Understanding the Formation and Mechanism of Anticipatory Responses in Escherichia coli
Abstract
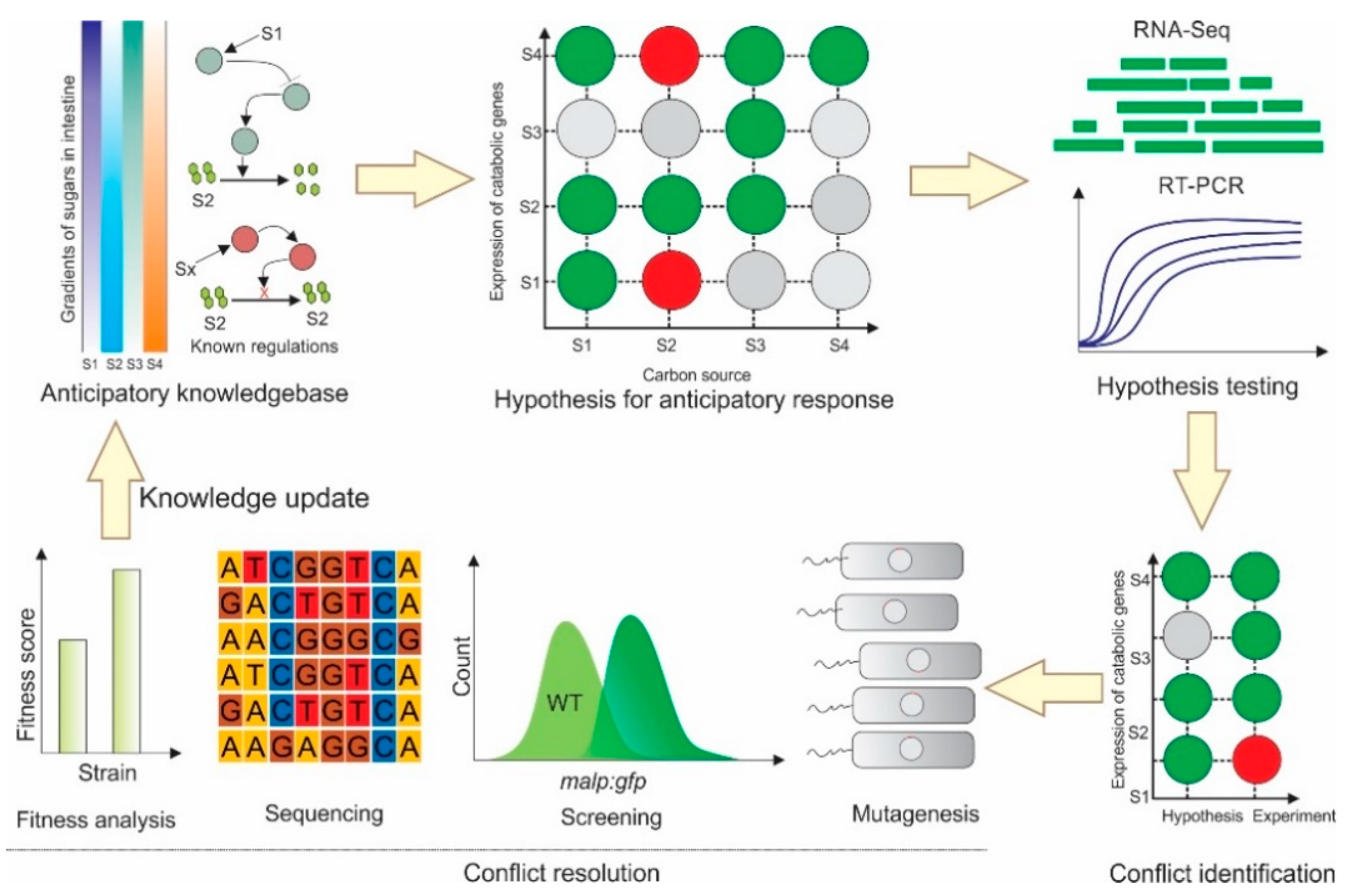
:1. Introduction
2. Results
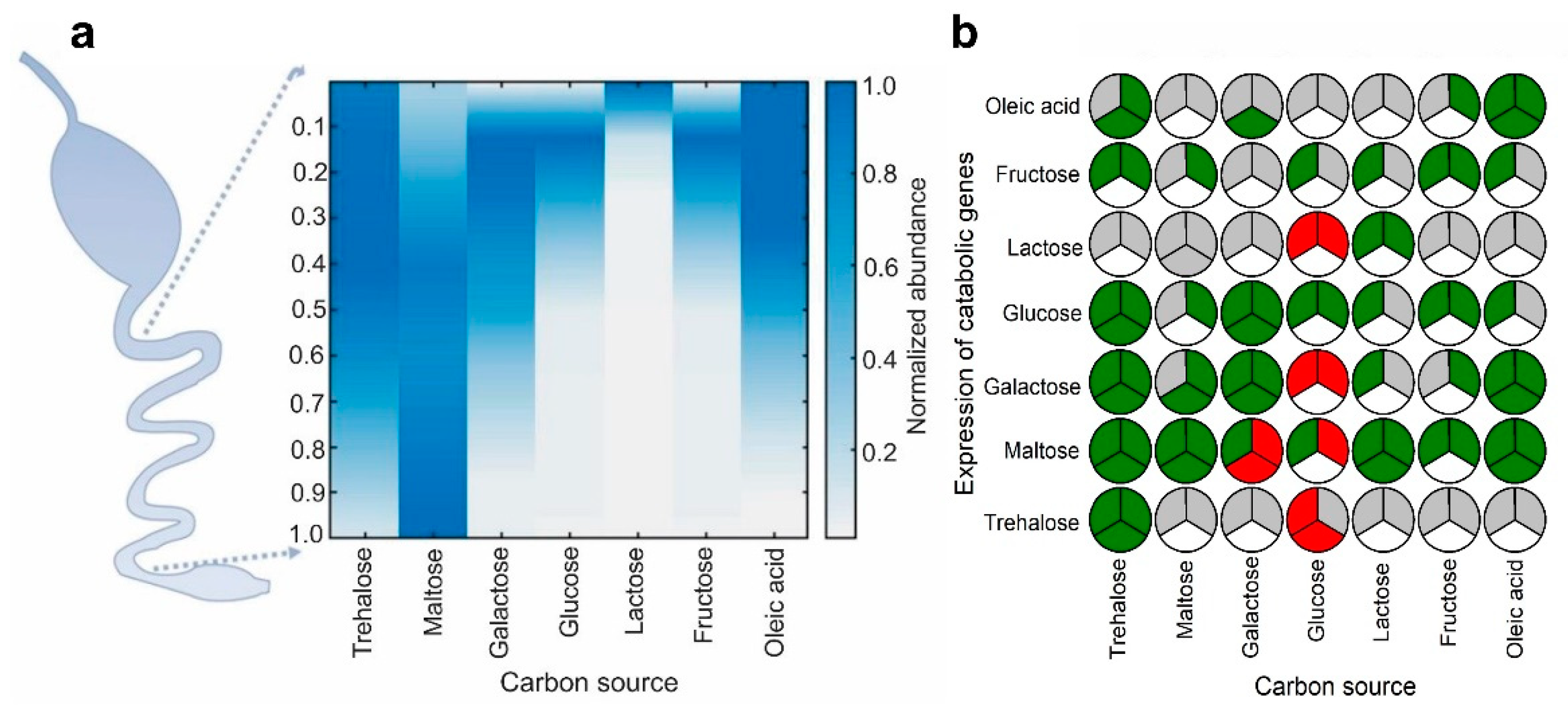
2.1. Identification of Carbon Sources with Spatial Concentration Gradients in the Mammalian Intestine
2.2. Several Instances of Anticipatory Responses Were Identified at the Gene Expression Level in E. coli
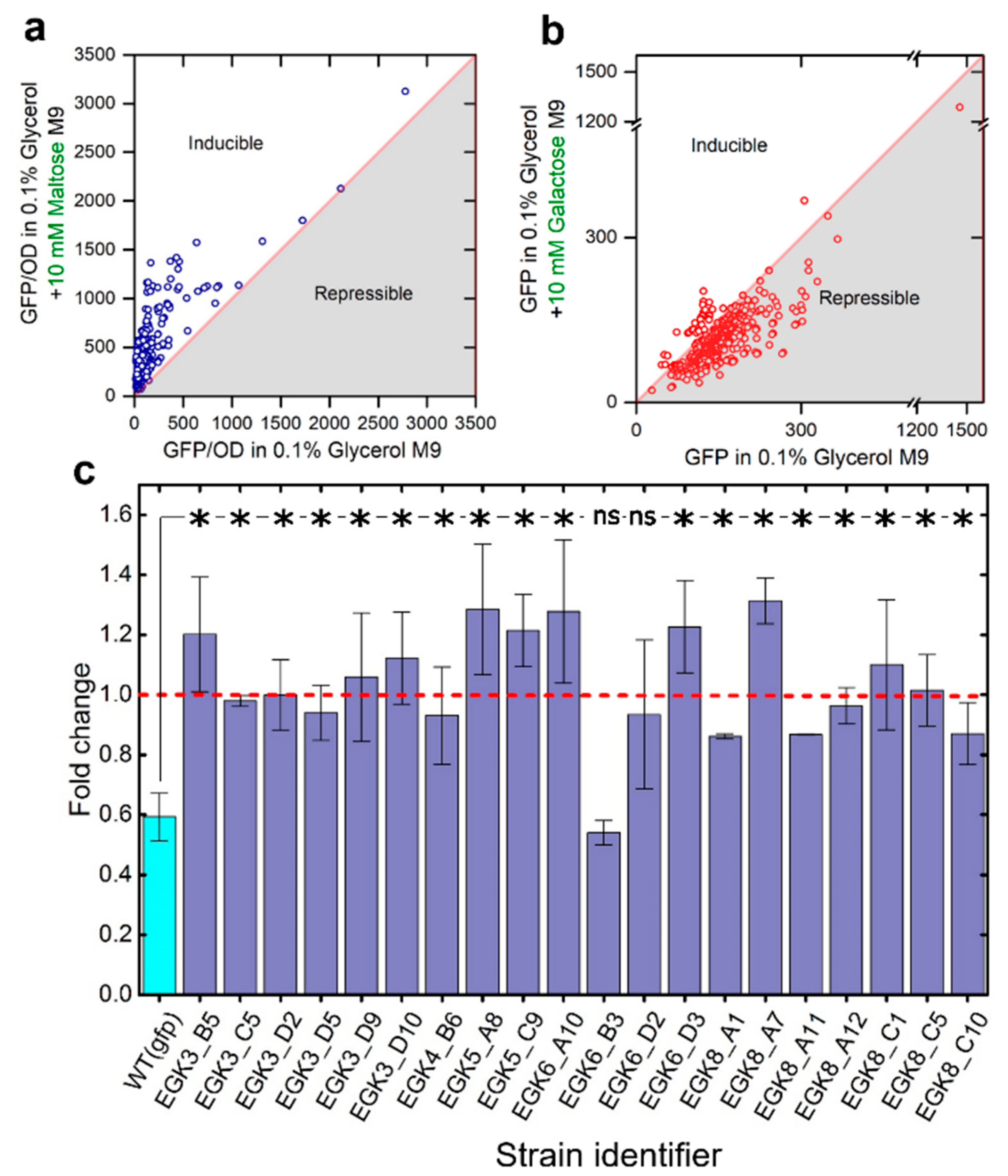
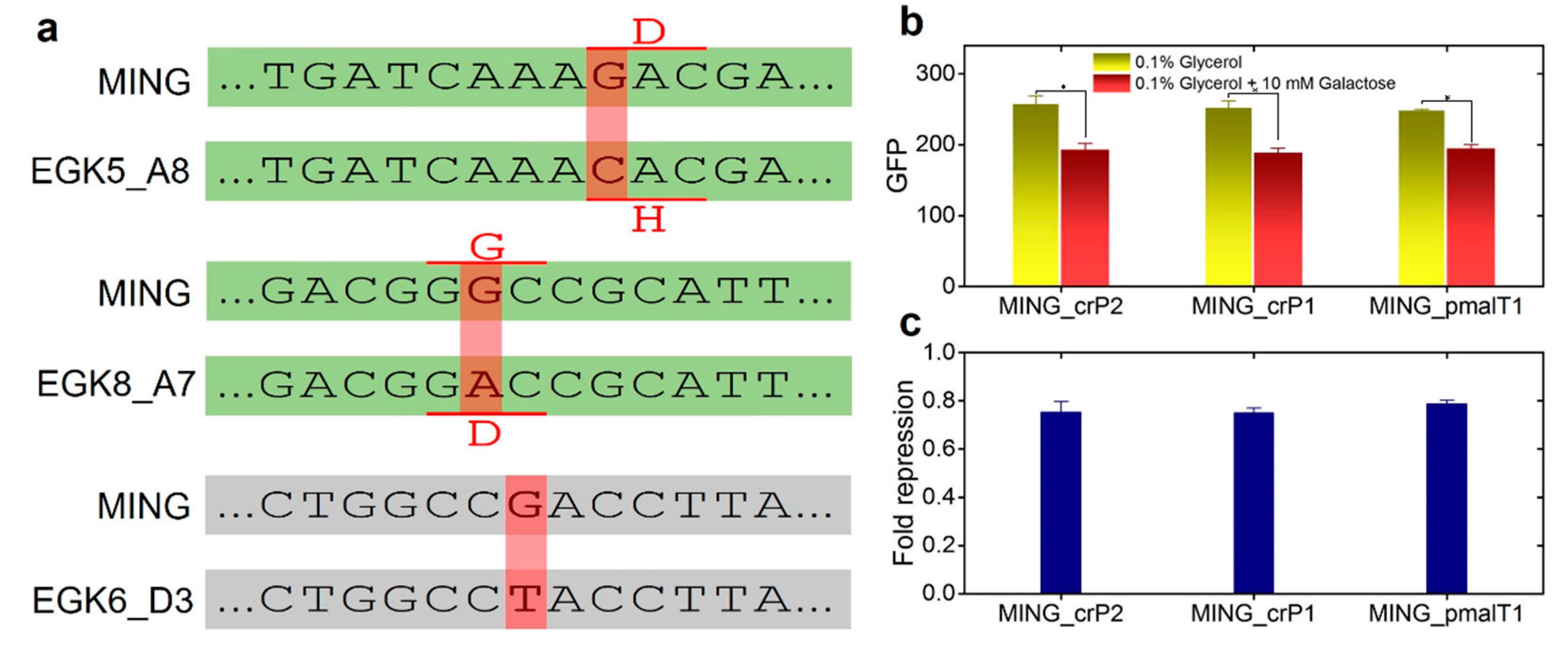
2.3. Anticipatory Responses in E. coli Are Re-Programmable
Repair Mutants Reverse the Response to D-Galactose
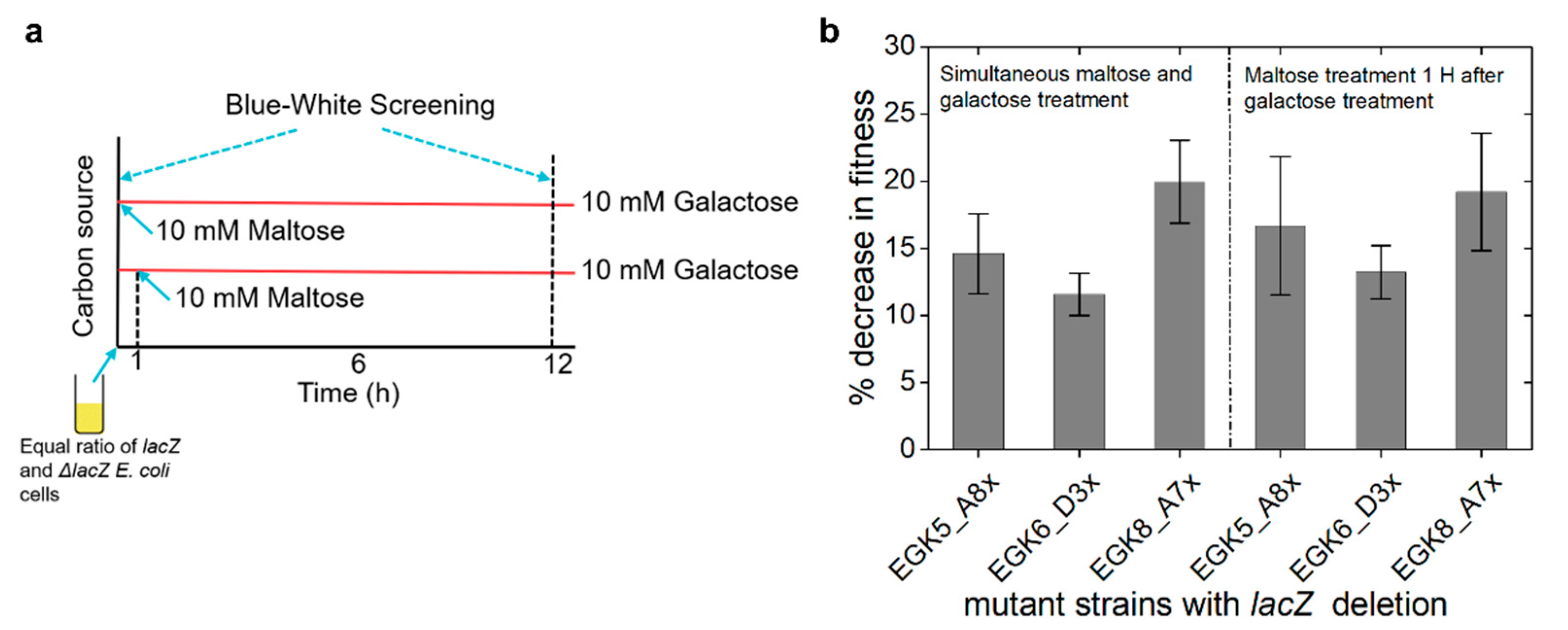
2.4. Expression of Maltose Operons in D-Galactose Is Costly
3. Discussion
4. Materials and Methods
4.1. Strains and Media
4.2. Growth Measurements at Different Concentrations of Carbon Sources
4.3. Treatment of E. coli MG1655 with Carbon Sources, RNA Isolation and Transcriptome Profiling
4.4. Real-Time Reverse Transcription PCR (RT-PCR)
4.5. Chromosomal Integration of gfp:kanR Cassette at the Downstream of malP
4.6. Ethyl Methanesulfonate (EMS) Driven Random Mutagenesis and Screening of the Desired Mutant
4.7. Mutation Repair
4.8. Competition Assays
