Effect of Low-Dose Ionizing Radiation on the Expression of Mitochondria-Related Genes in Human Mesenchymal Stem Cells
Abstract
:1. Introduction
2. Results
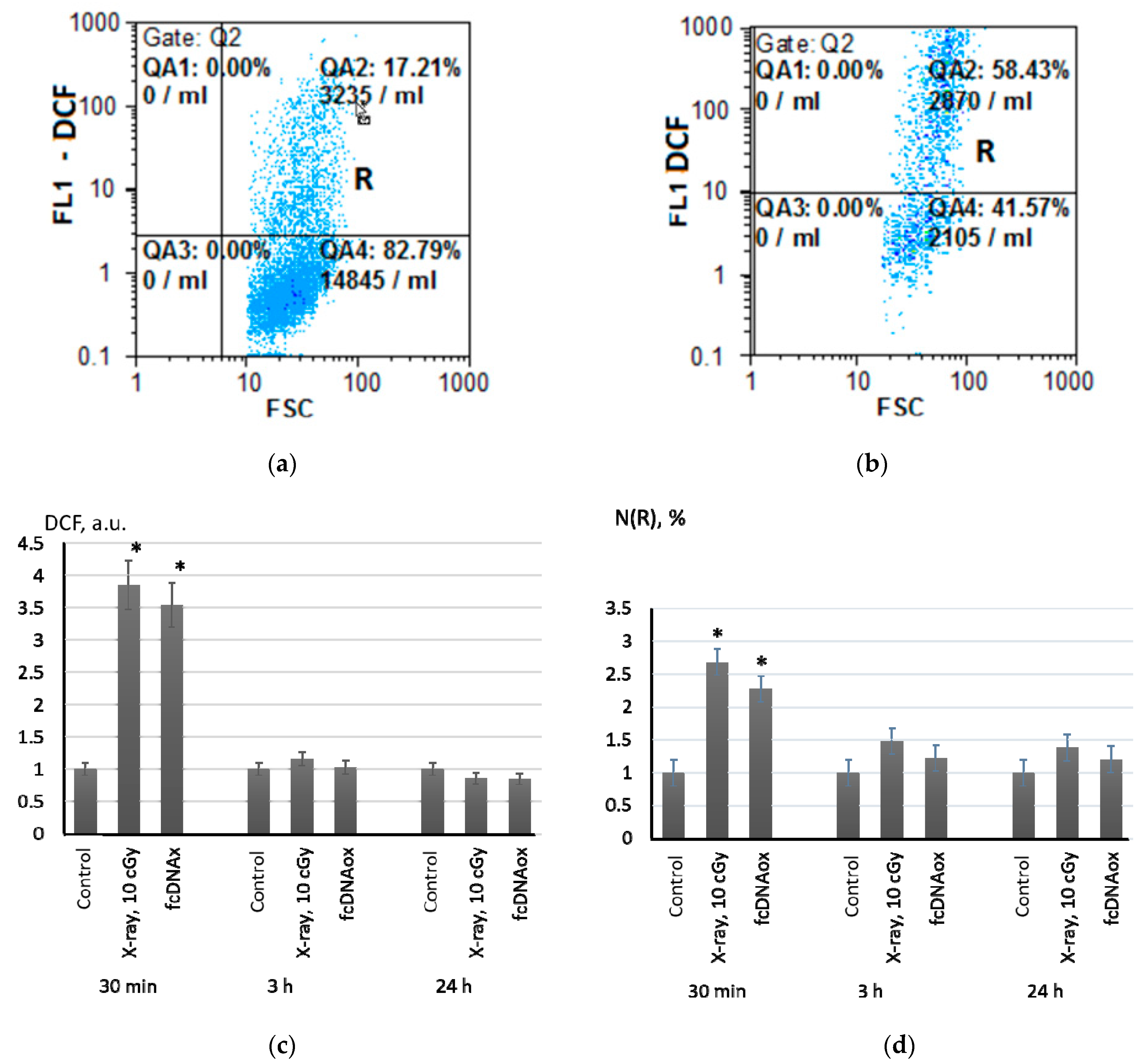
2.1. Low-Dose Ionizing Radiation Induce a Short-Term Increase in Intracellular ROS in MSCs
2.2. Effects of Low-Dose Radiation and Oxidized cfDNA on Mitochondrial Potential
2.3. Effects of Low-Dose Radiation and Oxidized fcDNA on The Transcriptional Activity of Genes Regulating Mitochondrial Functions
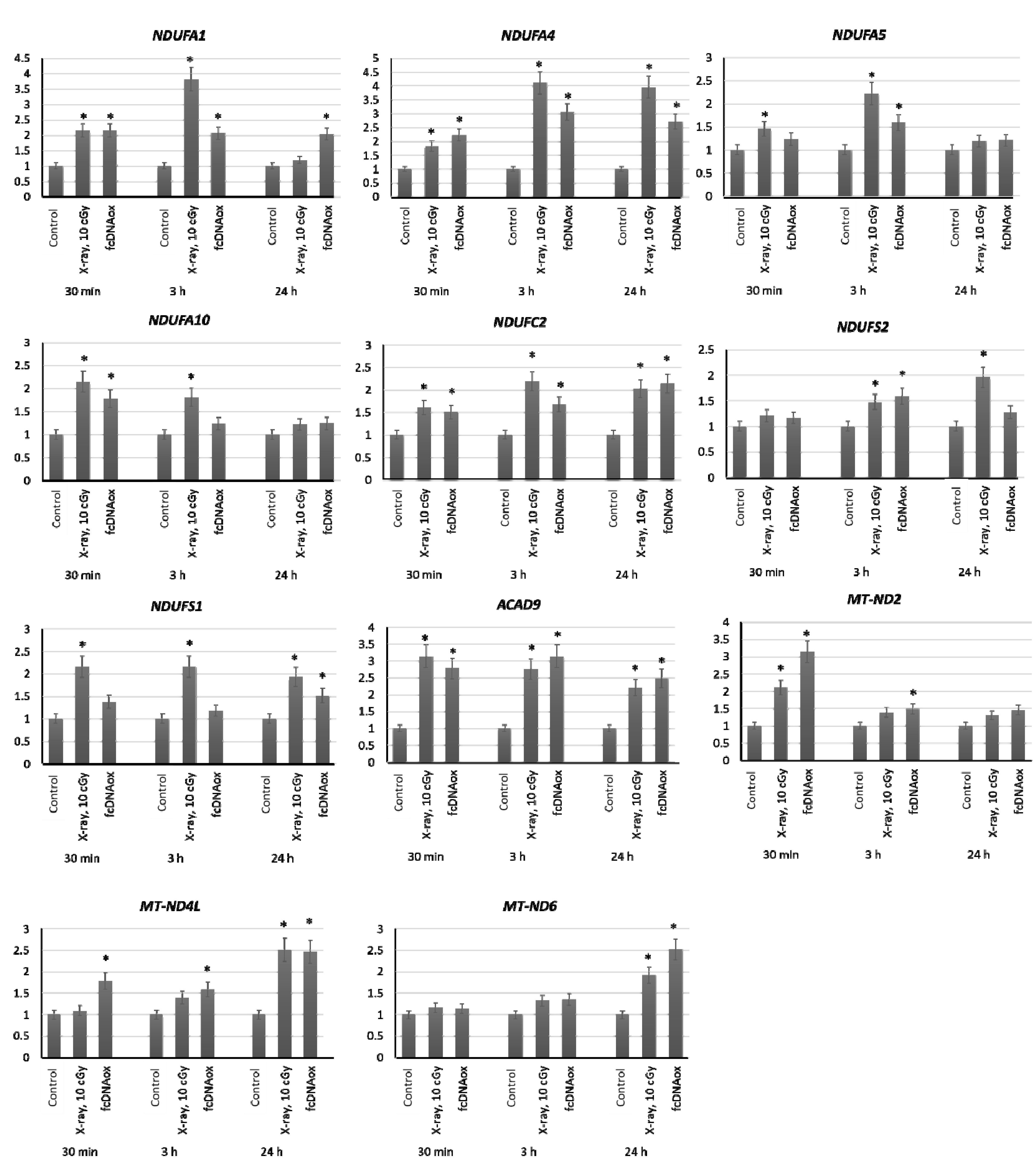
2.3.1. Mitochondrial Respiratory Complex I
2.3.2. Mitochondrial Respiratory Complex II
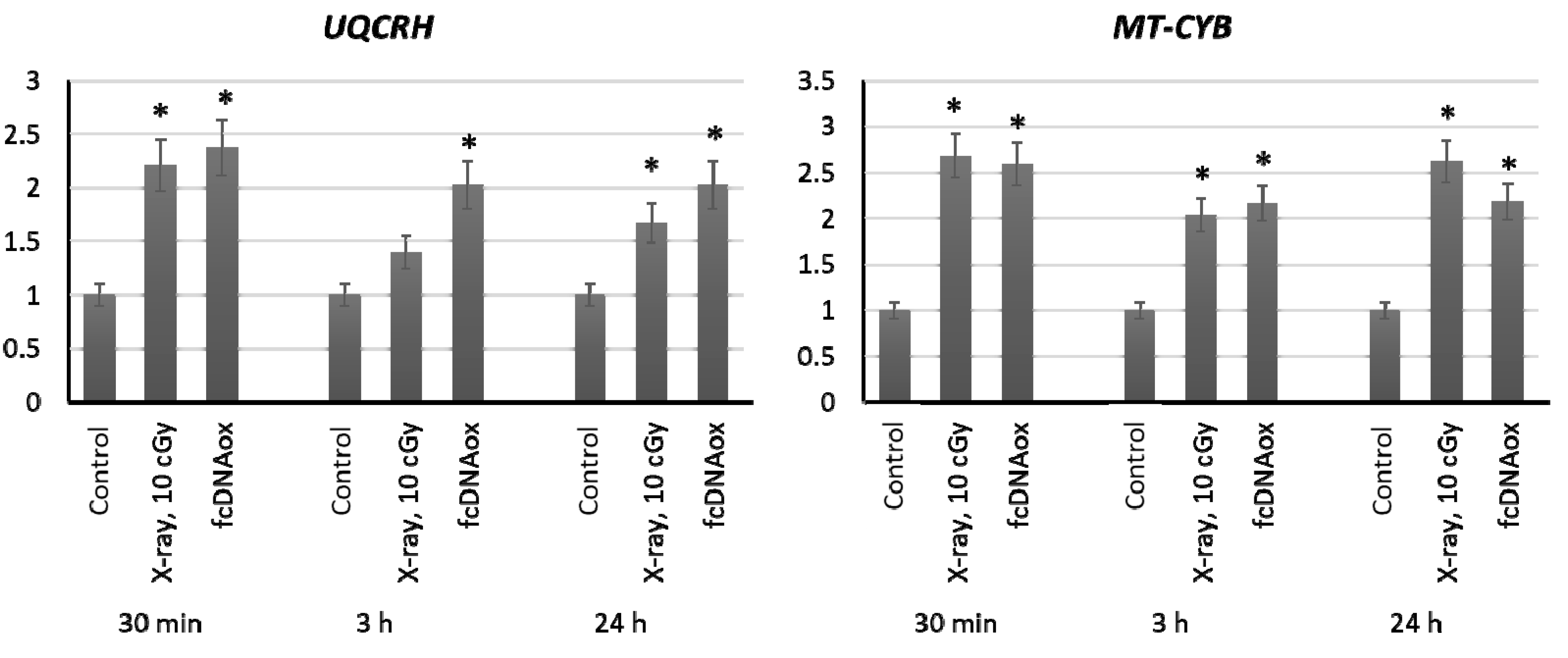
2.3.3. Mitochondrial Respiratory Complex III
2.3.4. Mitochondrial Respiratory Complex IV
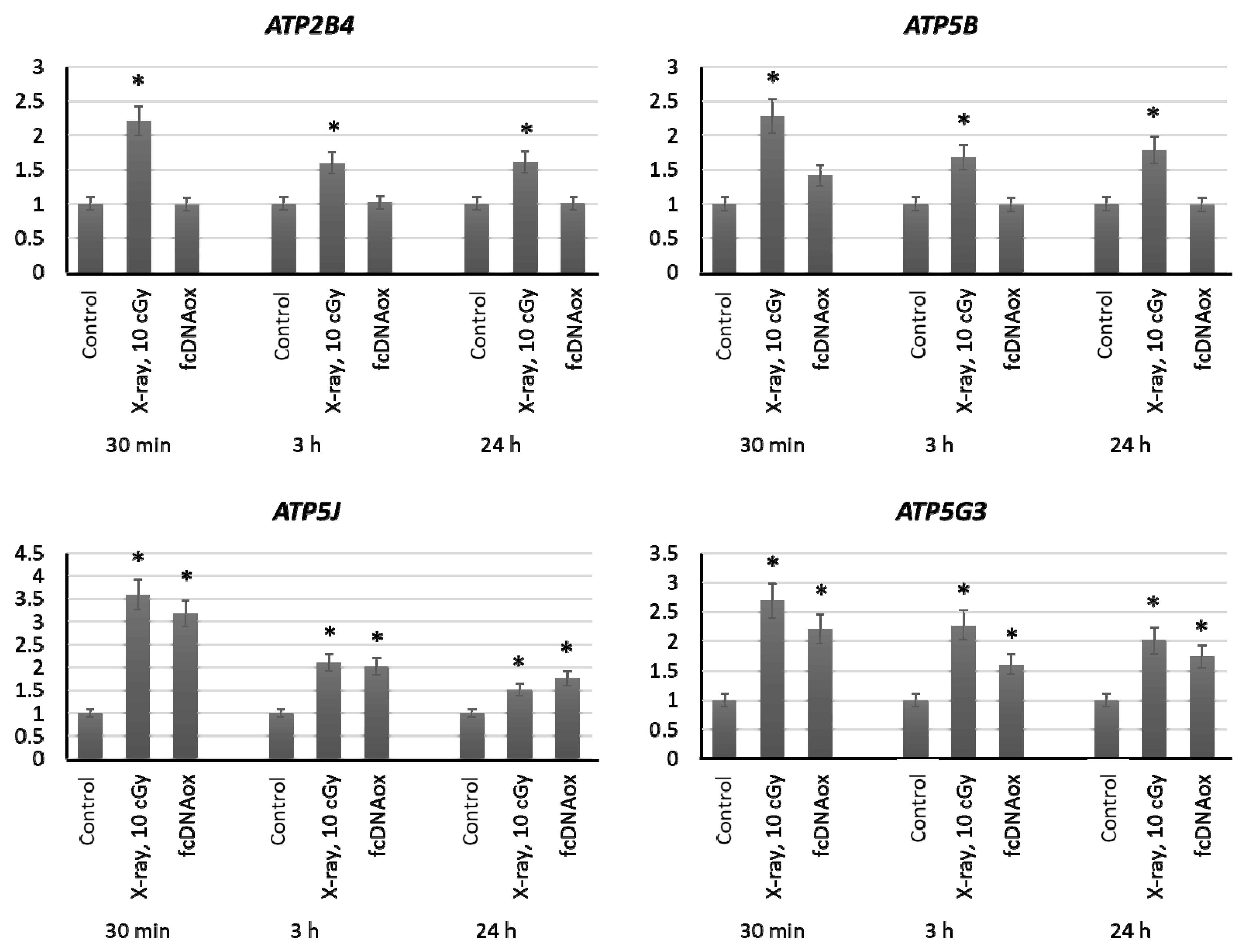
2.3.5. Mitochondrial Respiratory Complex V
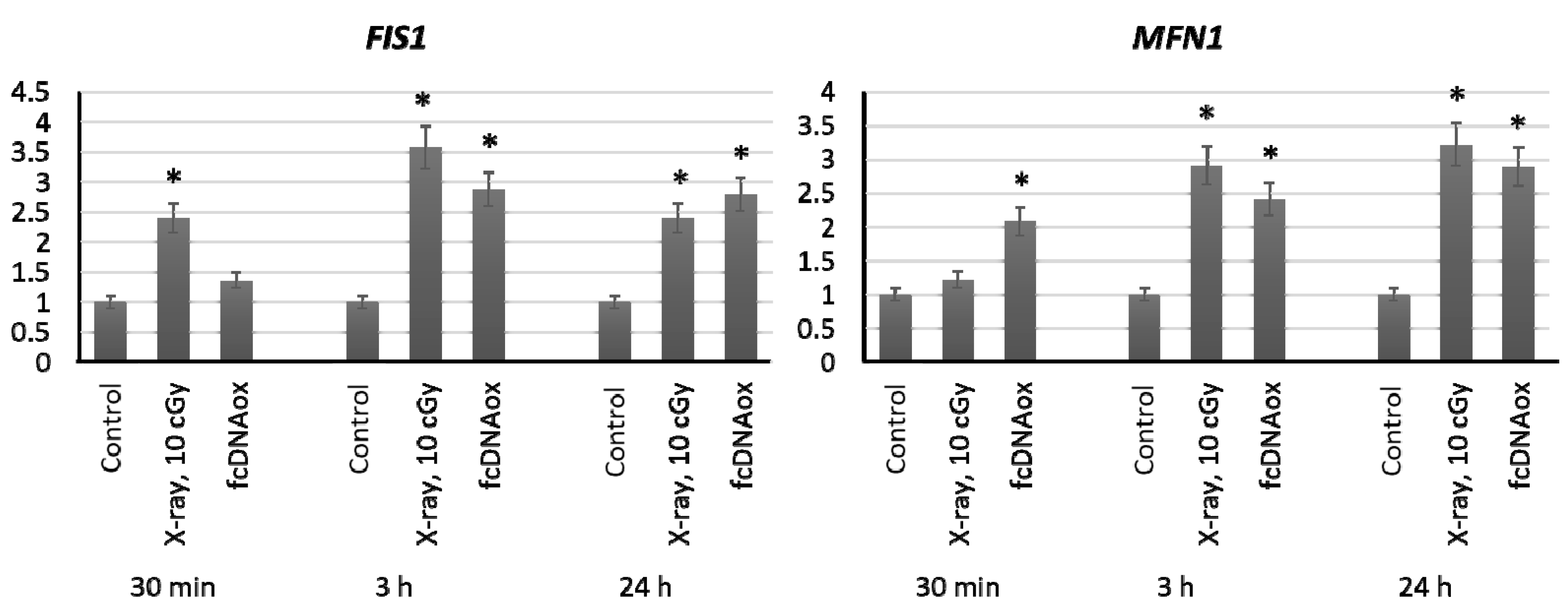
2.4. Genes of Mitochondrial Fusion and Fission
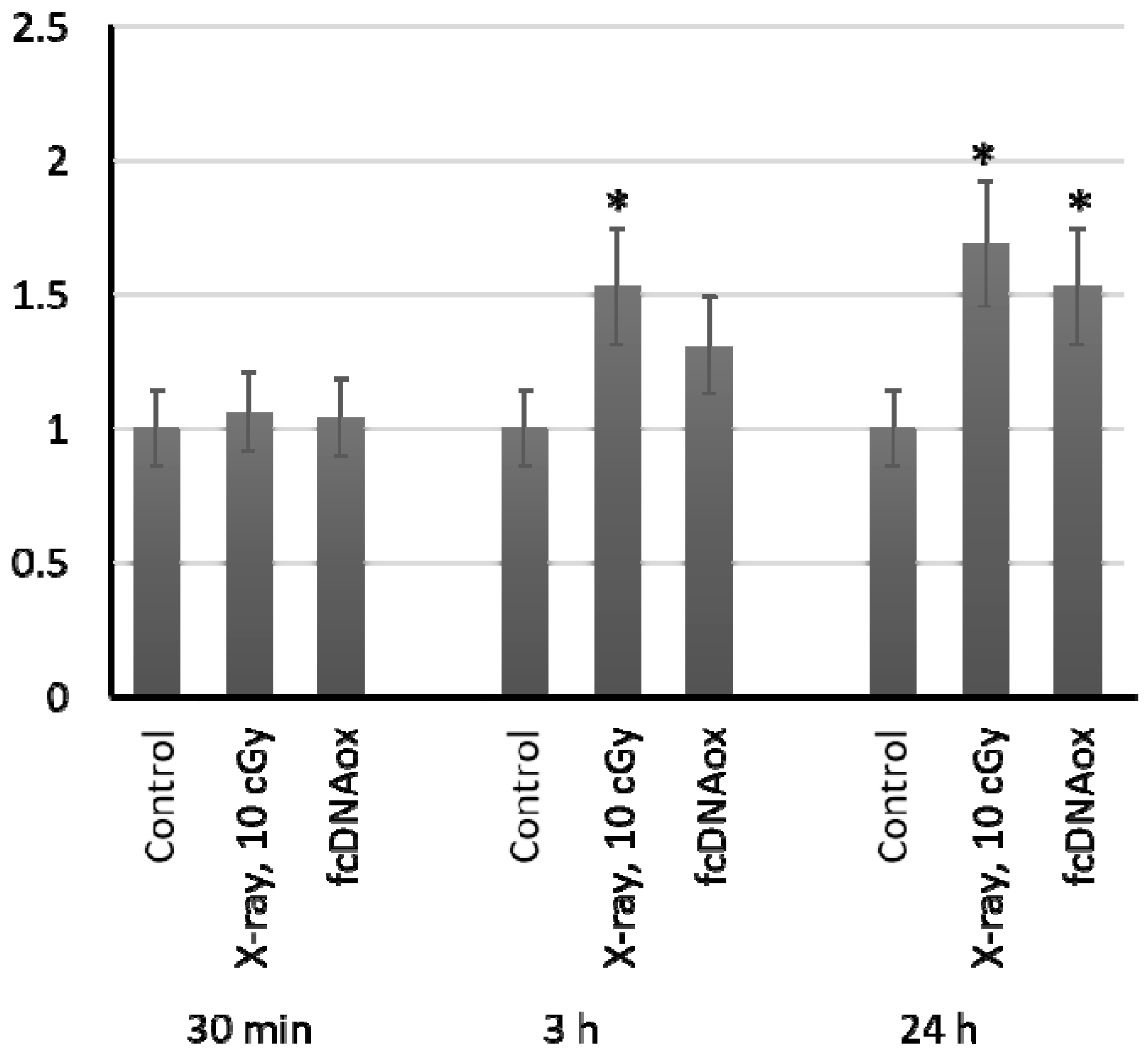
2.5. The Number of Copies of Mitochondrial DNA
2.6. Effect of Non-Oxidized DNA Fragments
3. Discussion
4. Materials and Methods
4.1. Mesenchymal Stem Cells Preparation and Characterizaton
4.2. Irradiation of Cells
4.3. Preparation of Standardized Oxidized Cell-Free DNA
4.4. Assessment of DNA Breaks with Comet Assay
4.5. Flow Cytometry
4.6. ROS Visualization with Fluorescence Microscopy
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brenner, D.J.; Shuryak, I.; Einstein, A.J. Impact of reduced patient life expectancy on potential cancer risks from radiologic imaging. Radiology 2011, 261, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabritius, G.; Brix, G.; Nekolla, E.; Klein, S.; Popp, H.D.; Meyer, M.; Glatting, G.; Hagelstein, C.; Hofmann, W.K.; Schoenberg, S.O.; et