee, M.; et al. MRE11 and ATM Expression Levels Predict Rectal Cancer Survival and Their Association with Radiotherapy Response. PLoS ONE 2016, 11, e0167675. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Ng, W.; Lee, M.; De Souza, P.; Shin, J.-S.; et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer 2018, 18, 869. [Google Scholar] [CrossRef]
- Bartucci, R.; Salvati, A.; Olinga, P.; Boersma, Y.L. Vanin 1: Its physiological function and role in diseases. Int. J. Mol. Sci. 2019, 20, 3891. [Google Scholar] [CrossRef]
- Chai, C.; Zhang, Y.; Song, J.; Lin, S.; Sun, S.; Chang, I. VNN1 overexpression is associated with poor response to preoperative chemoradiotherapy and adverse prognosis in patients with rectal cancers. Am. J. Transl. Res. 2016, 8, 4455–4463. [Google Scholar]
- Peng, H.; You, K.; Zhang, R.; Xi, S.; Zhang, T.; Dong, J.; Cai, M.; Wang, C.; Zhang, H.; Zhou, T.; et al. Predictive value of APAF-1 and COX-2 expression in pathologic complete response to neoadjuvant chemoradiotherapy for patients with locally advanced rectal adenocarcinoma. Oncotarget 2016, 7, 35233–35240. [Google Scholar] [CrossRef]
- Laurini, E.; Marson, D.; Fermeglia, A.; Aulic, S.; Fermeglia, M.; Pricl, S. Role of Rad51 and DNA repair in cancer: A molecular perspective. Pharmacol. Ther. 2020, 208, 107–492. [Google Scholar] [CrossRef]
- Tennstedt, P.; Fresow, R.; Simon, R.; Marx, A.; Terracciano, L.; Petersen, C.; Sauter, G.; Dikomey, E.; Borgmann, K. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int. J. Cancer 2012, 132, 2118–2126. [Google Scholar] [CrossRef] [PubMed]
- Jalan, M.; Olsen, K.S.; Powell, S.N. Emerging Roles of RAD52 in Genome Maintenance. Cancers 2019, 11, 1038. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Chua, W.; Ng, W.; Lee, M.; Roberts, T.L.; et al. Aberrant Expression of RAD52, Its Prognostic Impact in Rectal Cancer and Association with Poor Survival of Patients. Int. J. Mol. Sci. 2020, 21, 1768. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, K.; Li, B.; Massa, H.F.; Trask, B.J.; Date, T.; Fields, S. Stimulation of p53-mediated transcriptional activation by the p53- binding proteins, 53BP1 and 53BP2. J. Biol. Chem. 1998, 273, 26061–26068. [Google Scholar] [CrossRef]
- Huang, A.; Xiao, Y.; Peng, C.; Liu, T.; Lin, Z.; Yang, Q.; Zhang, T.; Liu, J.; Ma, H. 53BP1 expression and immunoscore are associated with the efficacy of neoadjuvant chemoradiotherapy for rectal cancer. Strahlenther. Onkol. 2020, 196, 465–473. [Google Scholar] [CrossRef]
- Barbacid, M. Ras genes. Annu. Rev. Biochem. 1987, 56, 779–827. [Google Scholar] [CrossRef]
- Garcia-Aguilar, J.; Chen, Z.; Smith, D.; Li, W.; Madoff, R.D.; Cataldo, P.; Marcet, J.; Pastor, C. Identification of a Biomarker Profile Associated With Resistance to Neoadjuvant Chemoradiation Therapy in Rectal Cancer. Ann. Surg. 2011, 254, 486–493. [Google Scholar] [CrossRef]
- Duldulao, M.P.; Lee, W.; Nelson, R.A.; Li, W.; Chen, Z.; Kim, J.; Garcia-Aguilar, J. Mutations in Specific Codons of the KRAS Oncogene are Associated with Variable Resistance to Neoadjuvant Chemoradiation Therapy in Patients with Rectal Adenocarcinoma. Ann. Surg. Oncol. 2013, 20, 2166–2171. [Google Scholar] [CrossRef]
- Martellucci, J.; Alemanno, G.; Castiglione, F.; Bergamini, C.; Valeri, A. Role of KRAS mutation as predictor of pathologic response after neoadjuvant chemoradiation therapy for rectal cancer. Updat. Surg. 2015, 67, 47–53. [Google Scholar] [CrossRef]
