Synthesis, Molecular Docking and Biological Characterization of Pyrazine Linked 2-Aminobenzamides as New Class I Selective Histone Deacetylase (HDAC) Inhibitors with Anti-Leukemic Activity
Abstract
:1. Introduction
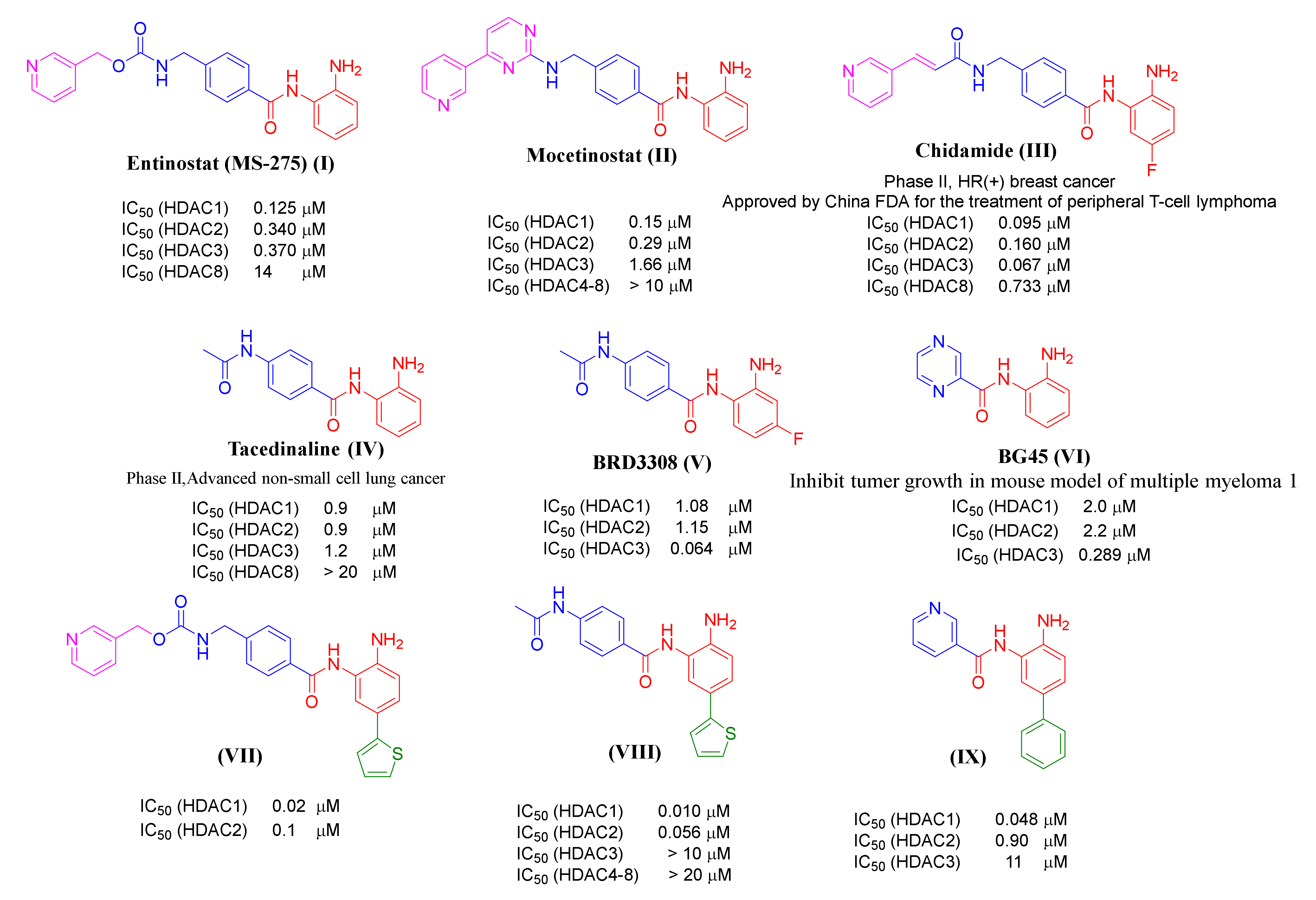
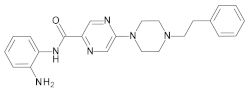
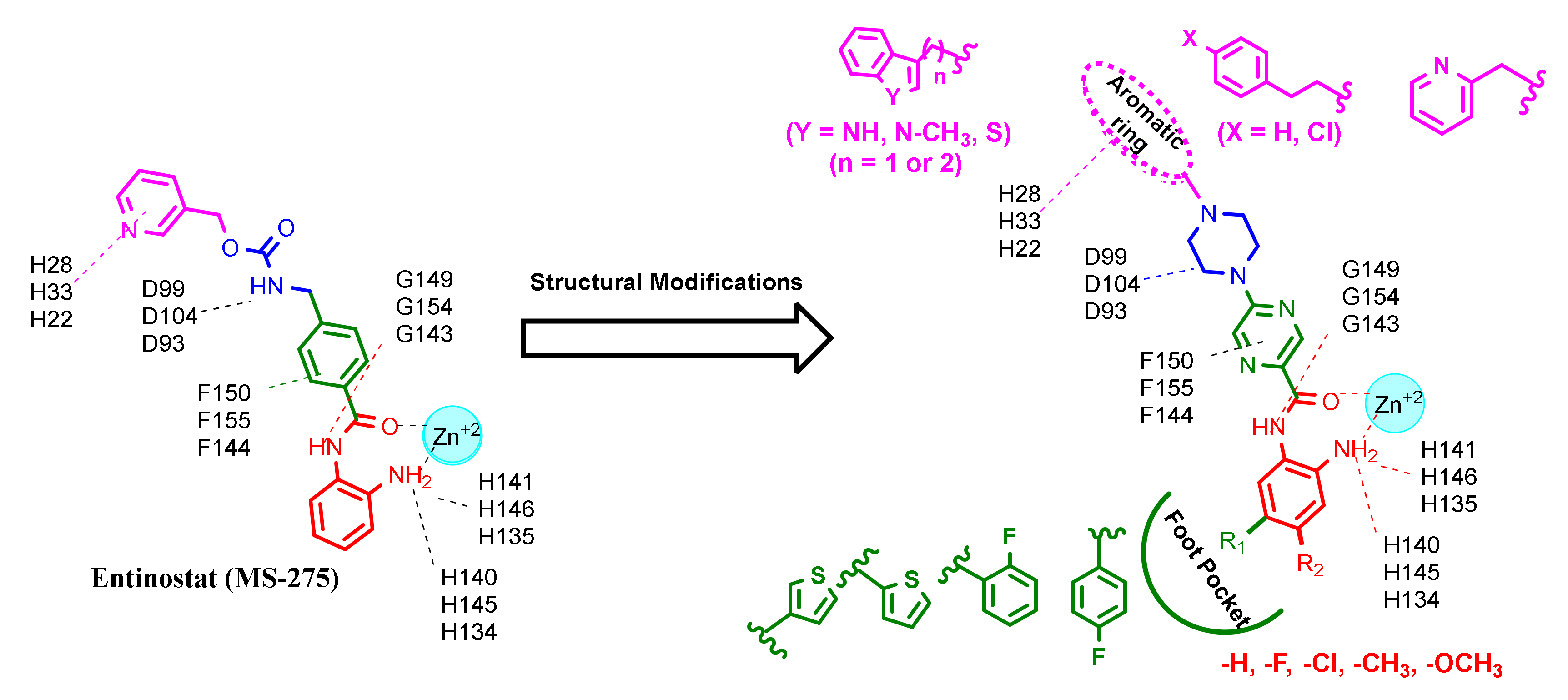
2. Results and Discussion
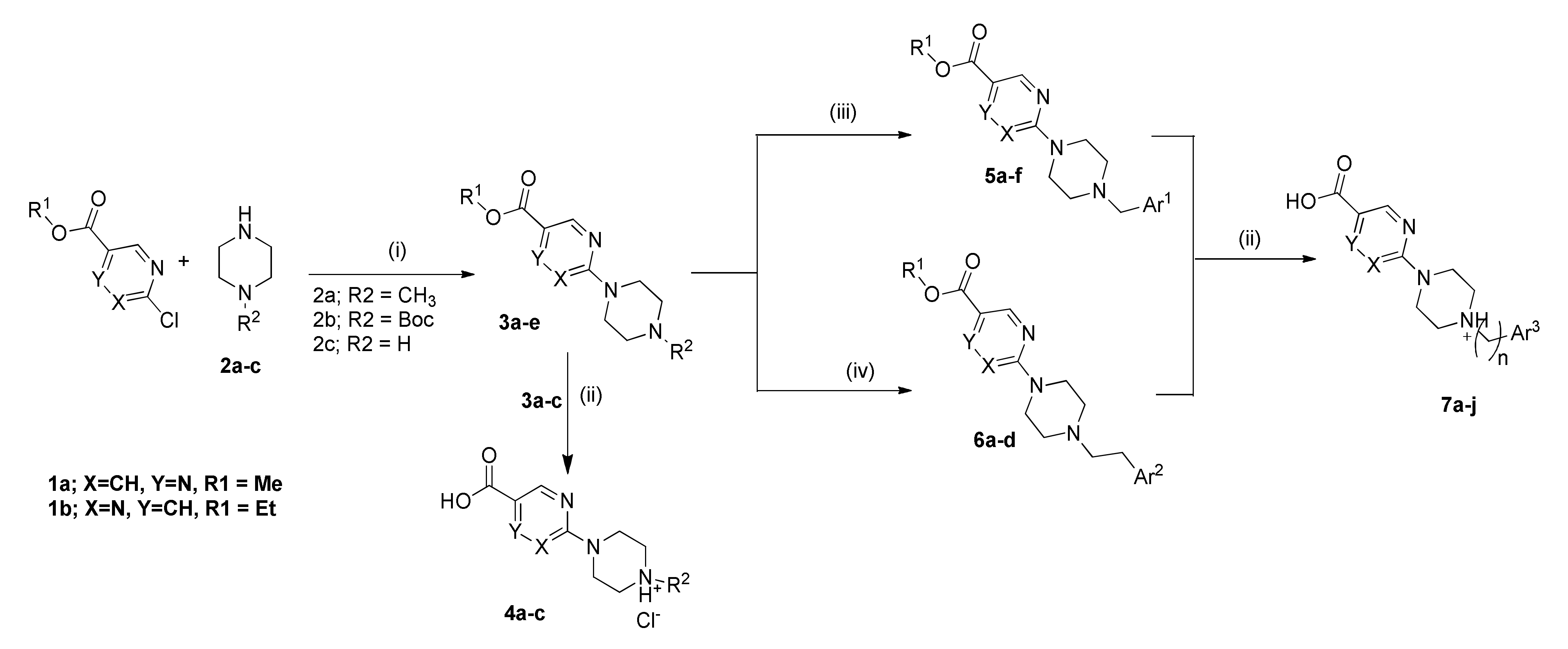
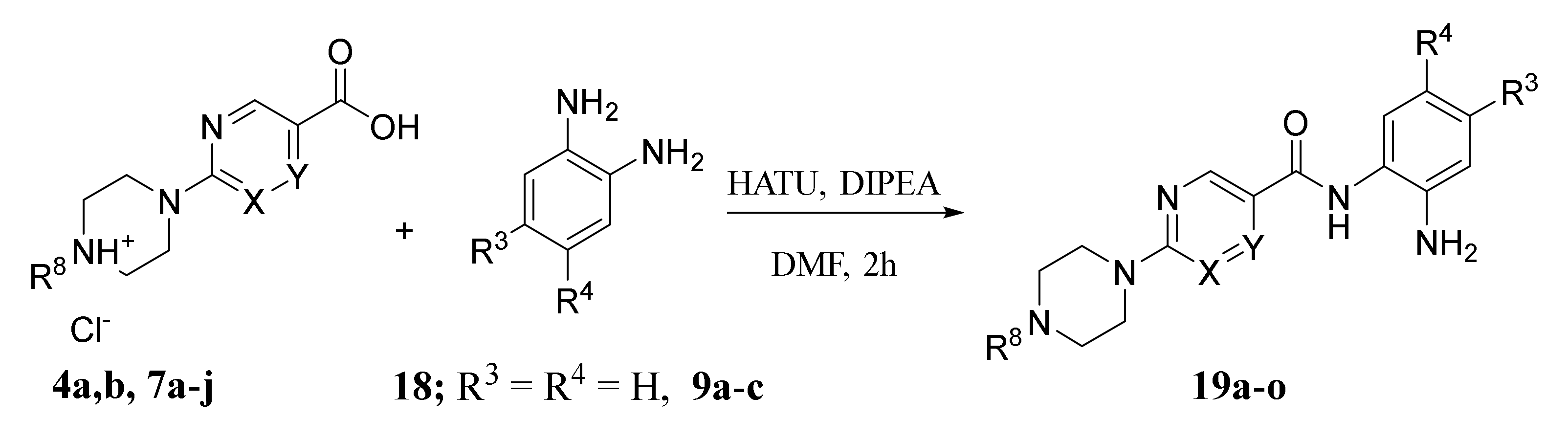
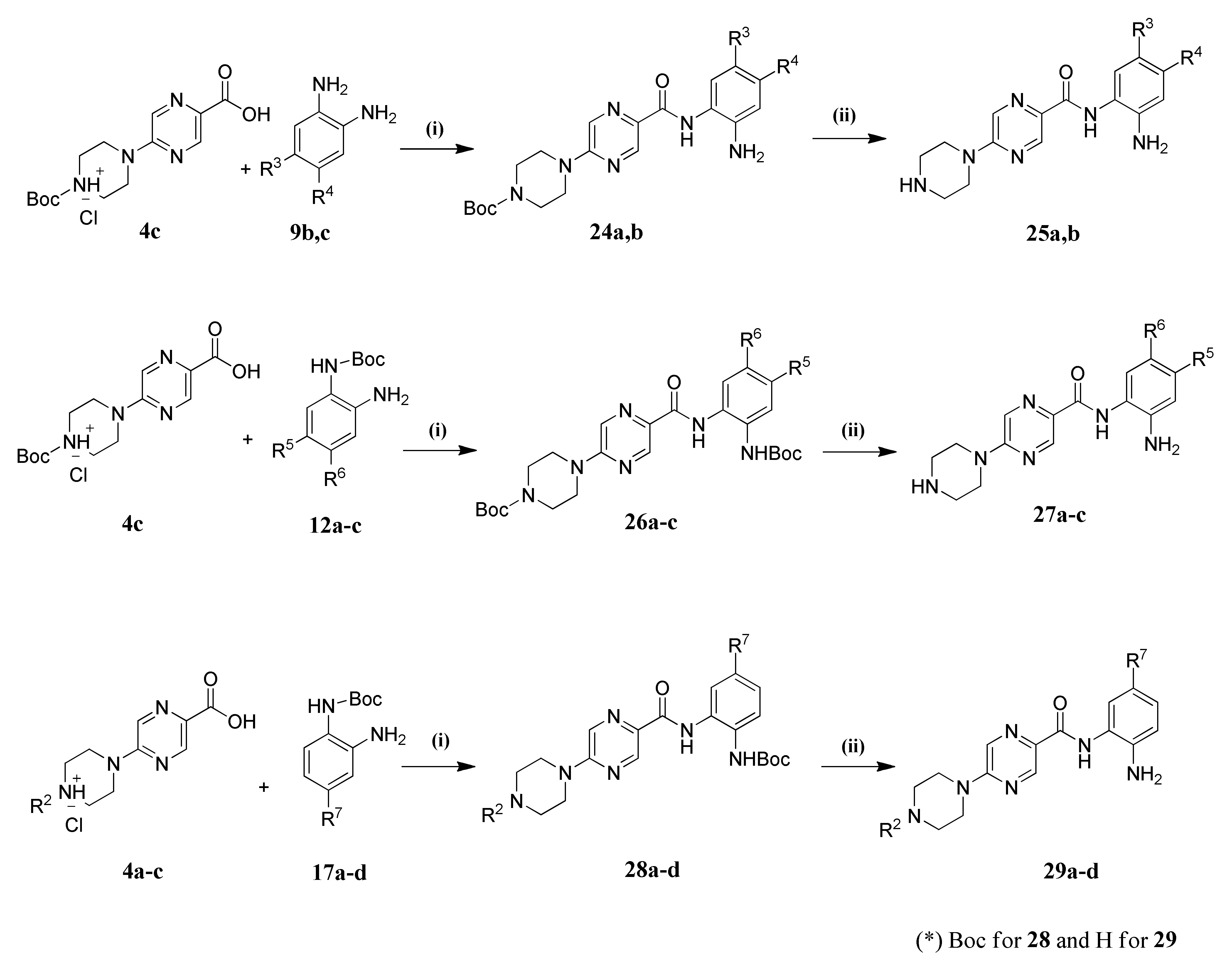
2.1. Chemistry
2.2. Biological Evaluation
In Vitro Testing of HDAC Inhibitory Activity
2.3. Molecular Docking Studies

2.4. Cellular Assays to Analyze Our New Inhibitors
2.4.1. Tests with Non-Transformed Cells
2.4.2. Biological Tests with Leukemic Cells
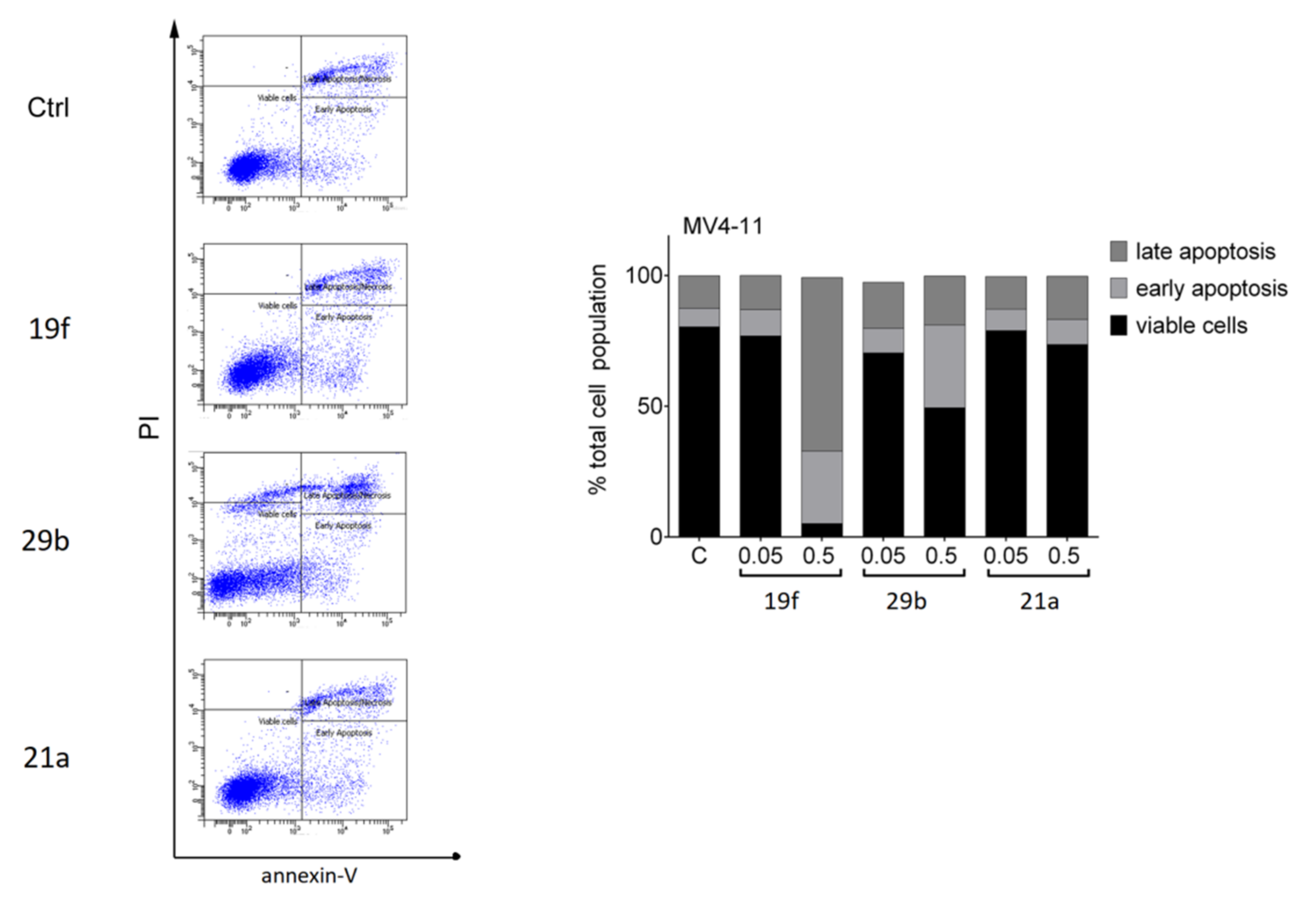
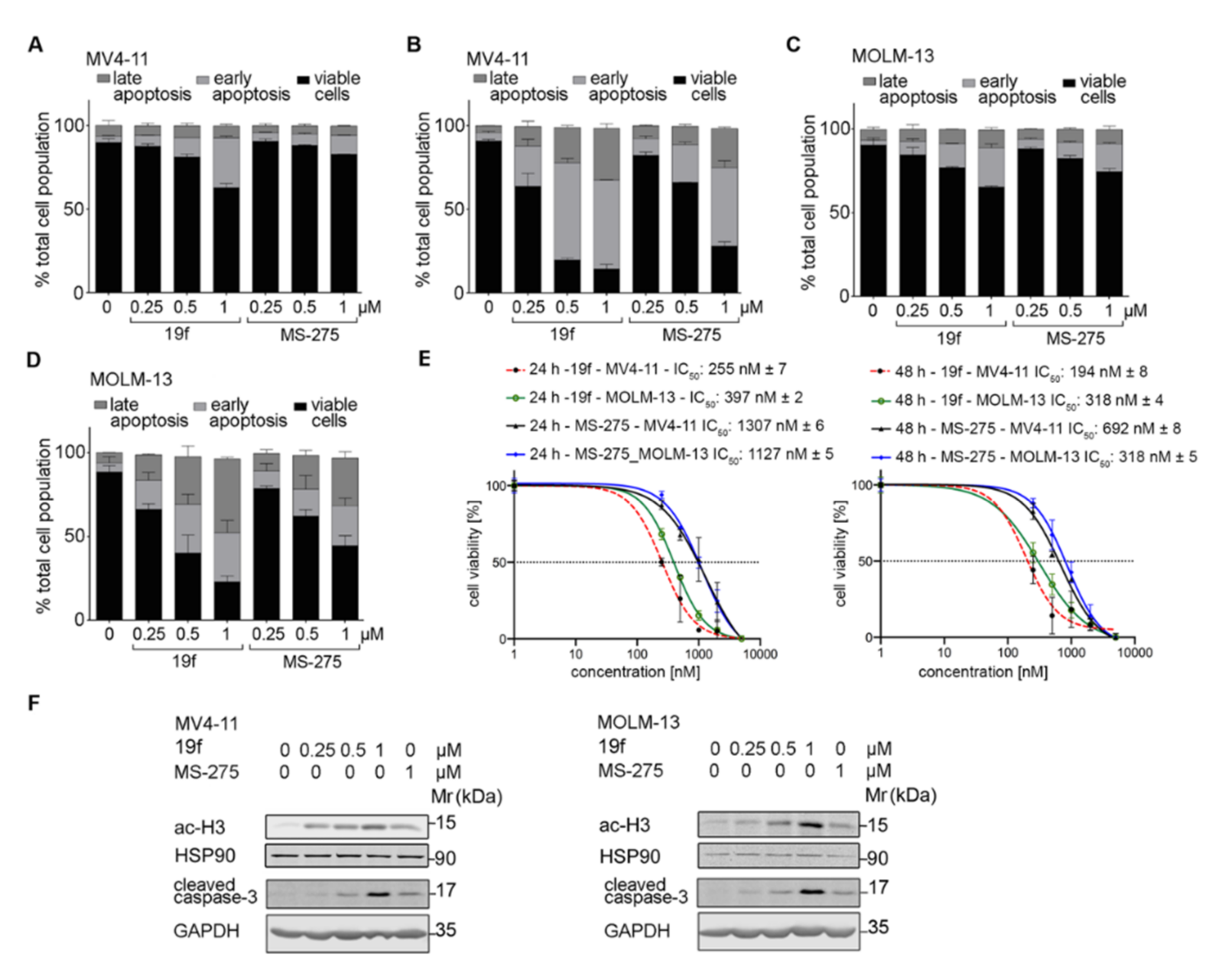

2.4.3. In Silico Prediction of Pharmacokinetic and Tox Data
3. Conclusions
4. Materials and Methods
4.1. Chemistry
4.1.1. General
4.1.2. General Procedure for Amide Coupling
4.1.3. General Procedure for the Preparation of Final Target Compounds X through N-Boc Cleavage
4.1.4. Spectral Analysis of Final Compounds
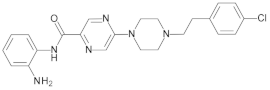
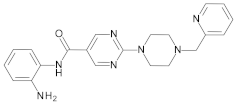
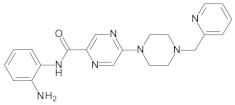
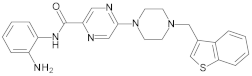
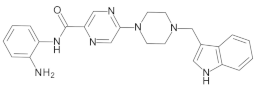
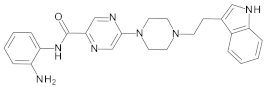
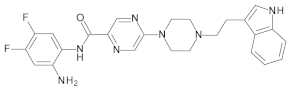
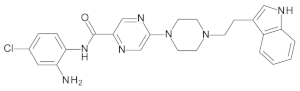
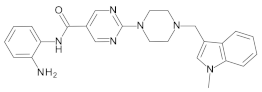
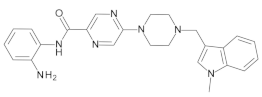
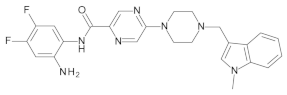
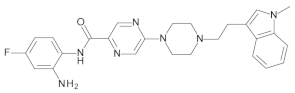
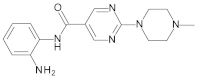
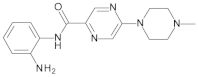
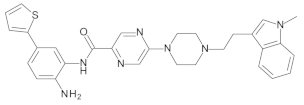
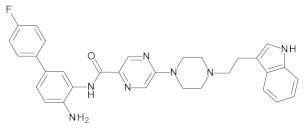
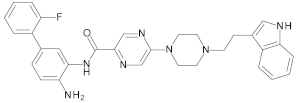
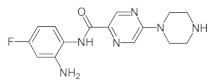
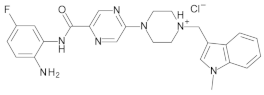
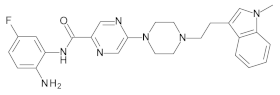
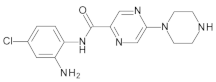
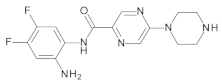

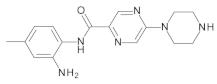
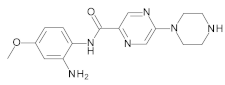
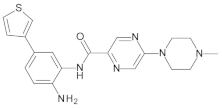
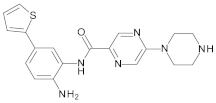
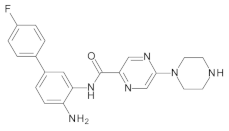
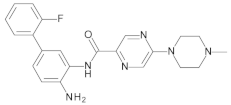
4.2. Biological Evaluation
4.2.1. In Vitro HDAC Inhibition Assay
4.2.2. Cellular Assay
4.3. Computational Studies
4.4. PAINS Filter
4.5. In Silico Prediction of Pharmacokinetic and Tox Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoo, C.B.; Jones, P.A. Epigenetic therapy of cancer: Past, present and future. Nat. Rev. Drug Discov. 2006, 5, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Sadakierska-Chudy, A.; Filip, M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Krautkramer, K.A.; Feldman, J.L.; Denu, J.M. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 2015, 10, 95–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chadha, S.; Wang, L.; Hancock, W.W.; Beier, U.H. Sirtuin-1 in immunotherapy: A Janus-headed target. J. Leukoc. Biol. 2019, 106, 337–343. [Google Scholar] [CrossRef]