al. Cumulative radiation exposure from imaging procedures and associated lifetime cancer risk for patients with lymphoma. Sci. Rep. 2016, 6, 35181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, F.R.; Loke, W.K. Molecular mechanisms of low dose ionizing radiation-induced hormesis, adaptive responses, radioresistance, bystander effects, and genomic instability. Int. J. Radiat. Biol. 2015, 91, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, M.; Neumann, R. Changes in gene expression as one of the key mechanisms involved in radiation-induced bystander effect (Review). Biomed. Rep. 2018, 9, 99–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squillaro, T.; Galano, G.; De Rosa, R.; Peluso, G.; Galderisi, U. Concise Review: The Effect of Low-Dose Ionizing Radiation on Stem Cell Biology: A Contribution to Radiation Risk. Stem. Cells 2018, 36, 1146–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rando, T.A. Stem cells, ageing and the quest for immortality. Nature 2006, 441, 1080–1086. [Google Scholar] [CrossRef]
- Alessio, N.; Del Gaudio, S.; Capasso, S.; Di Bernardo, G.; Cappabianca, S.; Cipollaro, M.; Peluso, G.; Galderisi, U. Low dose radiation induced senescence of human mesenchymal stromal cells and impaired the autophagy process. Oncotarget 2015, 6, 8155–8166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prise, K.M.; Saran, A. Concise review: Stem cell effects in radiation risk. Stem. Cells 2011, 29, 1315–1321. [Google Scholar] [CrossRef]
- Bevelacqua, J.J.; Mortazavi, S.M.J. Regarding “Concise Review: The Effect of Low-Dose Ionizing Radiation on Stem Cell Biology: A Contribution to Radiation Risk”. Stem. Cells 2018, 36, 1789. [Google Scholar] [CrossRef] [Green Version]