- Peng, J.; Lin, J.; Qiu, M.; Zhao, Y.; Deng, Y.; Shao, J.; Ding, P.; Zhang, H.; Wan, D.; Lu, Z.; et al. Oncogene mutation profile predicts tumor regression and survival in locally advanced rectal cancer patients treated with preoperative chemoradiotherapy and radical surgery. Tumor Biol. 2017, 39, 1010428317709638. [Google Scholar] [CrossRef]
- Gaedcke, J.; Grade, M.; Jung, K.; Schirmer, M.; Jo, P.; Obermeyer, C.; Wolff, H.A.; Herrmann, M.K.; Beissbarth, T.; Becker, H.; et al. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother. Oncol. 2010, 94, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lane, D.P. P53 in Health and Disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Chen MBin Wu, X.Y.; Yu, R.; Li, C.; Wang, L.Q.; Shen, W.; Lu, P.-H. P53 Status as a Predictive Biomarker for Patients Receiving Neoadjuvant Radiation-Based Treatment: A Meta-Analysis in Rectal Cancer. PLoS ONE 2012, 7, e45388. [Google Scholar]
- Lopezcrapez, E.; Bibeau, F.; Thezenas, S.; Ychou, M.; Simonylafontaine, J.; Thirion, A.; Azria, D.; Grenier, J.K.; Senesse, P. p53 status and response to radiotherapy in rectal cancer: A prospective multilevel analysis. Br. J. Cancer 2005, 92, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- El Otmani, I.; El Agy, F.; El Baradai, S.; Bouguenouch, L.; Lahmidani, N.; El Abkari, M.; Bemakah, D.A.; Toughrai, I.; El Bouhaddouti, H.; Mouaqit, O.; et al. Analysis of Molecular Pretreated Tumor Profiles as Predictive Biomarkers of Therapeutic Response and Survival Outcomes after Neoadjuvant Therapy for Rectal Cancer in Moroccan Population. Dis. Markers 2020, 2020, 8459303. [Google Scholar] [CrossRef]
- Hur, H.; Kim, N.K.; Min, B.S.; Baik, S.H.; Lee, K.Y.; Koom, W.S.; Ahn, J.B.; Kim, H. Can a biomarker-based scoring system predict pathologic complete response after preoperative chemoradiotherapy for rectal cancer? Dis. Colon Rectum 2014, 57, 592–601. [Google Scholar] [CrossRef]
- Kandioler, D.; Zwrtek, R.; Ludwig, C.; Janschek, E.; Ploner, M.; Hofbauer, F.; Kührer, I.; Kappel, S.; Wrba, F.; Horvath, M.; et al. TP53 Genotype but Not p53 Immunohistochemical Result Predicts Response to Preoperative Short-Term Radiotherapy in Rectal Cancer. Ann. Surg. 2002, 235, 493–498. [Google Scholar] [CrossRef]
- Rebischung, C.; Gérard, J.P.; Gayet, J.; Thomas, G.; Hamelin, R.; Laurent-Puig, P. Prognostic value of P53 mutations in rectal carcinoma. Int. J. Cancer 2002, 100, 131–135. [Google Scholar] [CrossRef]
- Gavioli, M.; Luppi, G.; Losi, L.; Bertolini, F.; Santantonio, M.; Falchi, A.M.; D’Amico, R.; Conte, P.; Natalini, G. Incidence and Clinical Impact of Sterilized Disease and Minimal Residual Disease After Preoperative Radiochemotherapy for Rectal Cancer. Dis. Colon Rectum 2005, 48, 1851–1857. [Google Scholar] [CrossRef]
- Sparano, J.A.; Paik, S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J. Clin. Oncol. 2008, 26, 721–728. [Google Scholar] [CrossRef]
- Van de Vijver, M.J.; He, Y.D.; van ‘t Veer, L.J.; Dai, H.; Hart, A.A.M.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. Faculty Opinions recommendation of A gene-expression signature as a predictor of survival in breast cancer. New Engl. J. Med. 2015, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Ghadimi, B.M.; Grade, M.; Difilippantonio, M.J.; Varma, S.; Simon, R.; Montagna, C.; Füzesi, L.; Langer, C.; Becker, H.; Liersch, T.; et al. Effectiveness of Gene Expression Profiling for Response Prediction of Rectal Adenocarcinomas to Preoperative Chemoradiotherapy. J. Clin. Oncol. 2005, 23, 1826–1838. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.K.; Callahan, R.E.; Hothem, Z.A.; Cousineau, C.S.; Kawak, S.; Thibodeau, B.J.; Bergeron, S.; Li, W.; Peeples, C.E.; Wasvary, H.J. Genomic variation as a marker of response to neoadjuvant therapy in locally advanced rectal cancer. Mol. Cell. Oncol. 2020, 7, 1716618. [Google Scholar] [CrossRef] [PubMed]
