Biological Implications of a Stroke Therapy Based in Neuroglobin Hyaluronate Nanoparticles. Neuroprotective Role and Molecular Bases
Abstract
:1. Introduction
2. Results
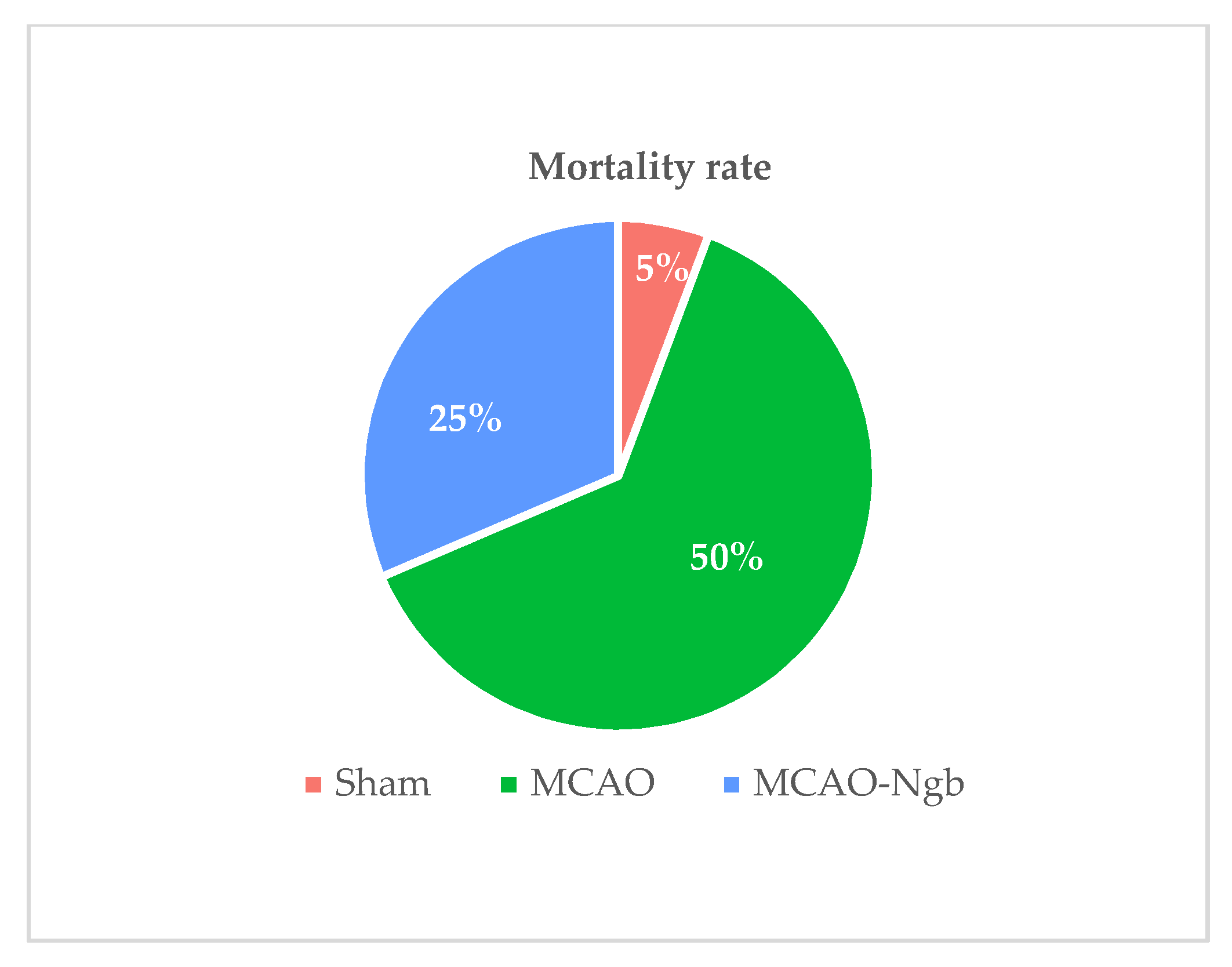
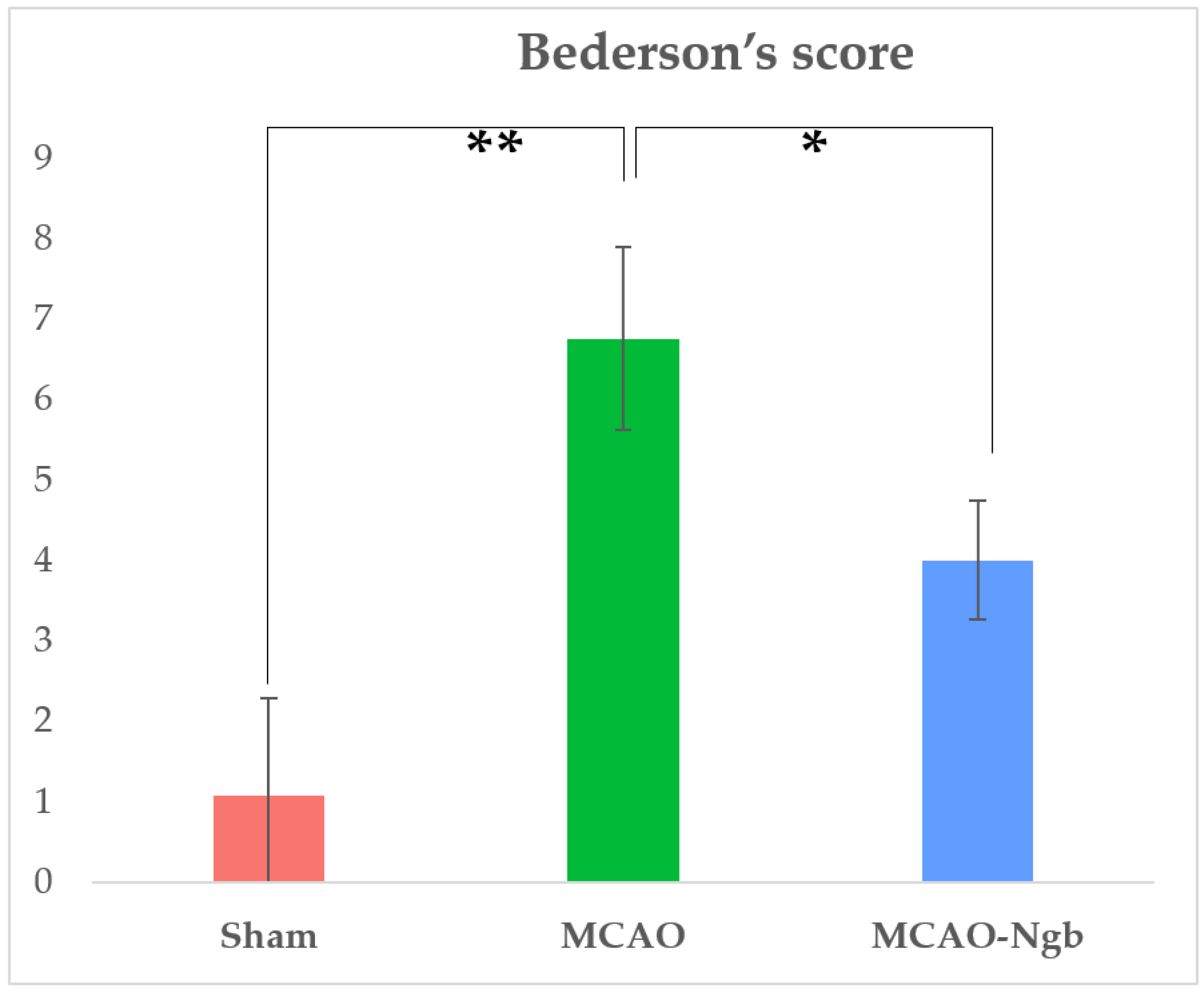
2.1. Ngb Improves the Survival and the Neurological Outcomes of the MCAO-Ngb Animals
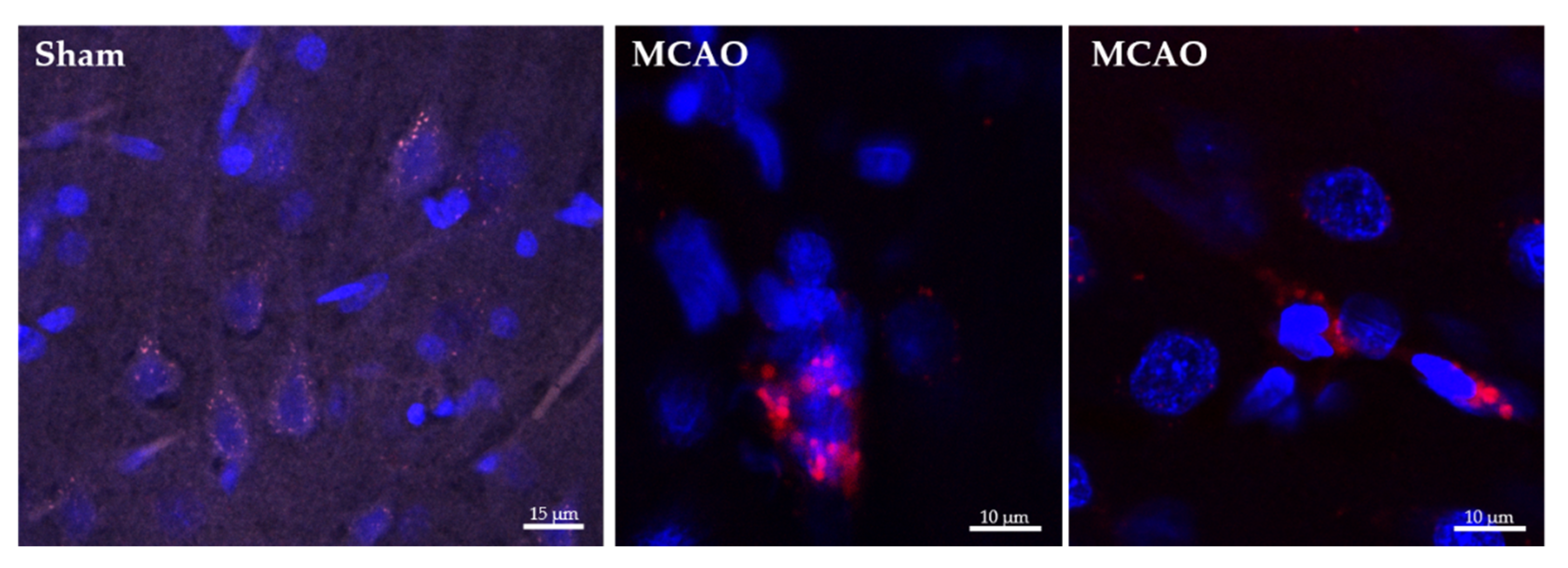
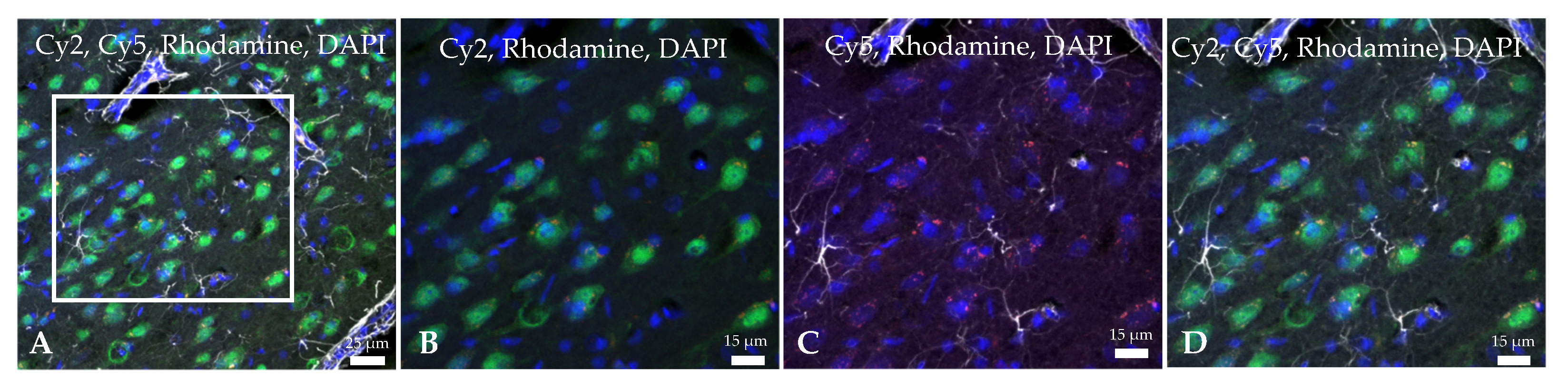
2.2. Ngb-NPs Cross the BBB, and are Endocytosed by Neurons
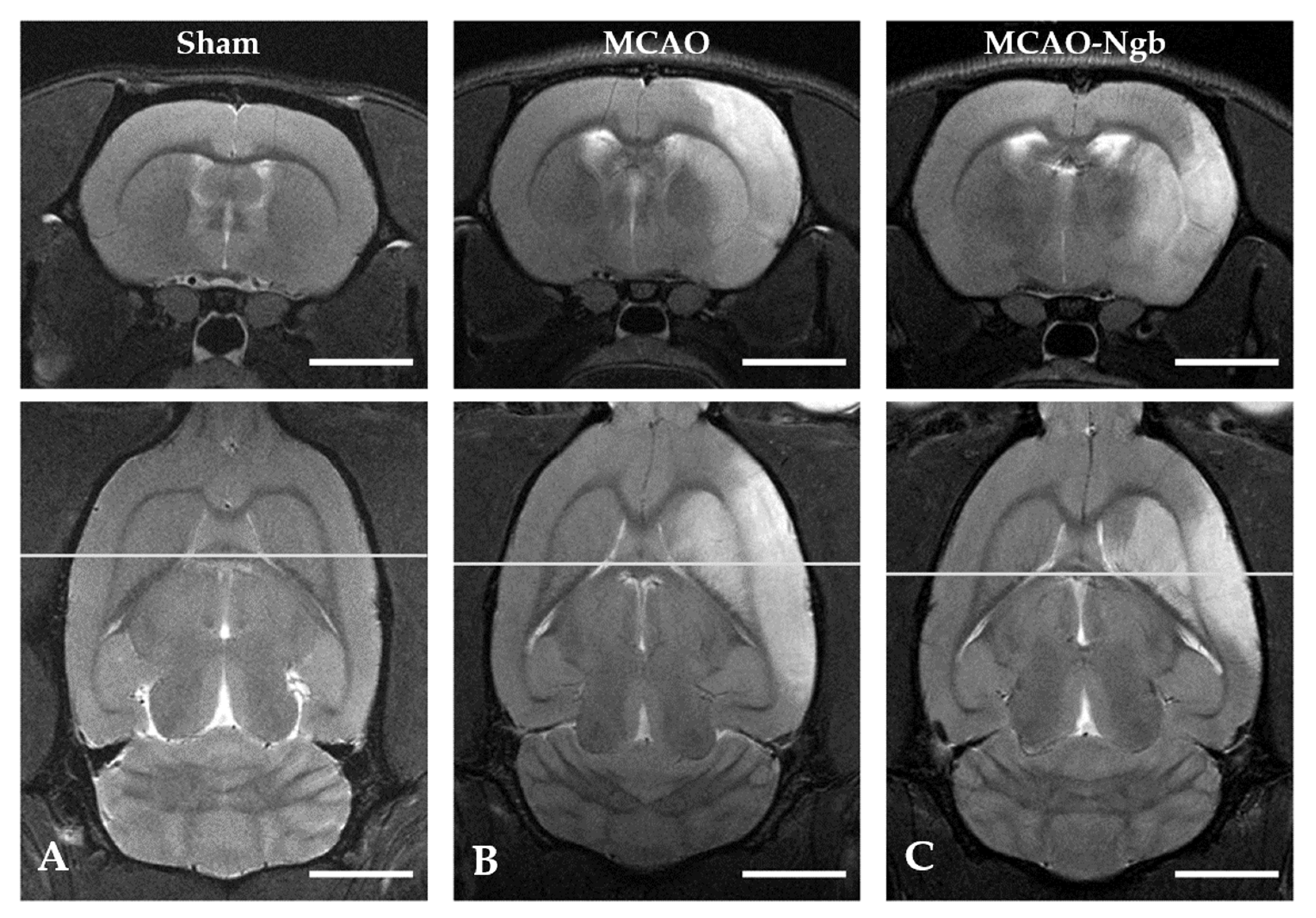
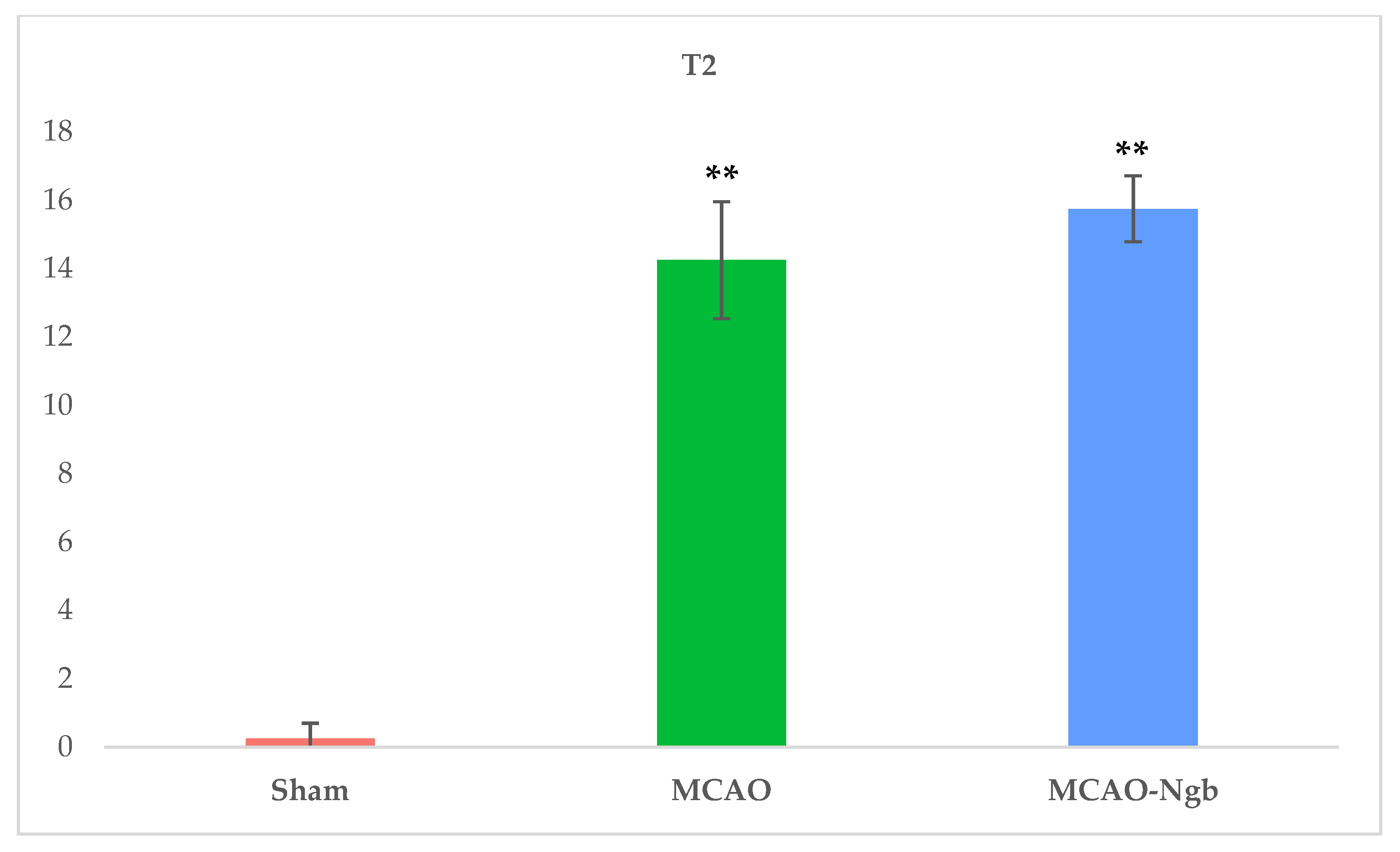
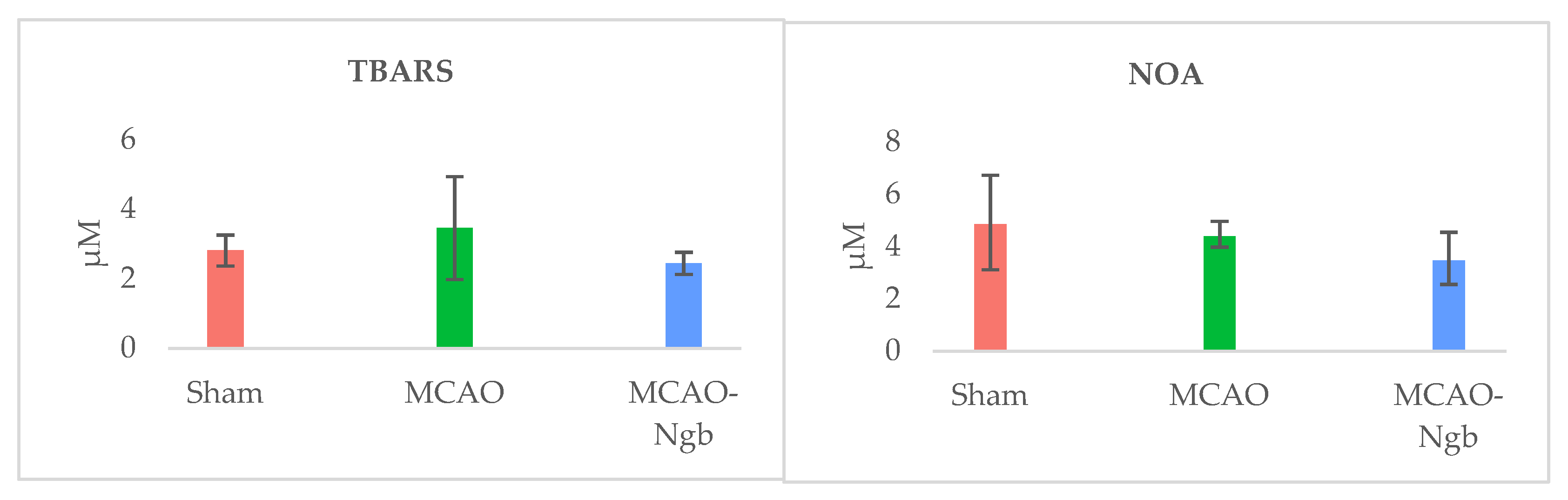
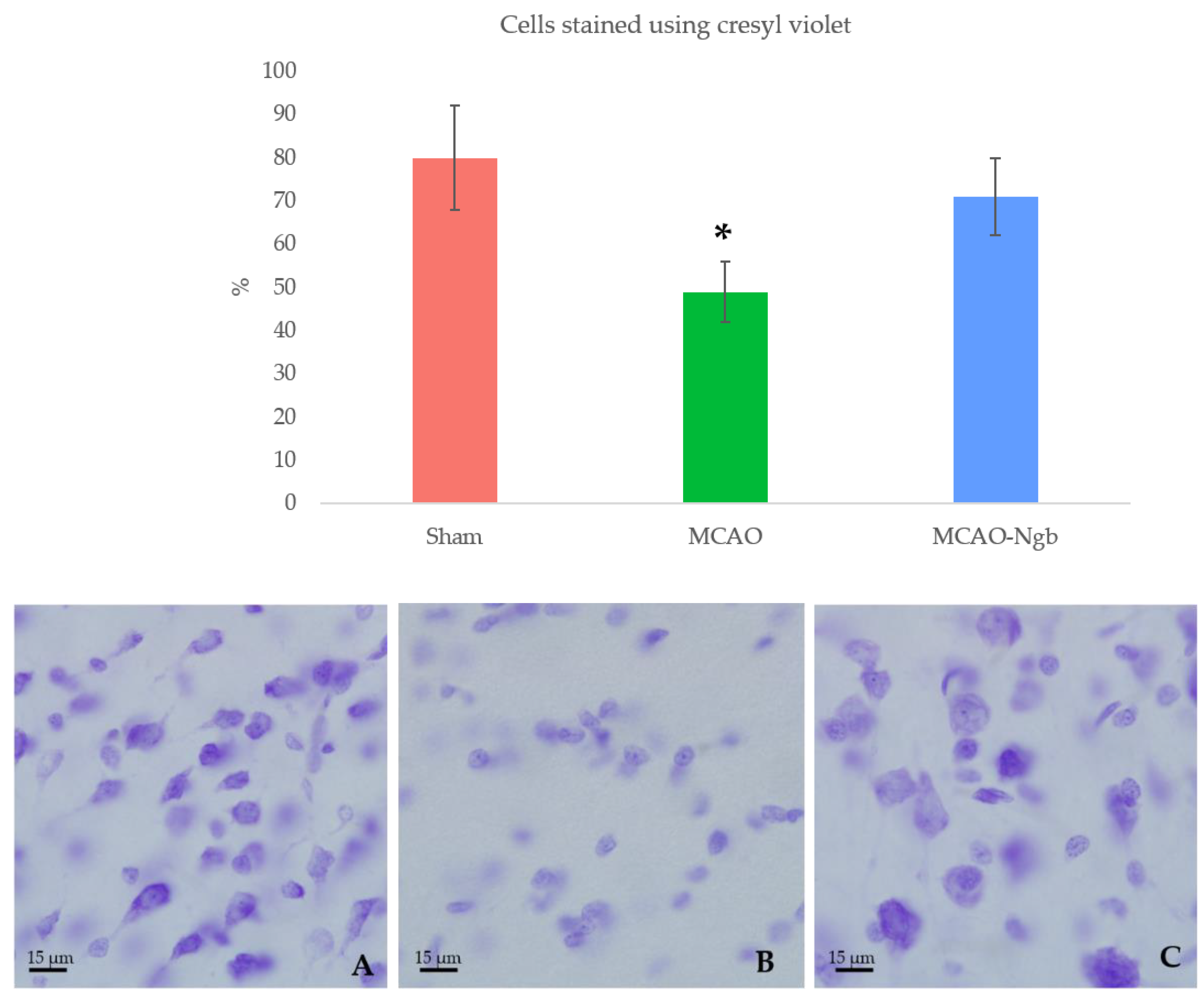
2.3. Ngb-NPs Treatment does not Affect the Infarct Size or the Oxidative/Nitrosative Stresses, but Improves the Histological Outcomes
2.4. Proteomic Findings Reveal Potential Biological Processes Involved in the Protective Action of Ngb
2.4.1. Protein Identification and Quantification
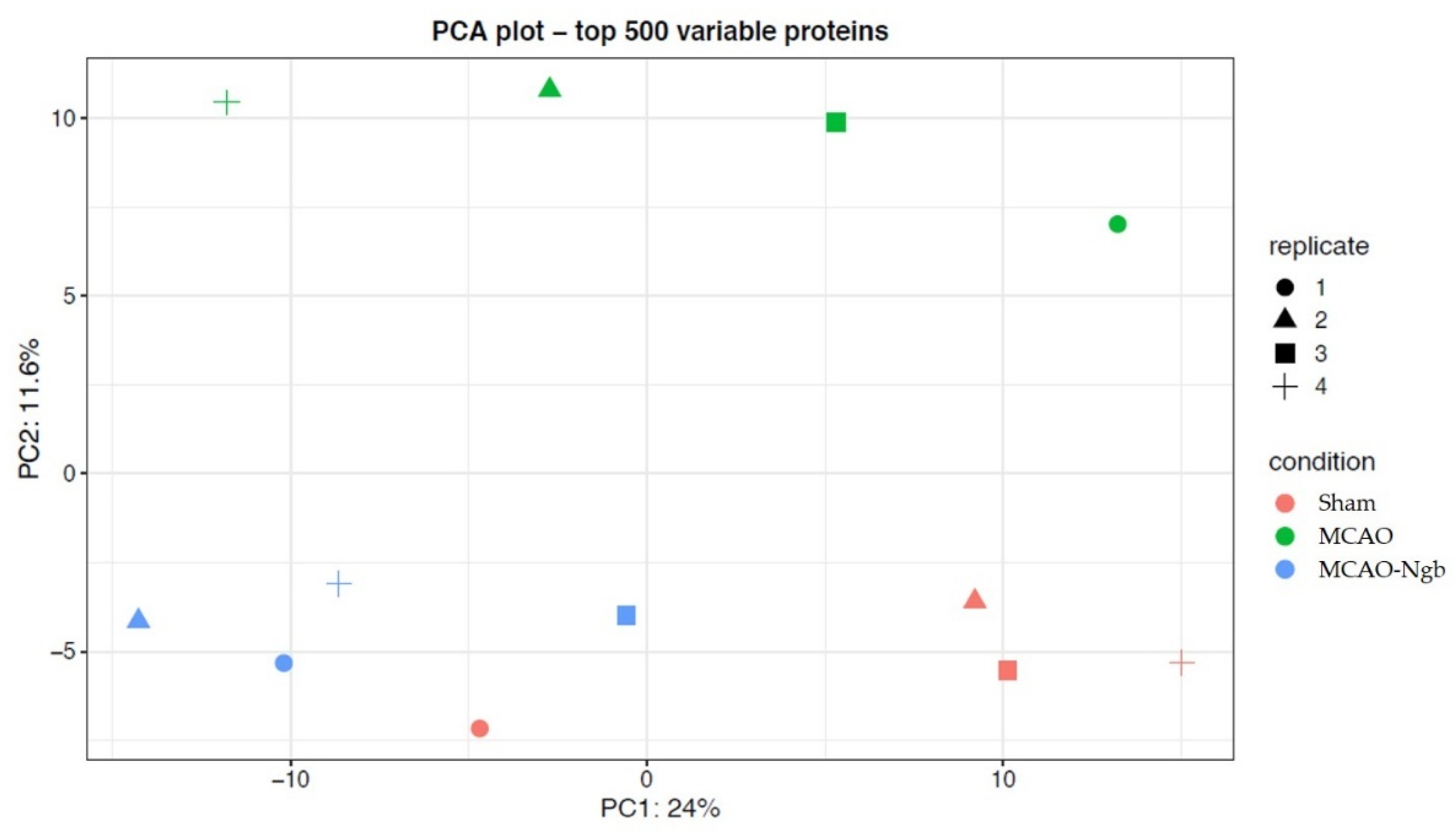
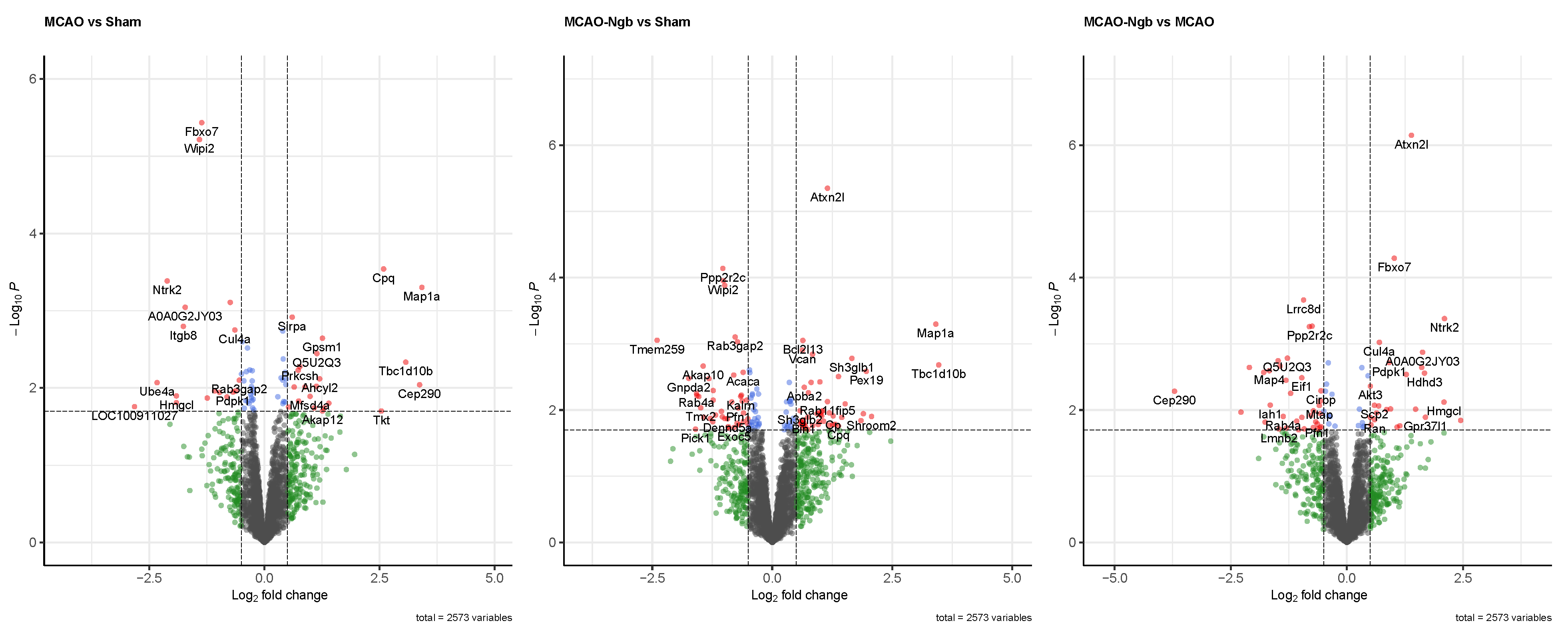
2.4.2. Quality Control and Differential Analysis
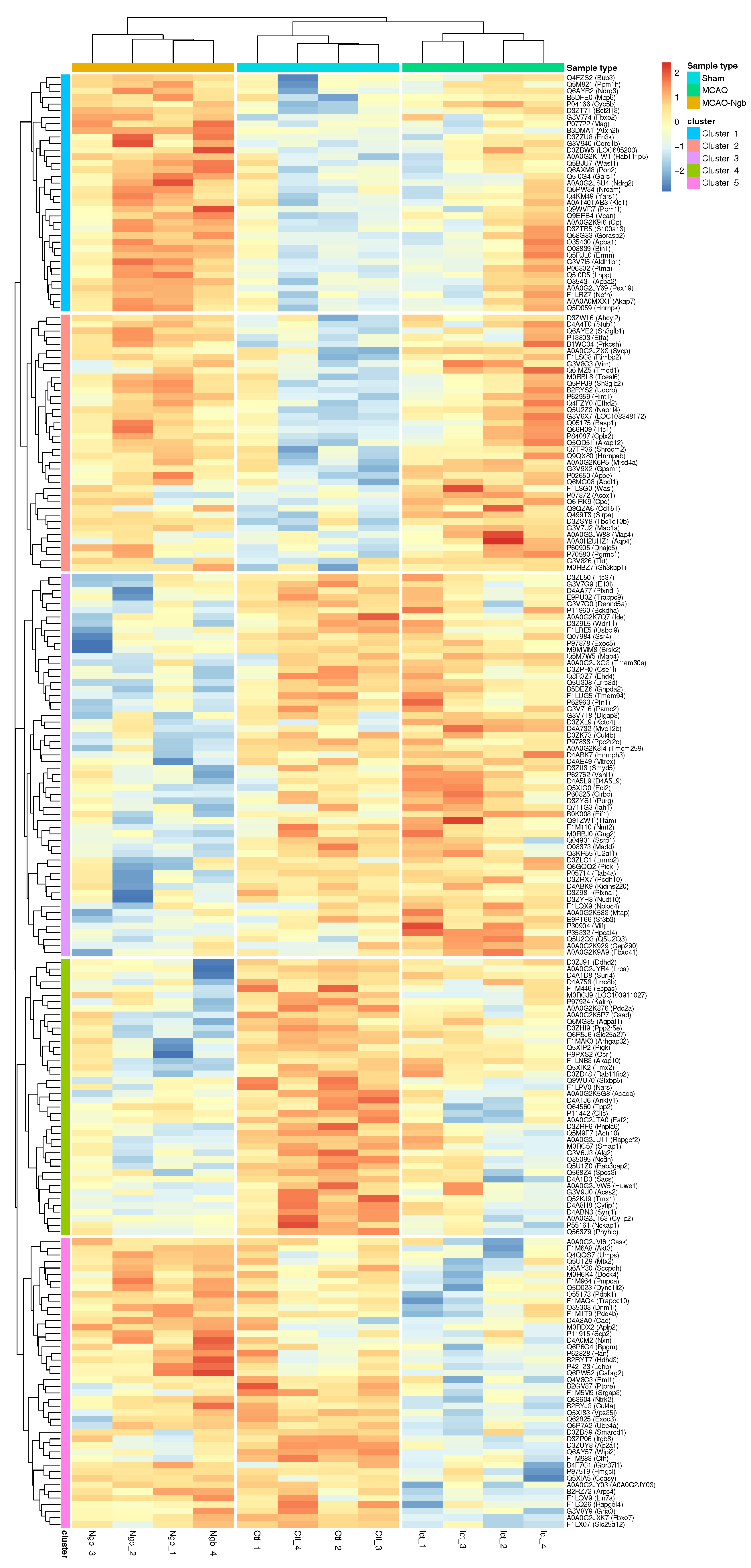
2.4.3. Hierarchical Clustering
2.4.4. GO and Pathways Enrichment
2.4.5. Individual Enrichments in Each of the Five Clusters
2.4.6. Global Enrichment Using Differentially Expressed Proteins
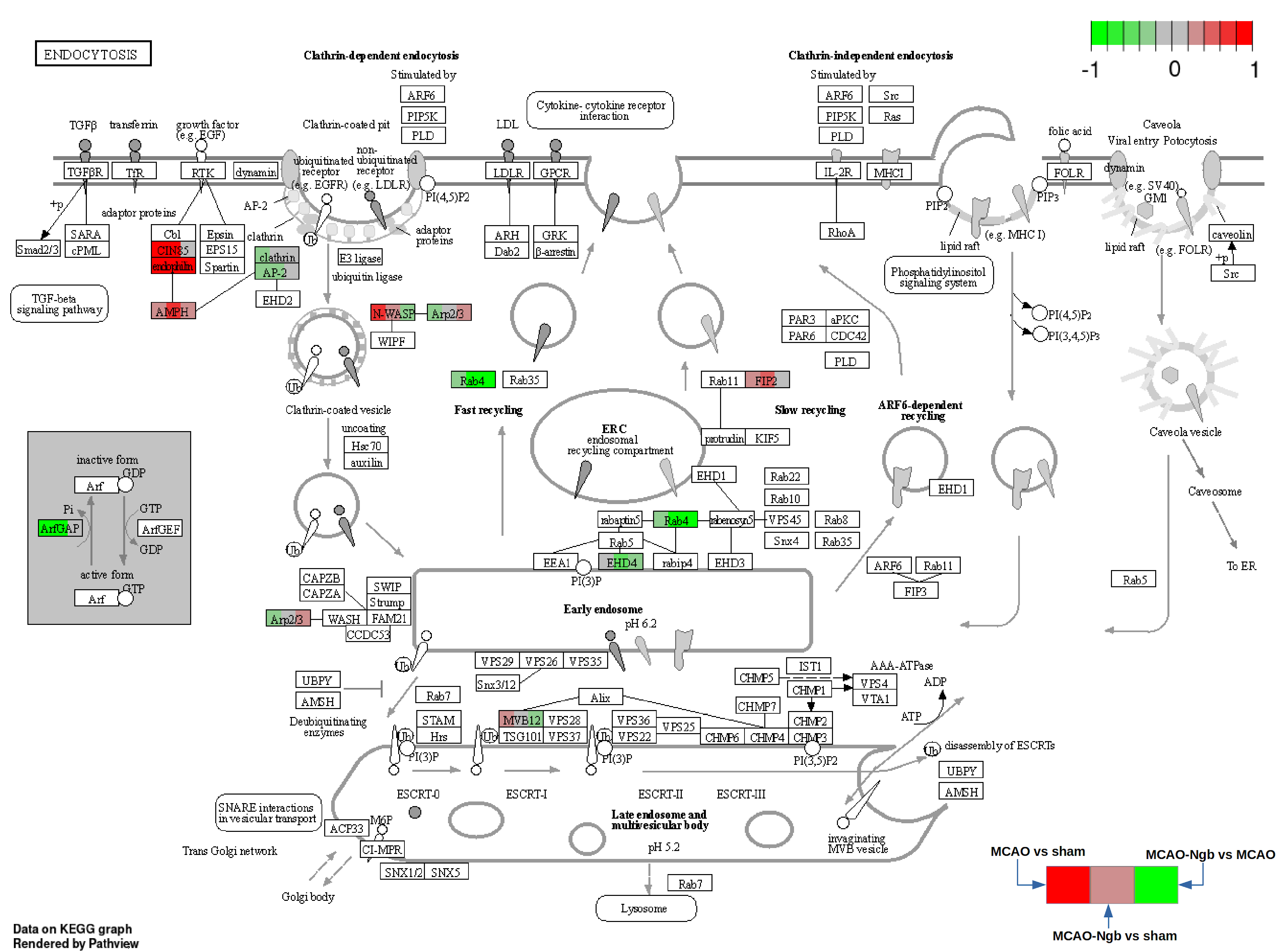
2.4.7. Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Biosynthesis of Ngb-NPs
4.2. Experimental Animals
4.3. Study Design
4.4. Stroke Model
4.5. Mortality Rate and Neurological Outcomes
4.6. MRI Neuroimage Study
4.7. Histological Studies
4.7.1. Cresyl-Violet (Nissl) Staining
4.7.2. Immunofluorescence
4.8. Oxidative and Nitrosative Stresses Determinations
4.9. Proteomics Analysis
4.9.1. Sample Preparation and Mass Spectrometry Analysis
4.9.2. Bioinformatics Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Feigin, V.L.; Norrving, B.; Mensah, G.A. Global burden of stroke. Circ. Res. 2017, 120, 439–448. [Google Scholar] [CrossRef]
- Pérez-Asensio, F.J.; Hurtado, O.; Burguete, M.C.; Moro, M.A.; Salom, J.B.; Lizasoain, I.; Torregrosa, G.; Leza, J.C.; Alborch, E.; Castillo, J.; et al. Inhibition of iNOS activity by 1400W decreases glutamate release and ameliorates stroke outcome after experimental ischemia. Neurobiol. Dis. 2005, 18, 375–384. [Google Scholar] [CrossRef]
- Beslow, L.A.; Smith, S.E.; Vossough, A.; Licht, D.J.; Kasner, S.E.; Favilla, C.; Halperin, A.R.; Gordon, D.M.; Jones, C.I.; Cucchiara, A.J.; et al. Hemorrhagic transformation of childhood arterial ischemic stroke. Stroke 2011, 42, 941–946. [Google Scholar] [CrossRef] [Green Version]
- Terruso, V.; D’Amelio, M.; Di Benedetto, N.; Lupo, I.; Saia, V.; Famoso, G.; Mazzola, M.A.; Aridon, P.; Sarno, C.; Ragonese, P.; et al. Frequency and determinants for hemorrhagic transformation of cerebral infarction. Neuroepidemiology 2009, 33, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Van Den Berg, S.A.; Kruyt, N.D.; Van Den Berg, J.S.P.; Caminada, K.; Hofmeijer, J.; Kerkhoff, H.; De Leeuw, F.E.; Van Der Worp, H.B.; Nederkoorn, P.J. Multicentre randomised trial of acute stroke treatment in the ambulance with a nitroglycerine patch (mr asap). Eur. Stroke J. 2017, 2. [Google Scholar]
- Di Domenico, F.; Casalena, G.; Jia, J.; Sultana, R.; Barone, E.; Cai, J.; Pierce, W.M.; Cini, C.; Mancuso, C.; Perluigi, M.; et al. Sex differences in brain proteomes of neuron-specific STAT3-null mice after cerebral ischemia/reperfusion. J. Neurochem. 2012, 121, 680–692. [Google Scholar] [CrossRef]
- Wu, J.; Jin, Z.; Yang, X.; Yan, L.-J. Post-ischemic administration of 5-methoxyindole-2-carboxylic acid at the onset of reperfusion affords neuroprotection against stroke injury by preserving mitochondrial function and attenuating oxidative stress. Biochem. Biophys. Res. Commun. 2018, 497, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Auriel, E.; Bornstein, N. Neuroprotection in acute ischemic stroke—Current status. J. Cell. Mol. Med. 2010, 14, 2200–2202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmichael, S.T.; Kathirvelu, B.; Schweppe, C.A.; Nie, E. Molecular, cellular and functional events in axonal sprouting after stroke. Exp. Neurol. 2016, 287, 384–394. [Google Scholar] [CrossRef] [Green Version]
- Detante, O.; Muir, K.; Jolkkonen, J. Cell therapy in stroke—Cautious steps towards a clinical treatment. Transl. Stroke Res. 2017, 9, 321–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Liu, N.; Liu, J.; Yang, K.; Wang, X. Neuroglobin, a novel target for endogenous neuroprotection against stroke and neurodegenerative disorders. Int. J. Mol. Sci. 2012, 13, 6995–7014. [Google Scholar] [CrossRef] [PubMed]