- Pant, K.; Peixoto, E.; Richard, S.; Gradilone, S.A. Role of Histone Deacetylases in Carcinogenesis: Potential Role in Cholangiocarcinoma. Cells 2020, 9, 780. [Google Scholar] [CrossRef] [Green Version]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Gryder, B.E.; Sodji, Q.H.; Oyelere, A.K. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med. Chem. 2012, 4, 505–524. [Google Scholar] [CrossRef] [Green Version]
- Cappellacci, L.; Perinelli, D.R.; Maggi, F.; Grifantini, M.; Petrelli, R. Recent progress in histone deacetylase inhibitors as anticancer agents. Curr. Med. Chem. 2020, 27, 2449–2493. [Google Scholar] [CrossRef]
- Wagner, F.F.; Weïwer, M.; Lewis, M.C.; Holson, E.B. Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics 2013, 10, 589–604. [Google Scholar] [CrossRef] [Green Version]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef]
- Chien, W.; Lee, D.H.; Zheng, Y.; Wuensche, P.; Alvarez, R.; Wen, D.L.; Aribi, A.M.; Thean, S.M.; Doan, N.B.; Said, J.W. Growth inhibition of pancreatic cancer cells by histone deacetylase inhibitor belinostat through suppression of multiple pathways including HIF, NFkB, and mTOR signaling in vitro and in vivo. Mol. Carcinog. 2014, 53, 722–735. [Google Scholar] [CrossRef] [Green Version]
- Sivaraj, D.; Green, M.M.; Gasparetto, C. Panobinostat for the management of multiple myeloma. Future Oncol. 2017, 13, 477–488. [Google Scholar] [CrossRef]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [Green Version]
- Chifotides, H.T.; Bose, P.; Verstovsek, S. Givinostat: An emerging treatment for polycythemia vera. Expert Opin. Investig. Drugs 2020, 29, 525–536. [Google Scholar] [CrossRef]
- Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Fritzsche, F.R.; Niesporek, S.; Denkert, C.; Dietel, M.; et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer 2008, 98, 604–610. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [Green Version]
- Fritzsche, F.R.; Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Scholman, K.; Denkert, C.; Dietel, M.; et al. Class I histone deacetylases 1, 2 and 3 are highly expressed in renal cell cancer. BMC Cancer 2008, 8, 381. [Google Scholar] [CrossRef] [Green Version]
- Stypula-Cyrus, Y.; Damania, D.; Kunte, D.P.; Cruz, M.D.; Subramanian, H.; Roy, H.K.; Backman, V. HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure. PLoS ONE 2013, 8, e64600. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.-L.; Yue, Z.; Liu, L.; Pei, L.; Yin, Y.; Qin, L.; Zhao, J.; Liu, H.; Wang, H.; Jia, M. The expression of histone deacetylase HDAC1 correlates with the progression and prognosis of gastrointestinal malignancy. Oncotarget 2017, 8, 39241–39253. [Google Scholar] [CrossRef] [Green Version]