- Nugent, S.M.; Mothersill, C.E.; Seymour, C.; McClean, B.; Lyng, F.M.; Murphy, J.E. Increased mitochondrial mass in cells with functionally compromised mitochondria after exposure to both direct gamma radiation and bystander factors. Radiat. Res. 2007, 168, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Bar-Ziv, R.; Bolas, T.; Dillin, A. Systemic effects of mitochondrial stress. EMBO Rep. 2020, 21, e50094. [Google Scholar] [CrossRef]
- Ristow, M.; Schmeisser, K. Mitohormesis: Promoting Health and Lifespan by Increased Levels of Reactive Oxygen Species (ROS). Dose Response 2014, 12, 288–341. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Baldwin, L.A. Defining hormesis. Hum. Exp. Toxicol. 2002, 21, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.C.; Guan, X.; Ryan, J.A.; Rivera, A.G.; Mock, C.; Agrawal, V.; Letai, A.; Lerou, P.H.; Lahav, G. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem. Cell 2013, 13, 483–491. [Google Scholar] [CrossRef] [Green Version]
- Lan, M.L.; Acharya, M.M.; Tran, K.K.; Bahari-Kashani, J.; Patel, N.H.; Strnadel, J.; Giedzinski, E.; Limoli, C.L. Characterizing the radioresponse of pluripotent and multipotent human stem cells. PLoS ONE 2012, 7, e50048. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, J.; Grantham, V. Radiation Hormesis: Historical and Current Perspectives. J. Nucl. Med. Technol. 2015, 43, 242–246. [Google Scholar] [CrossRef] [Green Version]
- Cardarelli, J.J., 2nd; Ulsh, B.A. It Is Time to Move Beyond the Linear No-Threshold Theory for Low-Dose Radiation Protection. Dose Response 2018, 16, 1559325818779651. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M. Hormesis: Insight into Adaptive Defense Mechanisms against Ionizing Radiation Established during Evolution of Life on the Earth. In Fukushima Nuclear Accident; Sutou, S., Ed.; Nova Publishers: New York, NY, USA, 2015; pp. 89–99. [Google Scholar]