- Burmester, T.; Weich, B.; Reinhardt, S.; Hankeln, T. A vertebrate globin expressed in the brain. Nature 2000, 407, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Gorabi, A.M.; Aslani, S.; Barreto, G.E.; Báez-Jurado, E.; Kiaie, N.; Jamialahmadi, T.; Sahebkar, A. The potential of mitochondrial modulation by neuroglobin in treatment of neurological disorders. Free. Radic. Biol. Med. 2020, 162, 471–477. [Google Scholar] [CrossRef]
- Fiocchetti, M.; De Marinis, E.; Ascenzi, P.; Marino, M. Neuroglobin and neuronal cell survival. Biochim. et Biophys. Acta (BBA) - Proteins Proteom. 2013, 1834, 1744–1749. [Google Scholar] [CrossRef]
- Fordel, E.; Geuens, E.; Dewilde, S.; De Coen, W.; Moens, L. Hypoxia/ischemia and the regulation of neuroglobin and cytoglobin expression. IUBMB Life 2004, 56, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Yu, Z.; Gao, X.; Song, Y.; Yuan, J.; Xun, Y.; Wang, T.; Yan, F.; Yuan, S.; Zhang, J.; et al. Establishment of cell-based neuroglobin promoter reporter assay for neuroprotective compounds screening. CNS Neurol. Disord. Drug Targets 2016, 15, 629–639. [Google Scholar] [CrossRef]
- Ren, C.; Wang, P.; Wang, B.; Li, N.; Li, W.; Zhang, C.; Jin, K.; Ji, X. Limb remote ischemic per-conditioning in combination with post-conditioning reduces brain damage and promotes neuroglobin expression in the rat brain after ischemic stroke. Restor. Neurol. Neurosci. 2015, 33, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Schmidt-Kastner, R.; Haberkamp, M.; Schmitz, C.; Hankeln, T.; Burmester, T. Neuroglobin mRNA expression after transient global brain ischemia and prolonged hypoxia in cell culture. Brain Res. 2006, 1103, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xu, R.; Xie, F.; Zhu, H.; Zhu, J.; Wang, X. Hemin offers neuroprotection through inducing exogenous neuroglobin in focal cerebral hypoxic-ischemia in rats. Int. J. Clin. Exp. Pathol. 2014, 7, 2163–2171. [Google Scholar]
- Barreto, G.; McGovern, A.; Garcia-Segura, L. Role of neuroglobin in the neuroprotective actions of estradiol and estrogenic compounds. Cells 2021, 10, 1907. [Google Scholar] [CrossRef]
- Cai, B.; Li, W.; Mao, X.; Winters, A.; Ryou, M.-G.; Liu, R.; Greenberg, D.A.; Wang, N.; Jin, K.; Yang, S.-H. Neuroglobin overexpression inhibits AMPK signaling and promotes cell anabolism. Mol. Neurobiol. 2015, 53, 1254–1265. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.; Wang, H.; Xu, S.-W.; Yang, L.-H.; Chen, W.; Zhao, S.-X.; Shen, H.; Liu, Q.; Yang, R.-M.; Wang, J. Variants in oxidative stress-related genes affect the chemosensitivity through Nrf2-mediated signaling pathway in biliary tract cancer. EBioMedicine 2019, 48, 143–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Liu, Q.R.; Xiong, X.X.; Liu, J.M.; Lai, X.J.; Cheng, C.; Pan, F.; Chen, Y.; Bin Yu, S.; Yu, A.C.H.; et al. Neuroglobin promotes neurite outgrowth via differential binding to PTEN and Akt. Mol. Neurobiol. 2013, 49, 149–162. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, C.; Liu, Y.; Liu, N.; Lo, E.H.; Wang, X. Neuroglobin promotes neurogenesis through Wnt signaling pathway. Cell Death Dis. 2018, 9, 945. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jin, K.; Mao, X.O.; Zhu, Y.; Greenberg, D.A. Neuroglobin is up-regulated by and protects neurons from hypoxic-ischemic injury. Proc. Natl. Acad. Sci. USA 2001, 98, 15306–15311. [Google Scholar] [CrossRef] [Green Version]
- Jin, K.; Mao, Y.; Mao, X.; Xie, L.; Greenberg, D.A. Neuroglobin expression in ischemic stroke. Stroke 2010, 41, 557–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hundahl, C.; Kelsen, J.; Kjær, K.; Rønn, L.C.B.; Weber, R.E.; Geuens, E.; Hay-Schmidt, A.; Nyengaard, J.R. Does neuroglobin protect neurons from ischemic insult? A quantitative investigation of neuroglobin expression following transient MCAo in spontaneously hypertensive rats. Brain Res. 2006, 1085, 19–27. [Google Scholar] [CrossRef]
- Raida, Z.; Hundahl, C.A.; Kelsen, J.; Nyengaard, J.R.; Hay-Schmidt, A. Reduced infarct size in neuroglobin-null mice after experimental stroke in vivo. Exp. Transl. Stroke Med. 2012, 4, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raida, Z.; Hundahl, C.A.; Nyengaard, J.R.; Hay-Schmidt, A. Neuroglobin over expressing mice: Expression pattern and effect on brain ischemic infarct size. PLoS ONE 2013, 8, e76565. [Google Scholar] [CrossRef] [Green Version]