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hughes, B.S.; Cullum, A.J.; Bennett, A.F. An Experimental Evolutionary Study on Adaptation to Temporally Fluctuating pH in Escherichia coli. Physiol. Biochem. Zool. 2007, 80, 406–421. [Google Scholar] [CrossRef] [Green Version]
- Roszak, D.B.; Colwell, R.R. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 1987, 51, 365–379. [Google Scholar] [CrossRef]
- Freter, R.; Brickner, H.; Botney, M.; Cleven, D.; Aranki, A. Mechanisms That Control Bacterial Populations in Continuous-Flow Culture Models of Mouse Large Intestinal Flora. Infect. Immun. 1983, 39, 676–685. [Google Scholar] [CrossRef] [Green Version]
- Faith, J.J.; McNulty, N.P.; Rey, F.E.; Gordon, J.I. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 2011, 333, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Cooper, V.S.; Lenski, R.E. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 2000, 407, 736. [Google Scholar] [CrossRef]
- Eetemadi, A.; Rai, N.; Pereira, B.M.P.; Kim, M.; Schmitz, H.; Tagkopoulos, I. The Computational Diet: A Review of Computational Methods Across Diet, Microbiome, and Health. Front. Microbiol. 2020, 11, 393. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Freter, R.; Brickner, H.; Fekete, J.; Vickerman, M.M.; Carey, K.E. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 1983, 39, 686–703. [Google Scholar]