- Kabilan, U.; Graber, T.E.; Alain, T.; Klokov, D. Ionizing Radiation and Translation Control: A Link to Radiation Hormesis? Int. J. Mol. Sci. 2020, 21, 6650. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, V.A.; Ershova, E.S.; Veiko, N.N.; Malinovskaya, E.M.; Kalyanov, A.A.; Kameneva, L.V.; Stukalov, S.V.; Dolgikh, O.A.; Konkova, M.S.; Ermakov, A.V.; et al. Low-Dose Ionizing Radiation Affects Mesenchymal Stem Cells via Extracellular Oxidized Cell-Free DNA: A Possible Mediator of Bystander Effect and Adaptive Response. Oxid. Med. Cell Longev. 2017, 2017, 9515809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergeeva, V.A.; Kostyuk, S.V.; Ershova, E.S.; Malinovskaya, E.M.; Smirnova, T.D.; Kameneva, L.V.; Veiko, N.N. GC-Rich DNA Fragments and Oxidized Cell-Free DNA Have Different Effects on NF-kB and NRF2 Signaling in MSC. Adv. Exp. Med. Biol. 2016, 924, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Konkova, M.; Abramova, M.; Kalianov, A.; Ershova, E.; Dolgikh, O.; Umriukhin, P.; Izhevskaya, V.; Kutsev, S.; Veiko, N.; Kostyuk, S. Mesenchymal Stem Cells Early Response to Low-Dose Ionizing Radiation. Front. Cell Dev. Biol. 2020, 8, 584497. [Google Scholar] [CrossRef] [PubMed]
- Scadulto, R.C.; Grotyohann, L.W. Measurement of Mitochondrial Membrane Potential Using Fluorescent Rhodamine Derivatives. Biophys. J. 1999, 76, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Iommarini, L.; Kurelac, I.; Capristo, M.; Calvaruso, M.A.; Giorgio, V.; Bergamini, C.; Ghelli, A.; Nanni, P.; De Giovanni, C.; Carelli, V.; et al. Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum. Mol. Genet. 2014, 23, 1453–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ristow, M.; Zarse, K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp. Gerontol. 2010, 45, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Vikash, V.; Ye, Q.; Wu, D.; Liu, Y.; Dong, W. ROS and ROS-Mediated Cellular Signaling. Oxid. Med. Cell Longev. 2016, 2016, 4350965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Schulz, T.J.; Zarse, K.; Voigt, A.; Urban, N.; Birringer, M.; Ristow, M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007, 6, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Riemer, J. The mitochondrial disulfide relay system: Roles in oxidative protein folding and beyond. Int. J. Cell Biol. 2013, 2013, 742923. [Google Scholar] [CrossRef] [Green Version]
- Sies, H.; Feinendegen, L.E. Radiation Hormesis: The Link to Nanomolar Hydrogen Peroxide. Antioxid. Redox Signal. 2017, 27, 596–598. [Google Scholar] [CrossRef]
- Loseva, P.; Kostyuk, S.; Malinovskaya, E.; Clement, N.; Dechesne, C.A.; Dani, C.; Smirnova, T.; Glebova, K.; Baidakova, G.; Baranova, A.; et al. Extracellular DNA oxidation stimulates activation of NRF2 and reduces the production of ROS in human mesenchymal stem cells. Expert Opin. Biol. Ther. 2012, 12 (Suppl. 1), S85–S97. [Google Scholar] [CrossRef]
- Chestkov, I.V.; Jestkova, E.M.; Ershova, E.S.; Golimbet, V.G.; Lezheiko, T.V.; Kolesina, N.Y.; Dolgikh, O.A.; Izhevskaya, V.L.; Kostyuk, G.P.; Kutsev, S.I.; et al. ROS-Induced DNA Damage Associates with Abundance of Mitochondrial DNA in White Blood Cells of the Untreated Schizophrenic Patients. Oxid. Med. Cell Longev. 2018, 2018, 8587475. [Google Scholar] [CrossRef] [Green Version]