- Shang, A.; Liu, K.; Wang, H.; Wang, J.; Hang, X.; Yang, Y.; Wang, Z.; Zhang, C.; Zhou, D. Neuroprotective effects of neuroglobin after mechanical injury. Neurol. Sci. 2011, 33, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Wang, Y.; Sun, Y.; Mao, X.O.; Xie, L.; Miles, E.; Graboski, J.; Chen, S.; Ellerby, L.M.; Jin, K.; et al. Neuroglobin-overexpressing transgenic mice are resistant to cerebral and myocardial ischemia. Proc. Natl. Acad. Sci. USA 2006, 103, 17944–17948. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, J.; Zhu, H.; Tejima, E.; Tsuji, K.; Murata, Y.; Atochin, D.N.; Huang, P.L.; Zhang, C.; Lo, E.H. Effects of neuroglobin overexpression on acute brain injury and long-term outcomes after focal cerebral ischemia. Stroke 2008, 39, 1869–1874. [Google Scholar] [CrossRef] [Green Version]
- Raychaudhuri, S.; Skommer, J.; Henty, K.; Birch, N.; Brittain, T. Neuroglobin protects nerve cells from apoptosis by inhibiting the intrinsic pathway of cell death. Apoptosis 2009, 15, 401–411. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Liu, J.; Guo, S.; Xing, C.; Fan, X.; Ning, M.; Yuan, J.; Lo, E.; Wang, X. Neuroglobin-overexpression alters hypoxic response gene expression in primary neuron culture following oxygen glucose deprivation. Neuroscience 2009, 162, 396–403. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, D.A.; Jin, K.; Khan, A.A. Neuroglobin: An endogenous neuroprotectant. Curr. Opin. Pharmacol. 2008, 8, 20–24. [Google Scholar] [CrossRef] [Green Version]
- Peroni, D.; Negro, A.; Bähr, M.; Dietz, G.P. Intracellular delivery of Neuroglobin using HIV-1 TAT protein transduction domain fails to protect against oxygen and glucose deprivation. Neurosci. Lett. 2007, 421, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-Y.; Zhou, S.-N.; Lou, Z.-Y.; Zhu, C.-S.; Zheng, X.-P.; Hu, X.-Q. Translocation and neuroprotective properties of transactivator-of-transcription protein-transduction domain–neuroglobin fusion protein in primary cultured cortical neurons. Biotechnol. Appl. Biochem. 2008, 49, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Belayev, L.; Busto, R.; Zhao, W.; Ginsberg, M.D. Quantitative evaluation of blood-brain barrier permeability following middle cerebral artery occlusion in rats. Brain Res. 1996, 739, 88–96. [Google Scholar] [CrossRef]
- Rosenberg, G.A.; Estrada, E.Y.; Dencoff, J.E. Matrix metalloproteinases and TIMPs are associated with blood-brain barrier opening after reperfusion in rat brain. Stroke 1998, 29, 2189–2195. [Google Scholar] [CrossRef] [Green Version]
- McColl, B.W.; Rothwell, N.J.; Allan, S. Systemic inflammation alters the kinetics of cerebrovascular tight junction disruption after experimental stroke in mice. J. Neurosci. 2008, 28, 9451–9462. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Lin, Y.; Xue, X.-H.; Fang, L.; Wang, N.; Wu, Z.-Y. TAT-mediated delivery of neuroglobin protects against focal cerebral ischemia in mice. Exp. Neurol. 2010, 227, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Dietz, G.P. Protection by neuroglobin and cell-penetrating peptide-mediated delivery in vivo: A decade of research: Comment on Cai et al.: TAT-mediated delivery of neuroglobin protects against focal cerebral ischemia in mice. Exp Neurol. 2011; 227(1): 224–31. Exp. Neurol. 2011, 231, 1–10. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, L.R.; Frey, W.H. Intranasal delivery bypasses the blood-brain barrier to target therapeutic agents to the central nervous system and treat neurodegenerative disease. BMC Neurosci. 2008, 9, S5. [Google Scholar] [CrossRef] [Green Version]
- Karatas, H.; Aktas, Y.; Gursoy-Ozdemir, Y.; Bodur, E.; Yemisci, M.; Caban, S.; Vural, A.; Pinarbasli, O.; Capan, Y.; Fernandez-Megia, E.; et al. A Nanomedicine transports a peptide caspase-3 inhibitor across the blood–brain barrier and provides neuroprotection. J. Neurosci. 2009, 29, 13761–13769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, M.K.; Labhasetwar, V. Nanoparticle-mediated delivery of superoxide dismutase to the brain: An effective strategy to reduce ischemia-reperfusion injury. FASEB J. 2009, 23, 1384–1395. [Google Scholar] [CrossRef] [PubMed]
- Peralta, S.; Blanco, S.; Hernández, R.; Castán, H.; Siles, E.; Martínez-Lara, E.; Morales, M.E.; Peinado, M.; Ruiz, M.A. Synthesis and characterization of different sodium hyaluronate nanoparticles to transport large neurotherapheutic molecules through blood brain barrier after stroke. Eur. Polym. J. 2019, 112, 433–441. [Google Scholar] [CrossRef]
- Blanco, S.; Peralta, S.; Morales, M.E.; Martínez-Lara, E.; Pedrajas, J.R.; Castán, H.; Peinado, M.; Ruiz, M.A. Hyaluronate nanoparticles as a delivery system to carry neuroglobin to the brain after stroke. Pharmaceutics 2020, 12, 40. [Google Scholar] [CrossRef] [Green Version]
- Pascovici, D.; Handler, D.C.L.; Wu, J.X.; Haynes, P.A. Multiple testing corrections in quantitative proteomics: A useful but blunt tool. Proteomics 2016, 16, 2448–2453. [Google Scholar] [CrossRef]