- Pereira, F.C.; Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 2017, 19, 1366–1378. [Google Scholar] [CrossRef] [Green Version]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Nunez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Tagkopoulos, I.; Liu, Y.-C.; Tavazoie, S. Predictive Behavior Within Microbial Genetic Networks. Science 2008, 320, 1313–1317. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, A.; Romano, G.H.; Groisman, B.; Yona, A.; Dekel, E.; Kupiec, M.; Dahan, O.; Pilpel, Y. Adaptive prediction of environmental changes by microorganisms. Nature 2009, 460, 220–224. [Google Scholar] [CrossRef]
- Kim, M.; Rai, N.; Zorraquino, V.; Tagkopoulos, I. Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat. Commun. 2016, 7, 13090. [Google Scholar] [CrossRef] [Green Version]
- Batt, R.M.; Peters, T.J. Absorption of galactose by the rat small intestine in vivo: Proximal-distal kinetic gradients and a new method to express absorption per enterocyte. Clin. Sci. Mol. Med. 1976, 50, 499–509. [Google Scholar] [CrossRef]
- Coombe, N.B.; Smith, R.H. Absorption of glucose and galactose and digestion and absorption of lactose by the prepruminant calf. Br. J. Nutr. 1973, 30, 331–344. [Google Scholar] [CrossRef] [Green Version]
- Ferraris, R.P.; Yasharpour, S.; Lloyd, K.C.; Mirzayan, R.; Diamond, J.M. Luminal glucose concentrations in the gut under normal conditions. Am. J. Physiol. 1990, 259, G822–G837. [Google Scholar] [CrossRef]
- Thymann, T.; Moller, H.K.; Stoll, B.; Stoy, A.C.; Buddington, R.K.; Bering, S.B.; Jensen, B.B.; Olutoye, O.O.; Siggers, R.H.; Molbak, L.; et al. Carbohydrate maldigestion induces necrotizing enterocolitis in preterm pigs. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 297, G1115–G1125. [Google Scholar] [CrossRef]
- Cleveland, W.S. Robust Locally Weighted Regression and Smoothing Scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Malakar, P.; Venkatesh, K.V. Effect of substrate and IPTG concentrations on the burden to growth of Escherichia coli on glycerol due to the expression of Lac proteins. Appl. Microbiol. Biotechnol. 2012, 93, 2543–2549. [Google Scholar] [CrossRef]