- Kostyuk, S.V.; Ermakov, A.V.; Alekseeva, A.Y.; Smirnova, T.D.; Glebova, K.V.; Efremova, L.V.; Baranova, A.; Veiko, N.N. Role of extracellular DNA oxidative modification in radiation induced bystander effects in human endotheliocytes. Mutat. Res. 2012, 729, 52–60. [Google Scholar] [CrossRef]
- Ermakov, A.V.; Konkova, M.S.; Kostyuk, S.V.; Izevskaya, V.L.; Baranova, A.; Veiko, N.N. Oxidized extracellular DNA as a stress signal in human cells. Oxid. Med. Cell Longev. 2013, 2013, 649747. [Google Scholar] [CrossRef]
- He, J.; Ford, H.C.; Carroll, J.; Douglas, C.; Gonzales, E.; Ding, S.; Fearnley, I.M.; Walker, J.E. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. USA 2018, 115, 2988–2993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Iborra, S.; Cuadros, T.; Parent, A.; Romero-Gimenez, J.; Vila, M.; Perier, C. Defective mitochondrial protein import contributes to complex I-induced mitochondrial dysfunction and neurodegeneration in Parkinson’s disease. Cell Death Dis. 2018, 9, 1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valera-Alberni, M.; Canto, C. Mitochondrial stress management: A dynamic journey. Cell Stress 2018, 2, 253–274. [Google Scholar] [CrossRef] [Green Version]
- Knowlton, A.A.; Liu, T.T. Mitochondrial Dynamics and Heart Failure. Compr. Physiol. 2015, 6, 507–526. [Google Scholar] [CrossRef] [Green Version]
- Bose, A.; Beal, M.F. Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 2016, 139 (Suppl. S1), 216–231. [Google Scholar] [CrossRef]