- Thompson, B.J.; Ronaldson, P.T. Drug Delivery to the Ischemic Brain. Adv. Pharmacol. 2014, 71, 165–202. [Google Scholar] [CrossRef] [Green Version]
- Cramer, S.C. Treatments to promote neural repair after stroke. J. Stroke 2018, 20, 57–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Worp, H.B.; Macleod, M.R.; Bath, P.; Demotes, J.; Durand-Zaleski, I.; Gebhardt, B.; Gluud, C.; Kollmar, R.; Krieger, D.W.; Lees, K.R.; et al. EuroHYP-1: European multicenter, randomized, phase III clinical trial of therapeutic hypothermia plus best medical treatment vs. best medical treatment alone for acute ischemic stroke. Int. J. Stroke 2014, 9, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Loza, M.I.; Mirelman, D.; Brea, J.; Blanco, M.; Sobrino, T.; Campos, F. A novel mechanism of neuroprotection: Blood glutamate grabber. J. Cereb. Blood Flow Metab. 2015, 36, 292–301. [Google Scholar] [CrossRef] [PubMed]
- A Donnan, G.; Fisher, M.; Macleod, M.R.; Davis, S.M. Stroke. Lancet 2008, 371, 1612–1623. [Google Scholar] [CrossRef]
- García-Bonilla, L.; Campos, M.; Giralt, D.; Salat, D.; Chacón, P.; Guillamon, M.M.H.; Rosell, A.; Montaner, J. Evidence for the efficacy of statins in animal stroke models: A meta-analysis. J. Neurochem. 2012, 122, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Calahorra, J.; Martínez-Lara, E.; Granadino-Roldán, J.M.; Martí, J.M.; Cañuelo, A.; Blanco, S.; Oliver, F.J.; Siles, E. Crosstalk between hydroxytyrosol, a major olive oil phenol, and HIF-1 in MCF-7 breast cancer cells. Sci. Rep. 2020, 10, 6361. [Google Scholar] [CrossRef]
- Popa-Wagner, A.; Dumbrava, D.-A.; Dumitrascu, D.I.; Capitanescu, B.; Petcu, E.B.; Surugiu, R.; Fang, W.-H. Dietary habits, lifestyle factors and neurodegenerative diseases. Neural Regen. Res. 2020, 15, 394–400. [Google Scholar] [CrossRef]
- Hong, T.; Zhou, Y.; Peng, L.; Wu, X.; Li, Y.; Li, Y.; Zhao, Y. Knocking down peroxiredoxin 6 aggravates cerebral ischemia-reperfusion injury by enhancing mitophagy. Neuroscience 2021, 482, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Gusel’Nikova, V.V.; Korzhevskiy, D.E. NeuN as a neuronal nuclear antigen and neuron differentiation marker. Acta Naturae 2015, 7, 42–47. [Google Scholar] [CrossRef]
- Venturini, A.; Passalacqua, M.; Pelassa, S.; Pastorino, F.; Tedesco, M.; Cortese, K.; Gagliani, M.C.; Leo, G.; Maura, G.; Guidolin, D.; et al. Exosomes from astrocyte processes: Signaling to neurons. Front. Pharmacol. 2019, 10, 1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, L.; Chen, H.; Lu, K.; Huang, J.; Duan, H.; Zhao, Y. Clinical significance of changes in serum neuroglobin and HIF-1α concentrations during the early-phase of acute ischemic stroke. J. Neurol. Sci. 2017, 375, 52–57. [Google Scholar] [CrossRef]
- Ord, E.; Shirley, R.; McClure, J.D.; McCabe, C.; Kremer, E.; Macrae, I.M.; Work, L.M. Combined antiapoptotic and antioxidant approach to acute neuroprotection for stroke in hypertensive rats. J. Cereb. Blood Flow Metab. 2013, 33, 1215–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Wu, M.; Yue, Z. Autophagy and Parkinson’s disease. Adv. Exp. Med. Biol. 2020, 1207, 21–51. [Google Scholar] [CrossRef]
- Spagnol, V.; Oliveira, C.A.; Randle, S.J.; Passos, P.M.; Correia, C.R.; Simaroli, N.B.; Oliveira, J.S.; Mevissen, T.E.; Medeiros, A.C.; Gomes, M.D.; et al. The E3 ubiquitin ligase SCF(Fbxo7) mediates proteasomal degradation of UXT isoform 2 (UXT-V2) to inhibit the NF-κB signaling pathway. Biochim. Biophys. Acta Gen. Subj. 2020, 1865, 129754. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, L.; Wang, C.; Song, Y.; Miao, J.; Miao, M. Protection against acute cerebral ischemia/reperfusion injury by Leonuri Herba Total Alkali via modulation of BDNF-TrKB-PI3K/Akt signaling pathway in rats. Biomed. Pharmacother. 2020, 133, 111021. [Google Scholar] [CrossRef] [PubMed]
- Maccioni, R.B.; Cambiazo, V. Role of microtubule-associated proteins in the control of microtubule assembly. Physiol. Rev. 1995, 75, 835–864. [Google Scholar] [CrossRef]
- Halpain, S.; Dehmelt, L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006, 7, 224. [Google Scholar] [CrossRef]
- Lu, G.; Yi, J.; Gubas, A.; Wang, Y.-T.; Wu, Y.; Ren, Y.; Wu, M.; Shi, Y.; Ouyang, C.; Tan, H.; et al. Suppression of autophagy during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2 polyubiquitination and proteasomal degradation. Autophagy 2019, 15, 1917–1934. [Google Scholar] [CrossRef] [Green Version]
- Wan, W.; Liu, W. MTORC1 regulates autophagic membrane growth by targeting WIPI2. Autophagy 2019, 15, 742–743. [Google Scholar] [CrossRef] [Green Version]