- Shehata, T.E.; Marr, A.G. Effect of Nutrient Concentration on the Growth of Escherichia coli. J. Bacteriol. 1971, 107, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Inada, T.; Kimata, K.; Aiba, H. Mechanism responsible for glucose–lactose diauxie in Escherichia coli: Challenge to the cAMP model. Genes Cells 1996, 1, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Kopp, J.; Slouka, C.; Ulonska, S.; Kager, J.; Fricke, J.; Spadiut, O.; Herwig, C. Impact of Glycerol as Carbon Source onto Specific Sugar and Inducer Uptake Rates and Inclusion Body Productivity in E. coli BL21(DE3). Bioengineering 2017, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Lin, E.C. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 1976, 30, 535–578. [Google Scholar] [CrossRef]
- Cupples, C.G.; Miller, J.H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 1989, 86, 5345–5349. [Google Scholar] [CrossRef] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Reddy, L.; Poncet, M.; Self, M.W.; Peters, J.C.; Douw, L.; van Dellen, E.; Claus, S.; Reijneveld, J.C.; Baayen, J.C.; Roelfsema, P.R. Learning of anticipatory responses in single neurons of the human medial temporal lobe. Nat. Commun. 2015, 6, 8556. [Google Scholar] [CrossRef] [Green Version]
- Rai, N.; Huynh, L.; Kim, M.; Tagkopoulos, I. Population collapse and adaptive rescue during long-term chemostat fermentation. Biotechnol. Bioeng. 2019, 116, 693–703. [Google Scholar] [CrossRef]