- Azad, N.S.; el-Khoueiry, A.; Yin, J.; Oberg, A.L.; Flynn, P.; Adkins, D.; Sharma, A.; Weisenberger, D.J.; Brown, T.; Medvari, P.; et al. Combination epigenetic therapy in metastatic colorectal cancer (mCRC) with subcutaneous 5-azacitidine and entinostat: A phase 2 consortium/stand Up 2 cancer study. Oncotarget 2017, 8, 35326–35338. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.C.; Yvette, K.; Bociek, R.G.; Peter, L.; Lia, G.; Amanda, C.; Rachel, S.; Peter, O.; Scott, C.; Lori, K.; et al. ENGAGE- 501: Phase II study of entinostat (SNDX-275) in relapsed and refractory Hodgkin lymphoma. Haematologica 2016, 101, 968–975. [Google Scholar]
- Batlevi, C.L.; Crump, M.; Andreadis, C.; Rizzieri, D.; Assouline, S.E.; Fox, S.; van der Jagt, R.H.C.; Copeland, A.; Potvin, D.; Chao, R.; et al. A phase 2 study of mocetinostat, a histone deacetylase inhibitor, in relapsed or refractory lymphoma. Br. J. Haematol. 2017, 178, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Kramer, O.H. Inhibitors of HDACs–Effective Drugs Against Cancer? Curr. Cancer Drug Targets 2010, 10, 210–228. [Google Scholar] [CrossRef] [PubMed]
- Ungerstedt, J.S. Epigenetic Modifiers in Myeloid Malignancies: The Role of Histone Deacetylase Inhibitors. Int. J. Mol. Sci. 2018, 19, 3091. [Google Scholar] [CrossRef] [Green Version]
- Gryder, B.E.; Wu, L.; Woldemichael, G.M.; Pomella, S.; Quinn, T.R.; Park, P.M.C.; Cleveland, A.; Stanton, B.Z.; Song, Y.; Rota, R.; et al. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat. Commun. 2019, 10, 3004–3004. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, J.; Jiang, Q.; Zhang, L.; Song, W. Zinc binding groups for histone deacetylase inhibitors. J. Enzym. Inhib. Med. Chem. 2018, 33, 714–721. [Google Scholar] [CrossRef]
- Micelli, C.; Rastelli, G. Histone deacetylases: Structural determinants of inhibitor selectivity. Drug Discov. Today 2015, 20, 718–735. [Google Scholar] [CrossRef]
- Zhan, P.; Itoh, Y.; Suzuki, T.; Liu, X. Strategies for the discovery of target-specific or isoform-selective modulators. J. Med. Chem. 2015, 58, 7611–7633. [Google Scholar] [CrossRef]
- Perrin, J.; Werner, T.; Kurzawa, N.; Rutkowska, A.; Childs, D.D.; Kalxdorf, M.; Poeckel, D.; Stonehouse, E.; Strohmer, K.; Heller, B.; et al. Identifying drug targets in tissues and whole blood with thermal-shift profiling. Nat. Biotechnol. 2020, 38, 303–308. [Google Scholar] [CrossRef]
- Becher, I.; Werner, T.; Doce, C.; Zaal, E.A.; Tögel, I.; Khan, C.A.; Rueger, A.; Muelbaier, M.; Salzer, E.; Berkers, C.R.; et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 2016, 12, 908–910. [Google Scholar] [CrossRef]