- Kaehler, C.; Isensee, J.; Nonhoff, U.; Terrey, M.; Hucho, T.; Lehrach, H.; Krobitsch, S. Ataxin-2-Like Is a Regulator of stress granules and processing bodies. PLoS ONE 2012, 7, e50134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.; Sun, F. Prognostic alternative mRNA splicing in adrenocortical carcinoma. Front. Endocrinol. 2021, 12, 538364. [Google Scholar] [CrossRef]
- Génier, S.; Létourneau, D.; Gauthier, E.; Picard, S.; Boisvert, M.; Parent, J.-L.; Lavigne, P. In-depth NMR characterization of Rab4a structure, nucleotide exchange and hydrolysis kinetics reveals an atypical GTPase profile. J. Struct. Biol. 2020, 212, 107582. [Google Scholar] [CrossRef]
- Genet, G.; Boyé, K.; Mathivet, T.; Ola, R.; Zhang, F.; Dubrac, A.; Li, J.; Genet, N.; Geraldo, L.H.; Benedetti, L.; et al. Endophilin-A2 dependent VEGFR2 endocytosis promotes sprouting angiogenesis. Nat. Commun. 2019, 10, 2350. [Google Scholar] [CrossRef]
- Boucrot, E.; Ferreira, A.P.A.; Almeida-Souza, L.; Debard, S.; Vallis, Y.; Howard, G.; Bertot, L.; Sauvonnet, N.; McMahon, H.T. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 2014, 517, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Naslavsky, N.; Caplan, S. A Role for EHD4 in the regulation of early endosomal transport. Traffic 2008, 9, 995–1018. [Google Scholar] [CrossRef] [Green Version]
- Winckler, B.; Yap, C.C. Endocytosis and endosomes at the crossroads of regulating trafficking of axon outgrowth-modifying receptors. Traffic 2011, 12, 1099–1108. [Google Scholar] [CrossRef] [Green Version]
- Katanov, C.; Novak, N.; Vainshtein, A.; Golani, O.; DuPree, J.L.; Peles, E. N-wasp regulates oligodendrocyte myelination. J. Neurosci. 2020, 40, 6103–6111. [Google Scholar] [CrossRef]
- Wegner, A.M.; Nebhan, C.A.; Hu, L.; Majumdar, D.; Meier, K.M.; Weaver, A.M.; Webb, D.J. N-WASP and the Arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J. Biol. Chem. 2008, 283, 15912–15920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.; Kronenberg, G.; Balkaya, M.; Färber, K.; Gertz, K.; Kettenmann, H.; Endres, M. Acute neuroprotection by pioglitazone after mild brain ischemia without effect on long-term outcome. Exp. Neurol. 2009, 216, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Longa, E.Z.; Weinstein, P.R.; Carlson, S.; Cummins, R. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke 1989, 20, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Campos, F.; Sobrino, T.; Ramos-Cabrer, P.; Argibay, B.; Agulla, J.; Pérez-Mato, M.; Rodríguez-González, R.; Brea, D.; Castillo, J. Neuroprotection by glutamate oxaloacetate transaminase in ischemic stroke: An experimental study. J. Cereb. Blood Flow Metab. 2011, 31, 1378–1386. [Google Scholar] [CrossRef] [Green Version]
- Bederson, J.B.; Pitts, L.H.; Tsuji, M.; Nishimura, M.C.; Davis, R.L.; Bartkowski, H. Rat middle cerebral artery occlusion: Evaluation of the model and development of a neurologic examination. Stroke 1986, 17, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Buege, J.A.; Aust, S.D. Microsomal lipid peroxidation. Methods Enzymol. 1978, 52, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Jr, W. Filter aided sample preparation (FASP) method. Methods Mol. Biol. 2018, 1841, 3–10. [Google Scholar] [CrossRef]
- Meier, F.; Beck, S.; Grassl, N.; Lubeck, M.; Park, M.A.; Raether, O.; Mann, M. Parallel accumulation–serial fragmentation (PASEF): Multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. J. Proteome Res. 2015, 14, 5378–5387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier, F.; Brunner, A.-D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M.A.; et al. Online Parallel Accumulation–Serial Fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 2018, 17, 2534–2545. [Google Scholar] [CrossRef] [Green Version]

| Cluster | Category | Term | NumGenes | Background | Genes | p-Value | FDR | Description |
|---|---|---|---|---|---|---|---|---|
| 1 | Function | GO.0004721 | 3 | 83 | Ppm1f, Ppm1h, Lhpp | 1.80 × 10−4 | 0.0113 | phosphoprotein phosphatase activity |
| 1 | Function | GO.0004812 | 2 | 21 | Yars, Gars | 3.90 × 10−4 | 0.0130 | aminoacyl-tRNA ligase activity |
| 1 | Function | GO.0005546 | 2 | 35 | Apba1, Apba2 | 1.00 × 10−4 | 0.0149 | phosphatidylinositol-4,5-bisphosphate binding |
| 1 | Function | GO.0004722 | 2 | 30 | Ppm1f, Ppm1h | 7.60 × 10−4 | 0.0149 | protein serine/threonine phosphatase activity |
| 1 | Function | GO.0001540 | 2 | 28 | Apba1, Apba2 | 6.70 × 10−4 | 0.0149 | amyloid-beta binding |
| 1 | Function | GO.0003779 | 3 | 189 | Ermn, Coro1b, Wasf1 | 1.80 × 10−4 | 0.0153 | actin binding |
| 1 | Function | GO.0033218 | 3 | 228 | Apba1, Apba2, Mag | 3.10 × 10−4 | 0.0218 | amide binding |
| 1 | Function | GO.0051015 | 2 | 76 | Ermn, Coro1b | 4.40 × 10−4 | 0.0269 | actin filament binding |