- Zauber, N.P.; Marotta, S.P.; Berman, E.; Grann, A.; Rao, M.; Komati, N.; Ribiero, K.; Bishop, D.T. Molecular Genetic Changes Associated with Colorectal Carcinogenesis Are Not Prognostic for Tumor Regression Following Preoperative Chemoradiation of Rectal Carcinoma. Int. J. Radiat. Oncol. 2009, 74, 472–476. [Google Scholar] [CrossRef]
- Gantt, G.A.; Chen, Y.; Dejulius, K.; Mace, A.G.; Barnholtz-Sloan, J.; Kalady, M.F. Gene expression profile is associated with chemoradiation resistance in rectal cancer. Colorectal Dis. 2014, 16, 57–66. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Wu, X.; Chi, P. A four gene-based risk score system associated with chemoradiotherapy response and tumor recurrence in rectal cancer by co-expression network analysis. Onco Targets Ther. 2020, 13, 6721–6733. [Google Scholar] [CrossRef]
- Lee, Y.-E.; He, H.-L.; Shiue, Y.-L.; Lee, S.-W.; Lin, L.-C.; Wu, T.-F.; Chang, I.-W.; Lee, H.-H.; Li, C.-F. The prognostic impact of lipid biosynthesis-associated markers, HSD17B2 and HMGCS2, in rectal cancer treated with neoadjuvant concurrent chemoradiotherapy. Tumor Biol. 2015, 36, 7675–7683. [Google Scholar] [CrossRef]
- Vihko, P.; Isomaa, V.; Ghosh, D. Structure and function of 17β-hydroxysteroid dehydrogenase type 1 and type 2. Mol. Cell Endocrinol. 2001, 171, 71–76. [Google Scholar] [CrossRef]
- Di Leva, G.; Croce, C.M. MiRNA profiling of cancer. Curr. Opin. Genet. Dev. 2013, 23, 3–11. [Google Scholar] [CrossRef]
- Zhou, K.; Liu, M.; Cao, Y. New insight into microRNA functions in cancer: Oncogene-microRNA-tumor suppressor gene network. Front. Mol. Biosci. 2017, 4, 46. [Google Scholar] [CrossRef]
- Caramés, C.; Cristóbal, I.; Moreno, V.; Del Puerto, L.; Moreno, I.; Rodriguez, M.; Marín, J.P.; Correa, A.V.; Hernández, R.; Zenzola, V.; et al. MicroRNA-21 predicts response to preoperative chemoradiotherapy in locally advanced rectal cancer. Int. J. Colorectal Dis. 2015, 30, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Caramés, C.; Cristobal, I.; Moreno, V.; Marín, J.P.; González-Alonso, P.; Torrejón, B.; Minguez, P.; Leon, A.; Martín, J.I.; Hernández, R.; et al. MicroRNA-31 Emerges as a Predictive Biomarker of Pathological Response and Outcome in Locally Advanced Rectal Cancer. Int. J. Mol. Sci. 2016, 17, 878. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, E.; Zanon, C.; Sensi, F.; Digito, M.; Rugge, M.; Fassan, M.; Scarpa, M.; Pucciarelli, S.; Nitti, D.; Agostini, M. MiR-194 as predictive biomarker of responsiveness to neoadjuvant chemoradiotherapy in patients with locally advanced rectal adenocarcinoma. J. Clin. Pathol. 2018, 71, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Hotchi, M.; Shimada, M.; Kurita, N.; Iwata, T.; Sato, H.; Morimoto, S.; Yoshikawa, K.; Higashijima, J.; Miyatani, T. microRNA expression is able to predict response to chemoradiotherapy in rectal cancer. Mol. Clin. Oncol. 2012, 1, 137–142. [Google Scholar] [CrossRef]
- Svoboda, M.; Sana, J.; Fabian, P.; Kocakova, I.; Gombosova, J.; Nekvindova, J.; Radova, L.; Vyzula, R.; Slaby, O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat. Oncol. 2012, 7, 195. [Google Scholar] [CrossRef]