- Shen, S.; Kozikowski, A.P. Why Hydroxamates May Not Be the Best Histone Deacetylase Inhibitors—What Some May Have Forgotten or Would Rather Forget? ChemMedChem 2016, 11, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Choi, S.-E.; Lee, H.B.; Song, M.-W.; Kim, Y.H.; Jeong, J.Y.; Kang, Y.; Kim, H.J.; Kim, T.H.; Jeon, J.Y.; et al. A Class I Histone Deacetylase Inhibitor Attenuates Insulin Resistance and Inflammation in Palmitate-Treated C2C12 Myotubes and Muscle of HF/HFr Diet Mice. Front. Pharmacol. 2020, 11, 601448. [Google Scholar] [CrossRef]
- Cai, J.; Wei, H.; Hong, K.H.; Wu, X.; Cao, M.; Zong, X.; Li, L.; Sun, C.; Chen, J.; Ji, M. Discovery and preliminary evaluation of 2-aminobenzamide and hydroxamate derivatives containing 1, 2, 4-oxadiazole moiety as potent histone deacetylase inhibitors. Eur. J. Med. Chem. 2015, 96, 1–13. [Google Scholar] [CrossRef]
- Ning, Z.Q.; Li, Z.B.; Newman, M.J.; Shan, S.; Wang, X.H.; Pan, D.S.; Zhang, J.; Dong, M.; Du, X.; Lu, X.P. Chidamide (CS055/HBI-8000): A new histone deacetylase inhibitor of the benzamide class with antitumor activity and the ability to enhance immune cell-mediated tumor cell cytotoxicity. Cancer Chemother. Pharmacol. 2012, 69, 901–909. [Google Scholar] [CrossRef]
- Pan, D.-S.; Yang, Q.-J.; Fu, X.; Shan, S.; Zhu, J.-Z.; Zhang, K.; Li, Z.-B.; Ning, Z.-Q.; Lu, X.-P. Discovery of an orally active subtype-selective HDAC inhibitor, chidamide, as an epigenetic modulator for cancer treatment. MedChemComm 2014, 5, 1789–1796. [Google Scholar] [CrossRef]
- Wagner, F.F.; Lundh, M.; Kaya, T.; McCarren, P.; Zhang, Y.L.; Chattopadhyay, S.; Gale, J.P.; Galbo, T.; Fisher, S.L.; Meier, B.C.; et al. An Isochemogenic Set of Inhibitors To Define the Therapeutic Potential of Histone Deacetylases in beta-Cell Protection. ACS Chem. Biol. 2016, 11, 363–374. [Google Scholar] [CrossRef]
- Weïwer, M.; Lewis, M.C.; Wagner, F.F.; Holson, E.B. Therapeutic potential of isoform selective HDAC inhibitors for the treatment of schizophrenia. Future Med. Chem. 2013, 5, 1491–1508. [Google Scholar] [CrossRef]
- Tang, D.; Xu, L.; Zhang, M.; Dorfman, R.G.; Pan, Y.; Zhou, Q.; Zhou, L.; Wang, Y.; Li, Y.; Yin, Y.; et al. Metformin facilitates BG45-induced apoptosis via an anti-Warburg effect in cholangiocarcinoma cells. Oncol. Rep. 2018, 39, 1957–1965. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yu, Y.; Kelly, J.; Sha, D.; Alhassan, A.-B.; Yu, W.; Maletic, M.M.; Duffy, J.L.; Klein, D.J.; Holloway, M.K.; et al. Discovery of Highly Selective and Potent HDAC3 Inhibitors Based on a 2-Substituted Benzamide Zinc Binding Group. ACS Med. Chem. Lett. 2020, 11, 2476–2483. [Google Scholar] [CrossRef]