| 2 | Function | GO.0003779 | 4 | 189 | Wasl, Tmod1, Map1a, Shroom2 | 2.40 × 10−4 | 0.0075 | actin binding |
| 2 | KEGG | rno04144 | 4 | 252 | Sh3kbp1, Wasl, Sh3glb1, Sh3glb2 | 6.90 × 10−4 | 0.0255 | Endocytosis |
| 2 | KEGG | rno04141 | 3 | 157 | Prkcsh, Dnajc5, Stub1 | 2.00 × 10−4 | 0.0378 | Protein processing in endoplasmic reticulum |
| 3 | Function | GO.0051020 | 5 | 181 | Pfn1, Pick1, Madd, Dennd5a, Exoc5 | 6.50 × 10−5 | 0.0114 | GTPase binding |
| 3 | Function | GO.0017112 | 2 | 8 | Madd, Dennd5a | 2.30 × 10−4 | 0.0198 | Rab guanyl-nucleotide exchange factor activity |
| 3 | Function | GO.0017016 | 4 | 136 | Pfn1, Madd, Dennd5a, Exoc5 | 2.90 × 10−4 | 0.0198 | Ras GTPase binding |
| 3 | Function | GO.0016863 | 2 | 9 | Mif, Eci2 | 2.80 × 10−4 | 0.0198 | intramolecular oxidoreductase activity, transposing C=C bonds |
| 3 | Function | GO.0035255 | 2 | 31 | Pick1, Rab4a | 2.60 × 10−3 | 0.0482 | ionotropic glutamate receptor binding |
| 4 | Function | GO.0052745 | 2 | 12 | Ocrl, Synj1 | 2.20 × 10−4 | 0.0044 | inositol phosphate phosphatase activity |
| 4 | Function | GO.0017016 | 4 | 136 | Rab3gap2, Ocrl, Kalrn, Stxbp5 | 6.89 × 10−5 | 0.0044 | Ras GTPase binding |
| 4 | Function | GO.0005096 | 3 | 90 | Rab3gap2, Ocrl, Stxbp5 | 4.30 × 10−4 | 0.0044 | GTPase activator activity |
| 4 | Function | GO.0004439 | 2 | 5 | Ocrl, Synj1 | 5.07 × 10−5 | 0.0044 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity |
| 4 | Function | GO.0005088 | 2 | 35 | Rab3gap2, Kalrn | 1.60 × 10−3 | 0.0103 | Ras guanyl-nucleotide exchange factor activity |
| 4 | Function | GO.0017048 | 2 | 55 | Ocrl, Kalrn | 3.70 × 10−3 | 0.0210 | Rho GTPase binding |
| 4 | Function | GO.0017137 | 2 | 57 | Rab3gap2, Stxbp5 | 3.90 × 10−3 | 0.0211 | Rab GTPase binding |
| Category | Term | NumGenes | Background | Genes |
|---|---|---|---|---|
| Function | GO.0051020 | 19 | 181 | Dnm1l, Rab3gap2, Ocrl, Pfn1, Arfgef1, Rapgef4, Vcl, Pick1, Madd, Dennd5a, Bin1, Ngef, Exoc8, Myo1c, Anxa2, Kalrn, Stxbp5, Exoc5, Wasf1 |
| Function | GO.0017016 | 16 | 136 | Dnm1l, Rab3gap2, Ocrl, Pfn1, Rapgef4, Vcl, Madd, Dennd5a, Ngef, Exoc8, Myo1c, Anxa2, Kalrn, Stxbp5, Exoc5, Wasf1 |
| Function | GO.0003779 | 18 | 189 | Marcks, Pfn1, Hpca, Wasl, Tmod1, Tmod2, Vcl, Cnn3, Pick1, Cap1, Map1a, Tpm5, Myo1c, Ermn, Coro1b, Anxa2, Shroom2, Wasf1 |
| Function | GO.0050662 | 16 | 192 | Abat, Dld, Cryl1, Hmgcl, Txnrd1, Ivd, Idh3a, Ldhb, Etfa, Tkt, Eci2, Got1, Th, Acox1, Phgdh, Fasn |
| Function | GO.0050839 | 11 | 96 | Calr, Grin2b, Nptn, Src, Peta3, Gfap, Cadm2, Lphn1, P4hb, Nlgn2, Cd151 |
| Function | GO.0032550 | 16 | 203 | Ran, Dnm1l, Arf2, Fkbp4, Hsp90aa1, Arf5, Ehd2, Rap2b, Gnal, Gnai1, Arl2, Prps1, LOC314140, Rab4a, Ehd1, Ak4 |
| Function | GO.0032561 | 16 | 212 | Ran, Dnm1l, Arf2, Fkbp4, Hsp90aa1, Arf5, Ehd2, Rap2b, Gnal, Gnai1, Arl2, Prps1, LOC314140, Rab4a, Ehd1, Ak4 |
| Function | GO.0015631 | 14 | 170 | Dnm1l, Dpysl2, Cnn3, Map1a, Stmn1, Tppp3, Kif5b, Prune, Myo1c, Map4, Chp1, Gphn, Mapre2, Eml1 |
| Function | GO.0033218 | 16 | 228 | Calr, Cltc, Dld, Rnpep, Gria3, Hmgcl, Ctsb, Tpp2, Apba1, Eci2, Apba2, Itm2b, Mag, Apoe, Hspd1, Fasn |
| Function | GO.0051015 | 9 | 76 | Tmod1, Vcl, Pick1, Tpm5, Myo1c, Ermn, Coro1b, Anxa2, Shroom2 |
| Function | GO.0000287 | 10 | 99 | Ran, Enoph1, Hmgcl, Prpsap1, Idh3a, Tkt, Lhpp, Ephx2, Prps1, LOC314140 |
| Function | GO.0009055 | 7 | 46 | Dld, Sdhb, Txnrd1, Cyb5b, Cyb5a, Etfa, Etfb |
| Function | GO.0005525 | 14 | 199 | Ran, Dnm1l, Arf2, Fkbp4, Hsp90aa1, Arf5, Ehd2, Rap2b, Gnal, Gnai1, Arl2, Rab4a, Ehd1, Ak4 |
| Function | GO.0031072 | 8 | 66 | Tfam, Cltc, Fkbp4, Sgtb, St13, Hnrpk, Hspa9, Bax |
| Function | GO.0005088 | 6 | 35 | Rab3gap2, Rapgef4, Madd, Dennd5a, Ngef, Kalrn |
| Function | GO.0051082 | 6 | 37 | Calr, Hsp90aa1, St13, Hspa9, Hsp90b1, Hspd1 |
| Function | GO.0016874 | 8 | 74 | Farsa, Yars, Gars, Uba5, Acsl3, Sae1, Pars2, Ctps2 |
| Function | GO.0005085 | 7 | 57 | Rab3gap2, Arfgef1, Rapgef4, Madd, Dennd5a, Ngef, Kalrn |
| Function | GO.0008017 | 10 | 126 | Dnm1l, Dpysl2, Cnn3, Map1a, Kif5b, Myo1c, Map4, Chp1, Mapre2, Eml1 |
| Function | GO.0004749 | 3 | 5 | Prpsap1, Prps1, LOC314140 |
| Function | GO.0019003 | 6 | 44 | Ran, Rap2b, Gnai1, Prps1, LOC314140, Rab4a |
| Function | GO.0001540 | 5 | 28 | Gria3, Apba1, Apba2, Itm2b, Apoe |
| Function | GO.0016616 | 8 | 84 | Cryl1, Akr1b1, Idh3a, Ldhb, Fam213b, Hsd17b4, Phgdh, Fasn |
| Function | GO.1902936 | 6 | 46 | Wipi2, Pfn1, Cadps, Apba1, Apba2, Anxa2 |
| Function | GO.0017160 | 3 | 6 | Exoc8, Myo1c, Exoc5 |
| Function | GO.0005080 | 5 | 30 | Src, Pick1, C1qbp, Akt3, Srsf2 |
| Function | GO.1901981 | 7 | 68 | Wipi2, Gap43, Pfn1, Cadps, Apba1, Apba2, Anxa2 |