- Seo, A.Y.; Joseph, A.M.; Dutta, D.; Hwang, J.C.; Aris, J.P.; Leeuwenburgh, C. New insights into the role of mitochondria in aging: Mitochondrial dynamics and more. J. Cell Sci. 2010, 123, 2533–2542. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, K.D.; Guzy, R.D.; Pan, Y.; Young, R.M.; Cash, T.P.; Schumacker, P.T.; Simon, M.C. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005, 1, 393–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeliovich, H.; Zarei, M.; Rigbolt, K.T.; Youle, R.J.; Dengjel, J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat. Commun. 2013, 4, 2789. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R.; Oscoz, A.A.; Brunborg, G.; Gaivao, I.; Giovannelli, L.; Kruszewski, M.; Smith, C.C.; Stetina, R. The comet assay: Topical issues. Mutagenesis 2008, 23, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]


| Gene | 30 min | 3 h | 24 h | Gene | 30 min | 3 h | 24 h |
|---|---|---|---|---|---|---|---|
| NDUFA1 | 1.0 ± 0.3 | 1.1 ± 0.3 | 1.3 ± 0.3 | UQCRH | 1.0 ± 0.3 | 1.2 ± 0.3 | 1.3 ± 0.3 |
| NDUFA4 | 1.0 ± 0.2 | 1.1± 0.2 | 1.3 ± 0.2 | MT-CYB | 0.9 ± 0.2 | 1.1 ± 0.2 | 1.0 ± 0.2 |
| NDUFA5 | 0.9 ± 0.3 | 1.0 ± 0.3 | 1.2 ± 0.3 | UQCRH | 1.1 ± 0.3 | 1.0 ± 0.3 | 1.3 ± 0.3 |
| NDUFA10 | 1.0 ± 0.3 | 0.9 ± 0.4 | 1.3 ± 0.4 | MT-CYB | 1.2 ± 0.3 | 1.2 ± 0.3 | 1.3 ± 0.3 |
| NDUFC2 | 1.2 ± 0.2 | 1.0 ± 0.2 | 1.3 ± 0.3 | SURF1 | 0.9 ± 0.2 | 1.0 ± 0.2 | 1.3 ± 0.3 |
| NDUFS2 | 1.1 ± 0.3 | 1.1 ± 0.3 | 1.1 ± 0.3 | SCO1 | 1.0 ± 0.3 | 0.9 ± 0.2 | 1.2 ± 0.3 |
| NDUFS1 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.3 ± 0.3 | COX15 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.3 ± 0.3 |
| ACAD9 | 1.1 ± 0.3 | 0.9± 0.3 | 1.2 ± 0.3 | COX5A | 0.9 ± 0.2 | 1.1 ± 0.2 | 1.3 ± 0.2 |
| MT-ND2 | 0.9 ± 0.2 | 1.2 ± 0.3 | 1.3 ± 0.3 | ATP2B4 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.2 ± 0.2 |