- Moradei, O.M.; Mallais, T.C.; Frechette, S.; Paquin, I.; Tessier, P.E.; Leit, S.M.; Fournel, M.; Bonfils, C.; Trachy-Bourget, M.-C.; Liu, J. Novel aminophenyl benzamide-type histone deacetylase inhibitors with enhanced potency and selectivity. J. Med. Chem. 2007, 50, 5543–5546. [Google Scholar] [CrossRef]
- Wagner, F.F.; Weiwer, M.; Steinbacher, S.; Schomburg, A.; Reinemer, P.; Gale, J.P.; Campbell, A.J.; Fisher, S.L.; Zhao, W.N.; Reis, S.A.; et al. Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors. Bioorg. Med. Chem. 2016, 24, 4008–4015. [Google Scholar] [CrossRef] [PubMed]
- Arts, J.; King, P.; Mariën, A.; Floren, W.; Beliën, A.; Janssen, L.; Pilatte, I.; Roux, B.; Decrane, L.; Gilissen, R.; et al. JNJ-26481585, a Novel “Second-Generation” Oral Histone Deacetylase Inhibitor, Shows Broad-Spectrum Preclinical Antitumoral Activity. Clin. Cancer Res. 2009, 15, 6841–6851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zessin, M.; Kutil, Z.; Meleshin, M.; Nováková, Z.; Ghazy, E.; Kalbas, D.; Marek, M.; Romier, C.; Sippl, W.; Bařinka, C.; et al. One-Atom Substitution Enables Direct and Continuous Monitoring of Histone Deacylase Activity. Biochemistry 2019, 58, 4777–4789. [Google Scholar] [CrossRef] [PubMed]
- Lauffer, B.E.L.; Mintzer, R.; Fong, R.; Mukund, S.; Tam, C.; Zilberleyb, I.; Flicke, B.; Ritscher, A.; Fedorowicz, G.; Vallero, R.; et al. Histone Deacetylase (HDAC) Inhibitor Kinetic Rate Constants Correlate with Cellular Histone Acetylation but not Transcription and Cell Viability. J. Biol. Chem. 2013, 288, 26926–26943. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Zwinderman, M.R.H.; Dekker, F.J. The Process and Strategy for Developing Selective Histone Deacetylase 3 Inhibitors. Molecules 2018, 23, 551. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.; Chen, Q.; Shimada, K.; Tang, M.; Li, H.; Gurumurthy, A.; Khoury, J.D.; Xu, B.; Huang, S.; Qiu, Y. Histone deacetylase inhibitor targets CD123/CD47-positive cells and reverse chemoresistance phenotype in acute myeloid leukemia. Leukemia 2019, 33, 931–944. [Google Scholar] [CrossRef]
- Mori, M.; Kaneko, N.; Ueno, Y.; Yamada, M.; Tanaka, R.; Saito, R.; Shimada, I.; Mori, K.; Kuromitsu, S. Gilteritinib, a FLT3/AXL inhibitor, shows antileukemic activity in mouse models of FLT3 mutated acute myeloid leukemia. Investig. New Drugs 2017, 35, 556–565. [Google Scholar] [CrossRef] [Green Version]
- Schlenk, R.F.; Kayser, S. Midostaurin: A Multiple Tyrosine Kinases Inhibitor in Acute Myeloid Leukemia and Systemic Mastocytosis. In Small Molecules in Hematology; Martens, U.M., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 199–214. [Google Scholar]
- Cioccio, J.; Claxton, D. Therapy of acute myeloid leukemia: Therapeutic targeting of tyrosine kinases. Expert Opin. Investig. Drugs 2019, 28, 337–349. [Google Scholar] [CrossRef]