| Function | GO.0042578 | 12 | 191 | Ppm1f, Enoph1, Ppm1e, Ocrl, Ppm1h, Pde4b, Ptpre, Lhpp, Ephx2, Prune, Synj1, Plcl1 |
| Function | GO.0016667 | 5 | 32 | Dld, Txnrd1, Pcyox1, Txnl1, P4hb |
| Function | GO.0016791 | 10 | 139 | Ppm1f, Enoph1, Ppm1e, Ocrl, Ppm1h, Ptpre, Lhpp, Ephx2, Prune, Synj1 |
| Function | GO.0043021 | 6 | 53 | Abcf1, Eif2s1, MGC94335, Hnrpk, C1qbp, ENSRNOG00000037607 |
| Function | GO.0005546 | 5 | 35 | Pfn1, Cadps, Apba1, Apba2, Anxa2 |
| Function | GO.0017112 | 3 | 8 | Rab3gap2, Madd, Dennd5a |
| Function | GO.0030165 | 7 | 75 | Lin7a, Gria3, Dlgap3, Apba1, Grm5, Mpp2, Kidins220 |
| Function | GO.0035091 | 8 | 98 | Wipi2, Gap43, Pfn1, Cadps, Apba1, Apba2, Anxa2, Ap2a2 |
| Function | GO.0031625 | 9 | 123 | Dnm1l, Calr, Sh3kbp1, Src, Ckb, Bag5, Tsg101, Vcp, Nploc4 |
| Function | GO.0030246 | 10 | 148 | Nptx2, Calr, Tkt, Pgls, Mag, Prps1, Man2c1, Lphn1, LOC314140, Vcan |
| Function | GO.0019888 | 5 | 36 | Ppp2r2c, Nsfl1c, rCG_47223, Ppp1r1b, Ppp2r5b |
| Function | GO.0017048 | 6 | 55 | Ocrl, Pfn1, Vcl, Ngef, Kalrn, Wasf1 |
| Function | GO.0016887 | 11 | 177 | Abcf1, Hsp90aa1, Psmc2, Pcyox1, Anxa1, Kif5b, Myo1c, Vcp, Snrnp200, Dync1h1, Lonp1 |
| Function | GO.0004812 | 4 | 21 | Farsa, Yars, Gars, Pars2 |
| Function | GO.0016668 | 3 | 9 | Dld, Txnrd1, Txnl1 |
| Function | GO.0015643 | 3 | 9 | Nefh, Lphn1, Nfm |
| Function | GO.0017137 | 6 | 57 | Dnm1l, Rab3gap2, Madd, Dennd5a, Anxa2, Stxbp5 |
| Function | GO.0005543 | 11 | 180 | Marcks, Wipi2, Gap43, Pfn1, Cadps, Apba1, Apba2, Anxa1, Anxa2, Apoe, Ap2a2 |
| Function | GO.0019905 | 6 | 58 | Cplx2, Stx6, Scfd1, Rab4a, Stxbp5, Cplx1 |
| Function | GO.0048306 | 5 | 39 | Cplx2, Tsg101, Anxa1, Anxa2, Chp1 |
| Function | GO.0042277 | 11 | 187 | Calr, Cltc, Rnpep, Gria3, Ctsb, Tpp2, Apba1, Apba2, Itm2b, Apoe, Hspd1 |
| Function | GO.0051087 | 5 | 41 | Bag5, Sgtb, St13, Hspa9, Bax |
| Function | GO.0043531 | 4 | 24 | Prps1, Vcp, LOC314140, Lonp1 |
| Function | GO.0000149 | 7 | 84 | Cplx2, Exoc3, Stx6, Scfd1, Rab4a, Stxbp5, Cplx1 |
| Function | GO.0004427 | 2 | 2 | Lhpp, Prune |
| Function | GO.0001018 | 2 | 2 | Tfam, Lonp1 |
| Function | GO.0043022 | 4 | 25 | Abcf1, Eif2s1, MGC94335, C1qbp |
| Function | GO.0051117 | 5 | 43 | Nsfl1c, Hnrpk, Rab4a, Nploc4, Ufd1l |
| Function | GO.0008022 | 8 | 115 | Cltc, Src, Pick1, Sgtb, Sae1, Myo1c, Synj1, Hpcal4 |
| Function | GO.0008233 | 14 | 299 | Dpp10, Ncstn, Cpq, Rnpep, Nrd1, Scrn1, Ctsb, Sec11a, Tpp2, Psmb2, Pmpca, MGC109340, Arxes2, Lonp1 |
| Function | GO.0016651 | 5 | 49 | Dld, Txnrd1, Nqo1, Txnl1, ND4 |
| Function | GO.0005178 | 5 | 49 | Calr, Peta3, Gfap, P4hb, Cd151 |
| Function | GO.0051287 | 5 | 50 | Dld, Cryl1, Idh3a, Ldhb, Phgdh |
| Function | GO.0016903 | 4 | 32 | Dld, Akr1b1, Aldh1b1, Bckdha |
| Function | GO.0032564 | 2 | 4 | Hsp90aa1, St13 |
| Function | GO.0030984 | 2 | 4 | Ctsb, C1qbp |
| Function | GO.0019834 | 2 | 4 | Anxa1, Anxa2 |
| Function | GO.0050660 | 5 | 54 | Dld, Txnrd1, Ivd, Etfa, Acox1 |
| Function | GO.0016829 | 7 | 105 | Cd38, Hmgcl, Echdc1, Hsd17b4, Got1, Adcy5, Fasn |
| Function | GO.0051400 | 2 | 5 | Dnm1l, Bax |
| Function | GO.0036435 | 2 | 5 | Vcp, Ufd1l |
| Function | GO.0017075 | 3 | 18 | Cplx2, Stxbp5, Cplx1 |
| Function | GO.0016627 | 4 | 36 | Sdhb, Ivd, Acox1, Fasn |
| Function | GO.0005030 | 2 | 5 | Ntrk2, Sort1 |
| Function | GO.0004439 | 2 | 5 | Ocrl, Synj1 |
| Function | GO.0003924 | 8 | 135 | Ran, Dnm1l, Rap2b, Gnal, Gnai1, Gng7, Arl2, Rab4a |
| Function | GO.0045182 | 5 | 58 | Abcf1, Eif2s1, Cirbp, Eef1d, Eif4e |
| Function | GO.0031406 | 8 | 139 | Grin2b, Hmgcl, Got1, Th, Mag, C1qbp, Vcan, Acox1 |
| KEGG | rno04144 | 28 | 252 | Vps37b, Cltc, Sh3kbp1, Snx6, Arfgef1, Wasl, Arf5, Ehd4, Arpc4, Rab11fip2, Src, Ehd2, Bin1, Sh3glb1, Tsg101, Ap2a1, Ist1, Mvb12b, |
| Sh3glb2, Kif5b, Vps35, Rab11fip5, Rab4a, Ehd1, Hgs, Ap2a2, Vps26b, Smap1 | ||||
| KEGG | rno04141 | 20 | 157 | Calr, Skp1, Hsp90aa1, Gcs1, Nsfl1c, Fbxo2, Eif2s1, Sec13, Prkcsh, Dnajc5, Stub1, Bax, Wfs1, Hsp90b1, Sec24b, Vcp, P4hb, Nploc4, Ssr4, Ufd1l |
| KEGG | rno04728 | 15 | 122 | Ppp2r5a, Ppp2r5e, Ppp2r2c, Gria3, Grin2b, Kif5b, Gnal, Gnai1, Gng7, Th, Ppp1r1b, Adcy5, Akt3, Gng4, Ppp2r5b |
| KEGG | rno01200 | 13 | 112 | Sdhc, Dld, Acss1, Sdhb, Idh3a, Tkt, Got1, Pgls, Prps1, Cs, LOC314140, Phgdh, Acss2 |
| KEGG | rno04727 | 11 | 86 | Abat, Gabrg2, Adcy1, Src, Gnai1, Gng7, Adcy5, Plcl1, Slc12a5, Gphn, Gng4 |
| KEGG | rno00640 | 7 | 30 | Abat, Dld, Acss1, Echdc1, Ldhb, Bckdha, Acss2 |
| KEGG | rno00970 | 8 | 45 | Farsa, Yars, Gars, Nars, Hars, Mars, Iars2, Pars2 |
| KEGG | rno05016 | 13 | 181 | Tfam, Sdhc, Cltc, Ndufa12, Sdhb, Grin2b, Ndufb9, Tgm2, Ap2a1, Grm5, Bax, LOC685596, Ap2a2 |