- Baek, D.W.; Kim, G.; Kang, B.W.; Kim, H.J.; Park, S.Y.; Park, J.S.; Choi, G.-S.; Kang, M.K.; Hur, K.; Kim, J.G. High expression of microRNA-199a-5p is associated with superior clinical outcomes in patients with locally advanced rectal cancer. J. Cancer Res. Clin. Oncol. 2019, 146, 105–115. [Google Scholar] [CrossRef]
- Yasuda, K.; Nirei, T.; Sunami, E.; Nagawa, H.; Kitayama, J. Density of CD4(+) and CD8(+) T lymphocytes in biopsy samples can be a predictor of pathological response to chemoradiotherapy (CRT) for rectal cancer. Radiat. Oncol. 2011, 6, 49. [Google Scholar] [CrossRef]
- Aras, S.; Raza Zaidi, M. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017, 10, 58. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, S.; Peng, Y.; Zhuang, J.; Yang, Y.; Xu, Y.; Guan, G. Construction of the Prediction Model for Locally Advanced Rectal Cancer Following Neoadjuvant Chemoradiotherapy Based on Pretreatment Tumor-Infiltrating Macrophage-Associated Biomarkers. OncoTargets Ther. 2021, 14, 2599–2610. [Google Scholar] [CrossRef]
- Saigusa, S.; Toiyama, Y.; Tanaka, K.; Inoue, Y.; Mori, K.; Ide, S.; Imaoka, H.; Kawamura, M.; Mohri, Y.; Kusunoki, M. Prognostic relevance of stromal CD26 expression in rectal cancer after chemoradiotherapy. Int. J. Clin. Oncol. 2015, 21, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Caputo, D.; Caricato, M.; Coppola, A.; La Vaccara, V.; Fiore, M.; Coppola, R. Neutrophil to lymphocyte ratio (NLR) and derived neutrophil to lymphocyte ratio (d-NLR) predict non-responders and postoperative complications in patients undergoing radical surgery after neo-adjuvant radio-chemotherapy for rectal adenocarcinoma. Cancer Investig. 2016, 34, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Andras, D.; Crisan, D.; Craciun, R.; Nemes, A.; Caziuc, A.; Drasovean, R.; Seicean, R.; Scurtu, R.; Bintintan, V.; Eniu, D.; et al. Neutrophil-to-lymphocyte ratio: A hidden gem in predicting neoadjuvant treatment response in locally advanced rectal cancer? J. Buon 2020, 25, 1436–1442. [Google Scholar] [PubMed]
- Tada, N.; Tsuno, N.H.; Kawai, K.; Murono, K.; Nirei, T.; Ishihara, S.; Sunami, E.; Kitayama, J.; Watanabe, T. Changes in the plasma levels of cytokines/chemokines for predicting the response to chemoradiation therapy in rectal cancer patients. Oncol. Rep. 2013, 31, 463–471. [Google Scholar] [CrossRef]
- Zezulová, M.; Bartoušková, M.; Hlídková, E.; Juráňová, J.; Červinková, B.; Kasalová, E.; Adam, T.; Krčmová, L.K.; Solichová, D.; Cwiertka, K.; et al. Prognostic significance of serum and urinary Neopterin concentrations in patients with rectal carcinoma treated with Chemoradiation. Anticancer Res. 2016, 36, 287–292. [Google Scholar]
- Angenete, E.; Langenskiöld, M.; Palmgren, I.; Falk, P.; Oresland, T.; Ivarsson, M.L. Transforming growth factor beta-1 in rectal tumour, mucosa and plasma in relation to radiotherapy and clinical outcome in rectal cancer patients. Int. J. Colorectal Dis. 2007, 22, 1331–1338. [Google Scholar] [CrossRef]
- Flem-Karlsen, K.; Fodstad, Y.; Nunes-Xavier, C.E. B7-H3 in Cancer—Beyond Immune Regulation. Trends Cancer 2018, 4, 401–404. [Google Scholar] [CrossRef]
- Wang, C.; Feng, H.; Cheng, X.; Liu, K.; Cai, D.; Zhao, R. Potential Therapeutic Targets of B7 Family in Colorectal Cancer. Front. Immunol. 2020, 11, 681. [Google Scholar] [CrossRef]