- Beyer, M.; Henninger, S.J.; Haehnel, P.S.; Mustafa, A.-H.M.; Gurdal, E.; Schubert, B.; Christmann, M.; Sellmer, A.; Mahboobi, S.; Drube, S.; et al. Identification of a highly efficient dual type I/II FMS-like tyrosine kinase inhibitor that disrupts the growth of leukemic cells. Cell Chem. Biol. 2021, 29, 1–14. [Google Scholar] [CrossRef]
- Beyer, M.; Romanski, A.; Mustafa, A.-H.M.; Pons, M.; Büchler, I.; Vogel, A.; Pautz, A.; Sellmer, A.; Schneider, G.; Bug, G.; et al. HDAC3 Activity is Essential for Human Leukemic Cell Growth and the Expression of β-catenin, MYC, and WT1. Cancers 2019, 11, 1436. [Google Scholar] [CrossRef] [Green Version]
- Kiweler, N.; Wünsch, D.; Wirth, M.; Mahendrarajah, N.; Schneider, G.; Stauber, R.H.; Brenner, W.; Butter, F.; Krämer, O.H. Histone deacetylase inhibitors dysregulate DNA repair proteins and antagonize metastasis-associated processes. J. Cancer Res. Clin. Oncol. 2020, 146, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Padrnos, L.; Mesa, R. Novel agents for the treatment of polycythemia vera: An insight into preclinical research and early phase clinical trials. Expert Opin. Investig. Drugs 2020, 29, 809–817. [Google Scholar] [CrossRef]
- Kutil, Z.; Novakova, Z.; Meleshin, M.; Mikesova, J.; Schutkowski, M.; Barinka, C. Histone Deacetylase 11 Is a Fatty-Acid Deacylase. ACS Chem. Biol. 2018, 13, 685–693. [Google Scholar] [CrossRef]
- Marek, M.; Shaik, T.B.; Heimburg, T.; Chakrabarti, A.; Lancelot, J.; Ramos-Morales, E.; Da Veiga, C.; Kalinin, D.; Melesina, J.; Robaa, D.; et al. Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants. J. Med. Chem. 2018, 61, 10000–10016. [Google Scholar] [CrossRef]
- Kutil, Z.; Mikešová, J.; Zessin, M.; Meleshin, M.; Nováková, Z.; Alquicer, G.; Kozikowski, A.; Sippl, W.; Bařinka, C.; Schutkowski, M. Continuous Activity Assay for HDAC11 Enabling Reevaluation of HDAC Inhibitors. ACS Omega 2019, 4, 19895–19904. [Google Scholar] [CrossRef] [Green Version]
- Heimburg, T.; Kolbinger, F.R.; Zeyen, P.; Ghazy, E.; Herp, D.; Schmidtkunz, K.; Melesina, J.; Shaik, T.B.; Erdmann, F.; Schmidt, M.; et al. Structure-Based Design and Biological Characterization of Selective Histone Deacetylase 8 (HDAC8) Inhibitors with Anti-Neuroblastoma Activity. J. Med. Chem. 2017, 60, 10188–10204. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Minami, J.; Suzuki, R.; Mazitschek, R.; Gorgun, G.; Ghosh, B.; Cirstea, D.; Hu, Y.; Mimura, N.; Ohguchi, H.; Cottini, F. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia 2014, 28, 680–689. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger Release 2019-1: Maestro, Protein Preparation Wizard, Prime, Epik, Ligprep, Confgen, Glide; Schrödinger LLC.: New York, NY, USA, 2019.
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE), 2019.01; Chemical Computing Group: Montreal, QC, Canada, 2019.