| KEGG | rno04723 | 11 | 144 | Gabrg2, Ndufa12, Gria3, Adcy1, Ndufb9, Grm5, Gnai1, Gng7, ND4, Adcy5, Gng4 |
| KEGG | rno05132 | 8 | 79 | Pfn1, Wasl, Arpc4, Dync1li2, Actb, Klc2, Wasf1, Dync1h1 |
| KEGG | rno05030 | 6 | 46 | Grin2b, Gnai1, Th, Ppp1r1b, Adcy5, Gpsm1 |
| KEGG | rno05012 | 10 | 134 | Sdhc, Ndufa12, Sdhb, Ndufb9, Gnal, Gnai1, Th, ND4, Adcy5, LOC685596 |
| KEGG | rno03050 | 6 | 46 | Psmd6, Psmb2, Psmc2, Psmd1, Psmd4, Psmd3 |
| KEGG | rno00190 | 10 | 130 | Ppa1, Atp6v1g2, Sdhc, Ndufa12, Sdhb, Ndufb9, Ppa2, Lhpp, ND4, LOC685596 |
| KEGG | rno00020 | 5 | 29 | Sdhc, Dld, Sdhb, Idh3a, Cs |
| KEGG | rno00280 | 6 | 51 | Abat, Dld, Hmgcl, Ivd, Aldh1b1, Bckdha |
| KEGG | rno05100 | 7 | 73 | Cltc, Wasl, Arpc4, Src, Vcl, Actb, Wasf1 |
| KEGG | rno01230 | 7 | 73 | Idh3a, Tkt, Got1, Prps1, Cs, LOC314140, Phgdh |
| KEGG | rno00620 | 5 | 35 | Dld, Acss1, Aldh1b1, Ldhb, Acss2 |
| KEGG | rno04721 | 6 | 60 | Cplx2, Atp6v1g2, Cltc, Ap2a1, Cplx1, Ap2a2 |
| KEGG | rno04261 | 9 | 133 | Ppp2r5a, Ppp2r5e, Ppp2r2c, Rapgef4, Adcy1, Gnai1, Adcy5, Akt3, Ppp2r5b |
| KEGG | rno04146 | 7 | 82 | Hmgcl, Scp2, Acsl3, Hsd17b4, Eci2, Ephx2, Acox1 |
| KEGG | rno04120 | 9 | 130 | Skp1, Fbxo2, Ube2o, Sae1, Ube4a, Cul4a, Stub1, Ddb1, Cul4b |
| KEGG | rno00010 | 6 | 59 | Dld, Acss1, Bpgm, Aldh1b1, Ldhb, Acss2 |
| KEGG | rno04724 | 8 | 109 | Gria3, Adcy1, Grin2b, Grm5, Gnai1, Gng7, Adcy5, Gng4 |
| KEGG | rno05010 | 10 | 164 | Sdhc, Ncstn, Ndufa12, Sdhb, Grin2b, Ndufb9, Lrp1, App, LOC685596, Apoe |
| KEGG | rno04714 | 12 | 221 | Sdhc, Ndufa12, Sdhb, Adcy1, Ndufb9, Acsl3, Slc25a20, ND4, Smarcd1, Actb, Adcy5, LOC685596 |
| KEGG | rno05032 | 7 | 88 | Gabrg2, Pde4b, Adcy1, Gnai1, Gng7, Adcy5, Gng4 |
| KEGG | rno04713 | 7 | 90 | Gria3, Adcy1, Grin2b, Gnai1, Gng7, Adcy5, Gng4 |
| KEGG | rno04071 | 8 | 116 | Ppp2r5a, Ppp2r5e, Ppp2r2c, Gnai1, Bax, Akt3, Pdpk1, Ppp2r5b |
| KEGG | rno00030 | 4 | 29 | Tkt, Pgls, Prps1, LOC314140 |
| KEGG | rno05169 | 11 | 206 | Ran, Cd38, Psmd6, Psmc2, Psmd1, Vim, Psmd4, Psmd3, Snw1, Akt3, ENSRNOG00000025731 |
| KEGG | rno04926 | 8 | 122 | Adcy1, Src, Gnai1, Gng7, Adcy5, Col4a1, Akt3, Gng4 |
| KEGG | rno04915 | 8 | 122 | Fkbp4, Hsp90aa1, Adcy1, Src, Gnai1, Hsp90b1, Adcy5, Akt3 |
| KEGG | rno05034 | 8 | 130 | Grin2b, Gnai1, Gng7, Th, Ppp1r1b, Ntrk2, Adcy5, Gng4 |
| KEGG | rno04024 | 10 | 188 | Pde4b, Gria3, Rapgef4, Adcy1, Grin2b, Gnai1, Ppp1r1b, Adcy5, Acox1, Akt3 |
| KEGG | rno01210 | 3 | 17 | Idh3a, Got1, Cs |
| Feature | Score |
|---|---|
| Spontaneous activity | Moving and exploring = 0 Moving without exploring = 1 No moving or moving only when pulled by the tail = 2 |
| Left drifting during displacement | None = 0 Drifting only when elevated by the tail and pushed or pulled = 1 Spontaneous drifting = 2 Circling without displacement, or spinning = 3 |
| Resistance to left forepaw stretching | Stretching not allowed= 0 Stretching allowed after some attempts = 1, No resistance = 2 |
| Parachute reflex | Symmetrical = 0 Asymmetrical = 1 Contralateral forelimb retracted = 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peinado, M.Á.; Ovelleiro, D.; del Moral, M.L.; Hernández, R.; Martínez-Lara, E.; Siles, E.; Pedrajas, J.R.; García-Martín, M.L.; Caro, C.; Peralta, S.; et al. Biological Implications of a Stroke Therapy Based in Neuroglobin Hyaluronate Nanoparticles. Neuroprotective Role and Molecular Bases. Int. J. Mol. Sci. 2022, 23, 247. https://doi.org/10.3390/ijms23010247
Peinado MÁ, Ovelleiro D, del Moral ML, Hernández R, Martínez-Lara E, Siles E, Pedrajas JR, García-Martín ML, Caro C, Peralta S, et al. Biological Implications of a Stroke Therapy Based in Neuroglobin Hyaluronate Nanoparticles. Neuroprotective Role and Molecular Bases. International Journal of Molecular Sciences. 2022; 23(1):247. https://doi.org/10.3390/ijms23010247
Chicago/Turabian StylePeinado, María Ángeles, David Ovelleiro, María Luisa del Moral, Raquel Hernández, Esther Martínez-Lara, Eva Siles, José Rafael Pedrajas, María Luisa García-Martín, Carlos Caro, Sebastián Peralta, and et al. 2022. "Biological Implications of a Stroke Therapy Based in Neuroglobin Hyaluronate Nanoparticles. Neuroprotective Role and Molecular Bases" International Journal of Molecular Sciences 23, no. 1: 247. https://doi.org/10.3390/ijms23010247
APA StylePeinado, M. Á., Ovelleiro, D., del Moral, M. L., Hernández, R., Martínez-Lara, E., Siles, E., Pedrajas, J. R., García-Martín, M. L., Caro, C., Peralta, S., Morales, M. E., Ruiz, M. A., & Blanco, S. (2022). Biological Implications of a Stroke Therapy Based in Neuroglobin Hyaluronate Nanoparticles. Neuroprotective Role and Molecular Bases. International Journal of Molecular Sciences, 23(1), 247. https://doi.org/10.3390/ijms23010247