- Ingebrigtsen, V.A.; Boye, K.; Nesland, J.M.; Nesbakken, A.; Flatmark, K.; Fodstad, Ø. B7-H3 expression in colorectal cancer: Associations with clinicopathological parameters and patient outcome. BMC Cancer 2014, 14, 602. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, Y.; Gao, M.; Huang, A.; Chi, P. A three-phase trans-ethnic study reveals B7-H3 expression is a significant and independent biomarker associated with colon cancer overall survival. J. Gastrointest Oncol. 2021, 12, 2891–2905. [Google Scholar] [CrossRef]
- Zhu, Z.; Dong, W. Overexpression of HHLA2, a member of the B7 family, is associated with worse survival in human colorectal carcinoma. Onco Targets Ther. 2018, 11, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.-J.; Wang, L.-J.; Wang, G.-D.; Guo, Z.-Y.; Wei, M.; Meng, Y.-L.; Yang, A.-G.; Wen, W.-H. B7-H1 Expression Is Associated with Poor Prognosis in Colorectal Carcinoma and Regulates the Proliferation and Invasion of HCT116 Colorectal Cancer Cells. PLoS ONE 2013, 8, e76012. [Google Scholar] [CrossRef] [PubMed]
- Yasui, K.; Kondou, R.; Iizuka, A.; Miyata, H.; Tanaka, E.; Ashizawa, T.; Nagashima, T.; Ohshima, K.; Urakami, K.; Kusuhara, M.; et al. Effect of preoperative chemoradiotherapy on the immunological status of rectal cancer patients. J. Radiat. Res. 2020, 61, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.Y.; Wu, C.-Y.; Yu, J. The role of gut microbiota in cancer treatment: Friend or foe? Gut 2020, 69, 1867–1876. [Google Scholar] [CrossRef]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. J. Clin. Epidemiol. 2009, 62, 1006–1012. [Google Scholar] [CrossRef]
| Category | Type | Parameter |
|---|---|---|
| Biopolymers of Cancer Cells | Proteins | SATB1 XRCC2, hPEBP4, PITPNC1, FOXK1, FOXK2, Bcl-2, Cox-2, VEGF, APAF-1, FGF8, FGFR4, Survivin, FAK, GOLPH3, PAF15, N-κB/p65, PLK1, ATM, MRE11, VNN1, VRK1, VRK2, RAD51, 53BP1, PITPNC1 |
| Tumour suppressors and oncogenes | TP53, XIAP, TCF4, RAD51 | |
| Transcriptome/Epigenome | Transcriptomic and epigenetic signatures, miR-21, miR-223, miR-31, miR-106a, miR-20b | |
| Immunological markers | Tissue-based immunological markers | TIL |
| Blood-based immunological markers | cytokines | |
| Immunological biomarkers within tumors | CD80 (B7-1), CD86 (B7-2), PD-L1 (B7-H1), PD-L2 (B7-DC or CD273), ICOSL (B7-H2), CD276 (B7-H3), B7S1 (B7-H4, B7x or Vtcn1), VISTA (B7-H5, GI24 or PD-1H), B7-H6 and B7-H7 (HHLA2) | |
| Other biomarkers | Blood-based cancer markers, gut microbiota | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolskas, E.; Mikulskytė, G.; Sileika, E.; Suziedelis, K.; Dulskas, A. Tissue-Based Markers as a Tool to Assess Response to Neoadjuvant Radiotherapy in Rectal Cancer—Systematic Review. Int. J. Mol. Sci. 2022, 23, 6040. https://doi.org/10.3390/ijms23116040
Smolskas E, Mikulskytė G, Sileika E, Suziedelis K, Dulskas A. Tissue-Based Markers as a Tool to Assess Response to Neoadjuvant Radiotherapy in Rectal Cancer—Systematic Review. International Journal of Molecular Sciences. 2022; 23(11):6040. https://doi.org/10.3390/ijms23116040
Chicago/Turabian StyleSmolskas, Edgaras, Goda Mikulskytė, Ernestas Sileika, Kestutis Suziedelis, and Audrius Dulskas. 2022. "Tissue-Based Markers as a Tool to Assess Response to Neoadjuvant Radiotherapy in Rectal Cancer—Systematic Review" International Journal of Molecular Sciences 23, no. 11: 6040. https://doi.org/10.3390/ijms23116040
APA StyleSmolskas, E., Mikulskytė, G., Sileika, E., Suziedelis, K., & Dulskas, A. (2022). Tissue-Based Markers as a Tool to Assess Response to Neoadjuvant Radiotherapy in Rectal Cancer—Systematic Review. International Journal of Molecular Sciences, 23(11), 6040. https://doi.org/10.3390/ijms23116040