- Case, D.; Betz, R.; Cerutti, D.; Cheatham, T., III; Darden, T.; Duke, R.; Giese, T.; Gohlke, H.; Goetz, A.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Jakalian, A.; Bush, B.L.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000, 21, 132–146. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.H.; Le, J.; Abraham, M.H.; Hersey, A.; Eddershaw, P.J.; Luscombe, C.N.; Boutina, D.; Beck, G.; Sherborne, B.; Cooper, I.; et al. Evaluation of human intestinal absorption data and subsequent derivation of a quantitative structure–activity relationship (QSAR) with the Abraham descriptors. J. Pharm. Sci. 2001, 90, 749–784. [Google Scholar] [CrossRef] [Green Version]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014, 42, W53–W58. [Google Scholar] [CrossRef] [Green Version]


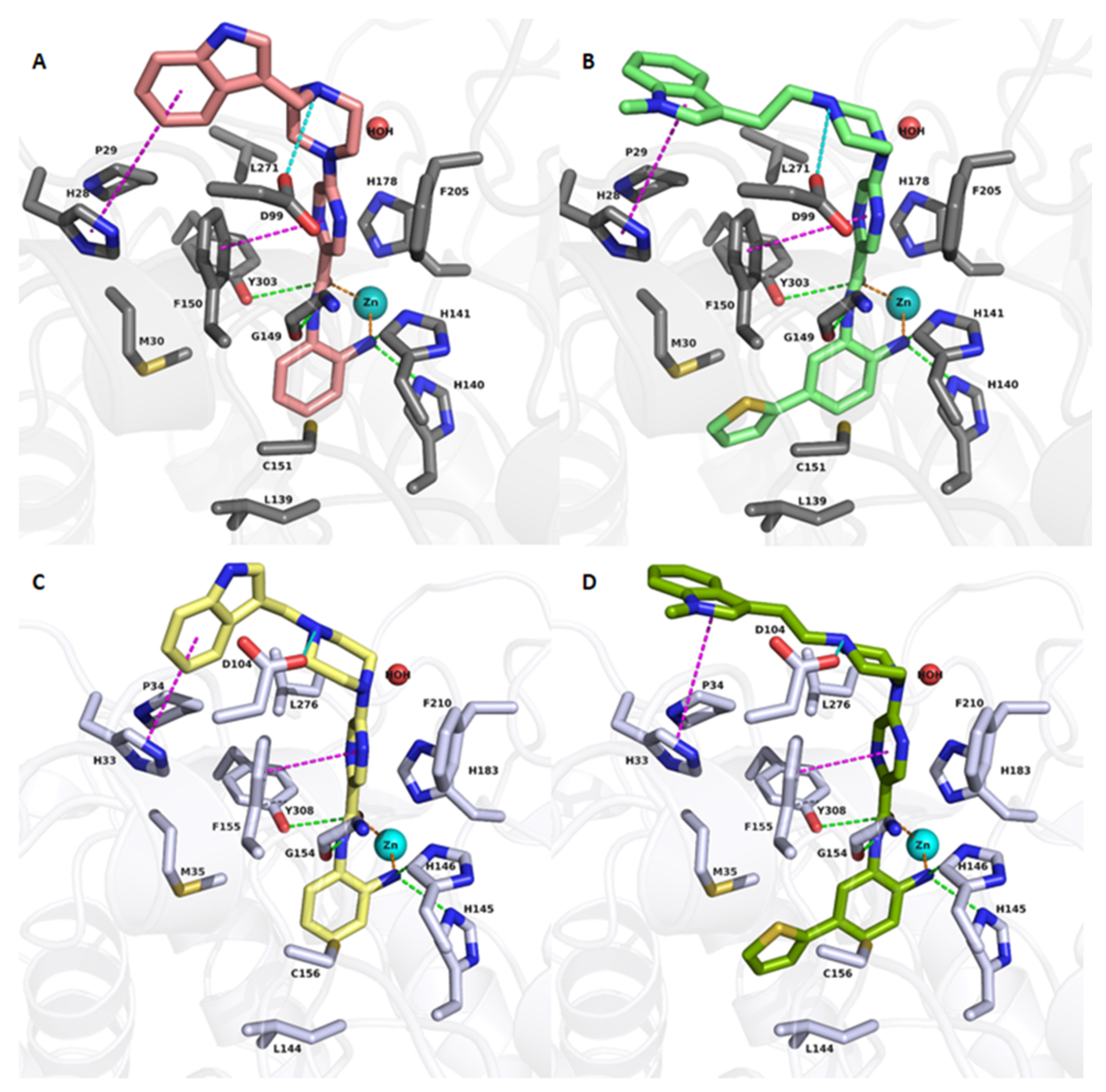

| Cpd. No. | X | Y | R 1 | R 2 | R 3 | HDAC1 (IC50 µM) | HDAC2 (IC50 µM) | HDAC3 (IC50 µM) | HDAC8 (% Inhib.) |
|---|---|---|---|---|---|---|---|---|---|
| 19a | CH | N |  | H | H | 0.51 ± 0.05 | 0.80 ± 0.07 | 1.12 ± 0.07 | 0% @ 1 µM |
| 19b | CH | N | 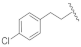 | H | H | 25.8% @ 2 µM | 30.3% @ 2 µM | 65.2% @2 µM | 3.4% @ 1 µM |
| 19c | N | CH | 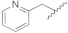 | H | H | 33.9% @ 2 µM | 20.1% @ 2 µM | 26.8% @2 µM | 0% @ 1 µM |
| 19d | CH | N | 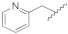 | H | H | 0.52 ± 0.07 | 1.43 ± 0.08 | 1.06 ± 0.04 | 0% @ 1 µM |
| 19e | CH | N |  | H | H | 0.21 ± 0.07 | 0.71 ± 0.04 | 0.84 ± 0.03 | 0% @ 1 µM |
| 19f | CH | N | 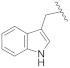 | H | H | 0.13 ± 0.01 | 0.28 ± 0.01 | 0.31 ± 0.01 | 0% @ 1 µM |
| 19g | CH | N | 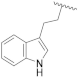 | H | H | 0.31 ± 0.03 | 0.96 ± 0.05 | 0.49 ± 0.06 | 0% @ 1 µM |
| 19h | CH | N |  | F | F | 0.81 ± 0.07 | 0.74 ± 0.03 | 0.57 ± 0.02 | n.d. |
| 19i | CH | N |  | Cl | H | 3.0 ± 0.2 | 2.7 ± 0.2 | 1.9 ± 0.1 | n.d. |
| 19j | N | CH | 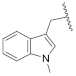 | H | H | 0.45 ± 0.06 | 0.93 ± 0.04 | 1.75 ± 0.06 | 0% @ 1 µM |
| 19k | CH | N | 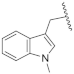 | H | H | 0.14 ± 0.02 | 0.56 ± 0.04 | 0.59 ± 0.03 | 0% @ 1 µM |
| 19l | CH | N |  | F | F | 0.29 ± 0.03 | 0.56 ± 0.02 | 0.81 ± 0.05 | n.d. |
| 19m | CH | N |  | F | H | 0.40 ± 0.06 | 1.48 ± 0.19 | 0.40 ± 0.02 | n.d. |
| 19n | N | CH | CH3 | H | H | 5% @ 1 µM | 7% @ 1 µM | 13% @ 1 µM | n.d. |
| 19o | CH | N | CH3 | H | H | 27% @ 1 µM | 15% @ 1 µM | 30% @ 1 µM | n.d. |
| 21a | CH | N | 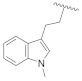 | H | 2-Thienyl | 0.26 ± 0.01 | 2.47 ± 0.22 | 0% @ 1 µM | n.d. |
| 21b | CH | N |  | H | 4-F-C6H4 | 0.70 ± 0.08 | 0.77 ± 0.06 | 0% @ 1 µM | 0% @ 1 µM |
| 21c | CH | N | 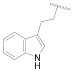 | H | 2-F-C6H4 | 0.76 ± 0.07 | 0.76 ± 0.04 | 15 ± 1 | 0% @ 1 µM |
| 23a | CH | N | H | F | H | 3.30 ± 0.18 | 2.20 ± 0.20 | 0.40 ± 0.01 | 0% @ 1 µM |
| 23b | CH | N | 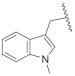 | H | F | 0.27 ± 0.03 | 0.50 ± 0.03 | 0.50 ± 0.02 | 38% @ 1 µM |
| 23c | CH | N | 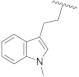 | H | F | 0.33 ± 0.02 | 1.37 ± 0.08 | 0.59 ± 0.04 | n.d. |
| 25a | CH | N | H | Cl | H | 0% @ 1 µM | 0% @ 1 µM | 8.7 ± 0.4 | n.d. |
| 25b | CH | N | H | F | F | 4.3 ± 0.3 | 4.2 ± 0.10 | 1.6 ± 0.1 | n.d. |
| 27a | CH | N | H | H | CF3 | 0% @ 1 µM | 0% @ 1 µM | 0% @ 1 µM | n.d. |
| 27b | CH | N | H | CH3 | H | 0% @ 1 µM | 0% @ 1 µM | 0% @ 1 µM | n.d. |
| 27c | CH | N | H | OCH3 | H | 20.0 ± 1.0 | 14.0 ± 2.0 | 14.0 ± 1.0 | n.d. |
| 29a | CH | N | CH3 | H | 3-Thienyl | 0.11 ± 0.01 | 0.18 ± 0.06 | 4.4 ± 0.1 | n.d. |
| 29b | CH | N | H | H | 2-Thienyl | 0.07 ± 0.01 | 0.26 ± 0.01 | 6.1 ± 0.7 | n.d. |
| 29c | CH | N | H | H | 4-F-C6H4 | 0.16 ± 0.03 | 0.34 ± 0.01 | 6.7 ± 0.5 | n.d. |
| 29d | CH | N | CH3 | H | 2-F-C6H4 | 0.18 ± 0.01 | 0.26 ± 0.07 | 12.0 ± 1.0 | n.d. |