- Sharp, S.J.; Schaack, J.; Cooley, L.; Burke, D.J.; Soll, D. Structure and transcription of eukaryotic tRNA genes. CRC Crit. Rev. Biochem. 1985, 19, 107–144. [Google Scholar] [CrossRef]
- Zhou, K.; Zhou, L.; Lim, Q.E.; Zou, R.; Stephanopoulos, G.; Too, H.-P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011, 12, 18. [Google Scholar] [CrossRef] [Green Version]
- Rai, N.; Ferreiro, A.; Neckelmann, A.; Soon, A.; Yao, A.; Siegel, J.; Facciotti, M.T.; Tagkopoulos, I. RiboTALE: A modular, inducible system for accurate gene expression control. Sci. Rep. 2015, 5, 10658. [Google Scholar] [CrossRef]
| Strain | Description | Reference |
|---|---|---|
| E. coli MG1655 | Wild-type | Laboratory stock |
| MING | MG1655 (malP:gfp:kanR) | This study |
| EGK5_A8 | MING with a point mutation in the coding region of crP | This study |
| EGK8_A7 | MING with a point mutation in the coding region of crP | This study |
| EGK6_D3 | MING with a point mutation in the promoter region of malT | This study |
| MING_crp1 | EGK8_A7 with a repaired point mutation in the coding region of crP | This study |
| MING_crp2 | EGK5_A8 with a repaired point mutation in the coding region of crP | This study |