| MT-ND4L | 1.0 ± 0.3 | 1.1 ± 0.3 | 1.1 ± 0.3 | ATP5B | 1.1 ± 0.2 | 1.1 ± 0.3 | 1.3 ± 0.3 |
| MT-ND6 | 1.1 ± 0.3 | 1.0 ± 0.2 | 1.2 ± 0.3 | ATP5J | 0.9± 0.2 | 1.1 ± 0.2 | 1.3 ± 0.2 |
| - | - | - | - | ATP5G3 | 1.0 ± 0.3 | 1.0 ± 0.3 | 1.1 ± 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostyuk, S.V.; Proskurnina, E.V.; Konkova, M.S.; Abramova, M.S.; Kalianov, A.A.; Ershova, E.S.; Izhevskaya, V.L.; Kutsev, S.I.; Veiko, N.N. Effect of Low-Dose Ionizing Radiation on the Expression of Mitochondria-Related Genes in Human Mesenchymal Stem Cells. Int. J. Mol. Sci. 2022, 23, 261. https://doi.org/10.3390/ijms23010261
Kostyuk SV, Proskurnina EV, Konkova MS, Abramova MS, Kalianov AA, Ershova ES, Izhevskaya VL, Kutsev SI, Veiko NN. Effect of Low-Dose Ionizing Radiation on the Expression of Mitochondria-Related Genes in Human Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2022; 23(1):261. https://doi.org/10.3390/ijms23010261
Chicago/Turabian StyleKostyuk, Svetlana V., Elena V. Proskurnina, Marina S. Konkova, Margarita S. Abramova, Andrey A. Kalianov, Elizaveta S. Ershova, Vera L. Izhevskaya, Sergey I. Kutsev, and Natalia N. Veiko. 2022. "Effect of Low-Dose Ionizing Radiation on the Expression of Mitochondria-Related Genes in Human Mesenchymal Stem Cells" International Journal of Molecular Sciences 23, no. 1: 261. https://doi.org/10.3390/ijms23010261
APA StyleKostyuk, S. V., Proskurnina, E. V., Konkova, M. S., Abramova, M. S., Kalianov, A. A., Ershova, E. S., Izhevskaya, V. L., Kutsev, S. I., & Veiko, N. N. (2022). Effect of Low-Dose Ionizing Radiation on the Expression of Mitochondria-Related Genes in Human Mesenchymal Stem Cells. International Journal of Molecular Sciences, 23(1), 261. https://doi.org/10.3390/ijms23010261