| CI994 | -- | -- | -- | -- | -- | 37% @ 1 µM | 36% @ 1 µM | 32% @ 1 µM | n.d. |
| RGFP-966 | -- | -- | -- | -- | -- | 16 ± 2 | 11 ± 1 | 1.3 ± 0.1 | n.d. |
| MS-275 | -- | -- | -- | -- | -- | 0.93 ± 0.1 | 0.95 ± 0.03 | 1.8 ± 0.1 | n.d. |
| Mocetinostat | -- | -- | -- | -- | -- | 0.33 ± 0.04 | 0.34 ± 0.01 | 0.93 ± 0.05 | n.d. |
| Cpd. No. | HDAC4 (IC50 µM) | HDAC5 (IC50 µM) | HDAC6 (IC50 µM) | HDAC7 (IC50 µM) | HDAC9 (IC50 µM) | HDAC11 (IC50 µM) |
|---|---|---|---|---|---|---|
| 19f | >20 | >20 | >20 | >20 | >20 | >20 |
| 21a | >20 | >20 | >20 | >20 | >20 | 7.5 ± 1 |
| Name | % Viability | SD | Name | % Viability | SD |
|---|---|---|---|---|---|
| 19a | 73.1 | 3.4 | 19m | 52.5 | 0.5 |
| 19b | 78.6 | 6.8 | 21a | 94.4 | 5.5 |
| 19c | 78.7 | 7.5 | 21b | 71.0 | 4.1 |
| 19d | 67.7 | 7.3 | 21c | 72.3 | 5.8 |
| 19e | 50.9 | 4.0 | 23a | 67.0 | 3.4 |
| 19f | 76.1 | 8.0 | 23b | 58.7 | 6.5 |
| 19g | 71.8 | 3.0 | 23c | 55.7 | 1.5 |
| 19h | 54.5 | 2.8 | 25b | 83.7 | 4.4 |
| 19i | 47.5 | 1.5 | 29a | 65.8 | 6.8 |
| 19j | 62.5 | 5.8 | 29b | 2.4 | 0.0 |
| 19k | 71.4 | 3.0 | 29c | 2.2 | 0.1 |
| 19l | 20.1 | 0.4 | 29d | 54.0 | 2.2 |
| MS-275 | 19f | 21a | |
|---|---|---|---|
| Human intestinal absorption% | 92.54 | 93.53 | 96.14 |
| AlogP98 value | 2.07 | 0.78 | 3.11 |
| Plasma Protein Binding% | 91.07 | 34.78 | 77.36 |
| Pure water solubility mg/L | 19.92 | 434.20 | 3.69 |
| CYP_2C19_inhibition | None | None | None |
| CYP_2C9_inhibition | Inhibitor | Inhibitor | Inhibitor |
| CYP_2D6_inhibition | None | None | Non |
| CYP_2D6_substrate | None | Substrate | Substrate |
| CYP_3A4_inhibition | Inhibitor | None | None |
| CYP_3A4_substrate | Substrate | Substrate | Substrate |
| hERG_inhibition | high risk | moderate risk | high risk |
| Pgp inhibition | None | None | None |
| Predicted LD50 | 22 mg/kg | 1500 mg/kg | 600 mg/kg |
| Toxicity prediction ProTox-II | 0/17 | 0/17 | 1/17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, H.S.; Abdelsalam, M.; Zeyn, Y.; Zessin, M.; Mustafa, A.-H.M.; Fischer, M.A.; Zeyen, P.; Sun, P.; Bülbül, E.F.; Vecchio, A.; et al. Synthesis, Molecular Docking and Biological Characterization of Pyrazine Linked 2-Aminobenzamides as New Class I Selective Histone Deacetylase (HDAC) Inhibitors with Anti-Leukemic Activity. Int. J. Mol. Sci. 2022, 23, 369. https://doi.org/10.3390/ijms23010369
Ibrahim HS, Abdelsalam M, Zeyn Y, Zessin M, Mustafa A-HM, Fischer MA, Zeyen P, Sun P, Bülbül EF, Vecchio A, et al. Synthesis, Molecular Docking and Biological Characterization of Pyrazine Linked 2-Aminobenzamides as New Class I Selective Histone Deacetylase (HDAC) Inhibitors with Anti-Leukemic Activity. International Journal of Molecular Sciences. 2022; 23(1):369. https://doi.org/10.3390/ijms23010369
Chicago/Turabian StyleIbrahim, Hany S., Mohamed Abdelsalam, Yanira Zeyn, Matthes Zessin, Al-Hassan M. Mustafa, Marten A. Fischer, Patrik Zeyen, Ping Sun, Emre F. Bülbül, Anita Vecchio, and et al. 2022. "Synthesis, Molecular Docking and Biological Characterization of Pyrazine Linked 2-Aminobenzamides as New Class I Selective Histone Deacetylase (HDAC) Inhibitors with Anti-Leukemic Activity" International Journal of Molecular Sciences 23, no. 1: 369. https://doi.org/10.3390/ijms23010369
APA StyleIbrahim, H. S., Abdelsalam, M., Zeyn, Y., Zessin, M., Mustafa, A.-H. M., Fischer, M. A., Zeyen, P., Sun, P., Bülbül, E. F., Vecchio, A., Erdmann, F., Schmidt, M., Robaa, D., Barinka, C., Romier, C., Schutkowski, M., Krämer, O. H., & Sippl, W. (2022). Synthesis, Molecular Docking and Biological Characterization of Pyrazine Linked 2-Aminobenzamides as New Class I Selective Histone Deacetylase (HDAC) Inhibitors with Anti-Leukemic Activity. International Journal of Molecular Sciences, 23(1), 369. https://doi.org/10.3390/ijms23010369