| MING_pmalT1 | EG6_D3 with a repaired point mutation in the promoter region of malT. | This study |
| EGK5_A8x | EGK5_A8 (ΔlacZ) | This study |
| EGK8_A7x | EGK8_A7x (ΔlacZ) | This study |
| EGK6_D3x | EGK6_D3x (ΔlacZ) | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, N.; Kim, M.; Tagkopoulos, I. Understanding the Formation and Mechanism of Anticipatory Responses in Escherichia coli. Int. J. Mol. Sci. 2022, 23, 5985. https://doi.org/10.3390/ijms23115985
Rai N, Kim M, Tagkopoulos I. Understanding the Formation and Mechanism of Anticipatory Responses in Escherichia coli. International Journal of Molecular Sciences. 2022; 23(11):5985. https://doi.org/10.3390/ijms23115985
Chicago/Turabian StyleRai, Navneet, Minseung Kim, and Ilias Tagkopoulos. 2022. "Understanding the Formation and Mechanism of Anticipatory Responses in Escherichia coli" International Journal of Molecular Sciences 23, no. 11: 5985. https://doi.org/10.3390/ijms23115985
APA StyleRai, N., Kim, M., & Tagkopoulos, I. (2022). Understanding the Formation and Mechanism of Anticipatory Responses in Escherichia coli. International Journal of Molecular Sciences, 23(11), 5985. https://doi.org/10.3390/ijms23115985






