The Study of Cerebrospinal Fluid microRNAs in Spinal Cord Injury and Neurodegenerative Diseases: Methodological Problems and Possible Solutions
Abstract
:1. Introduction
2. Detection of miRNAs in Cerebrospinal Fluid of Patients with Neurological Disorders
2.1. Spinal Cord Injury (SCI)
2.2. Neurodegenerative Diseases Exemplified by Alzheimer’s and Parkinson’s Disease
3. Methods for Isolation of miRNAs from Cerebrospinal Fluid
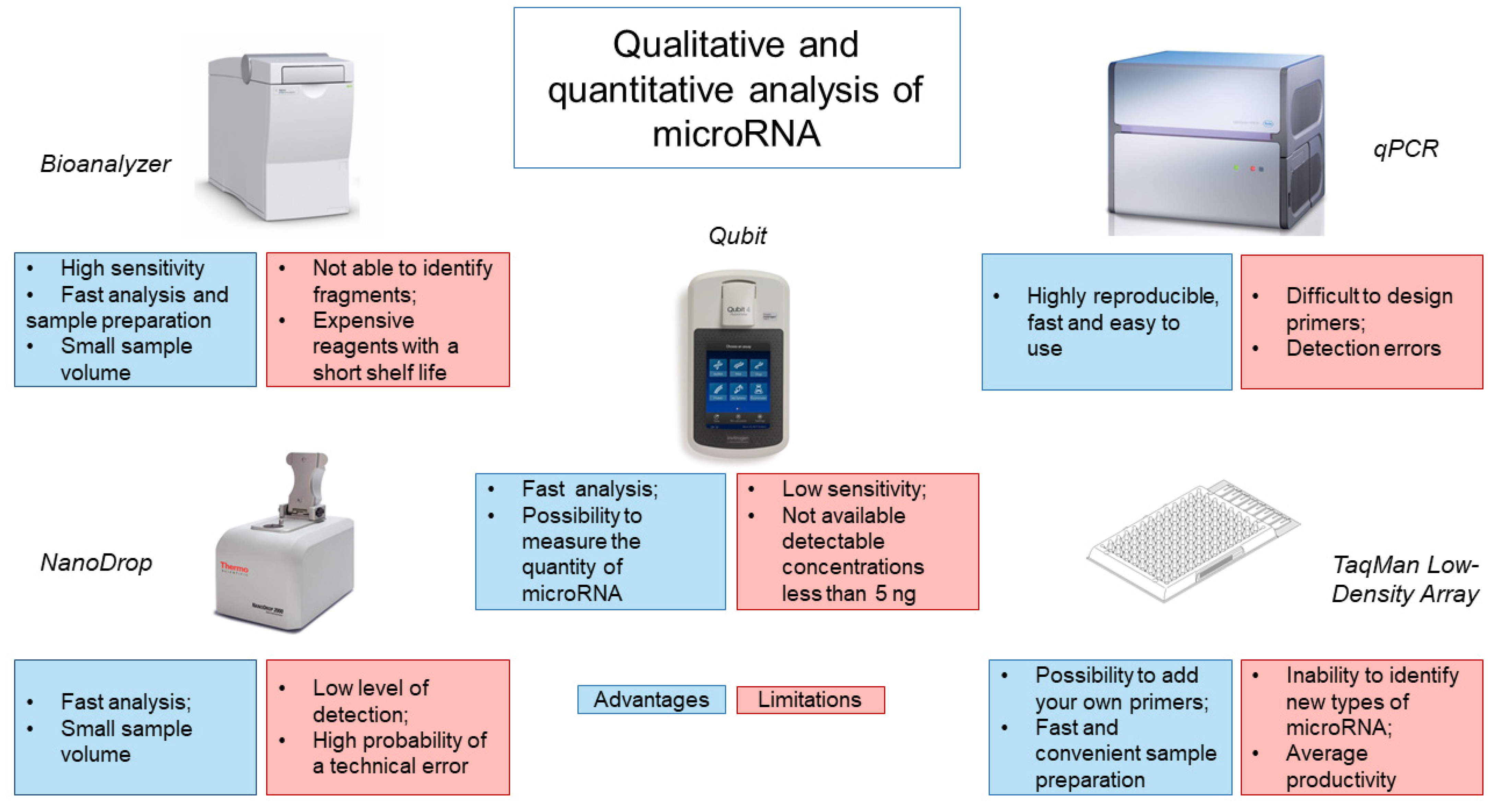
4. Assessment of the Quality of the Obtained miRNA
5. Methods for Studying the miRNA Expression Profile
6. Methods for Control and Normalization of miRNA Expression
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rao, P.; Benito, E.; Fischer, A. MicroRNAs as biomarkers for CNS disease. Front. Mol. Neurosci. 2013, 6, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, B.; Peplow, P.V. MicroRNAs as diagnostic and therapeutic tools for Alzheimer’s disease: Advances and limitations. Neural Regen. Res. 2019, 14, 242–255. [Google Scholar] [CrossRef]
- Batistela, M.S.; Josviak, N.D.; Sulzbach, C.D.; de Souza, R.L.R. An overview of circulating cell-free microRNAs as putative biomarkers in Alzheimer’s and Parkinson’s Diseases. Int. J. Neurosci. 2017, 127, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Tetreault, L.; Kalsi-Ryan, S.; Nouri, A.; Fehlings, M.G. Global prevalence and incidence of traumatic spinal cord injury. Clin. Epidemiol. 2014, 6, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.Z.; Ma, Z.J.; Li, J.R.; Kang, X.W. Mesenchymal stem cell-derived exosomes: Therapeutic opportunities and challenges for spinal cord injury. Stem Cell Res. Ther. 2021, 12, 102. [Google Scholar] [CrossRef] [PubMed]
- Noonan, V.K.; Fingas, M.; Farry, A.; Baxter, D.; Singh, A.; Fehlings, M.G.; Dvorak, M.F. Incidence and Prevalence of Spinal Cord Injury in Canada: A National Perspective. Neuroepidemiology 2012, 38, 219–226. [Google Scholar] [CrossRef]
- Curt, A.; Van Hedel, H.J.A.; Klaus, D.; Dietz, V.; Grp, E.-S.S. Recovery from a spinal cord injury: Significance of compensation, neural plasticity, and repair. J. Neurotrauma 2008, 25, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Steeves, J.D.; Kramer, J.K.; Fawcett, J.W.; Cragg, J.; Lammertse, D.P.; Blight, A.R.; Marino, R.J.; Ditunno, J.F.; Coleman, W.P.; Geisler, F.H.; et al. Extent of spontaneous motor recovery after traumatic cervical sensorimotor complete spinal cord injury. Spinal Cord 2011, 49, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Sandwell, S.; Markandaya, M. Neurotrauma, Prognosis and Outcome Predictions BT—Encyclopedia of Trauma Care; Papadakos, P.J., Gestring, M.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1079–1082. [Google Scholar] [CrossRef]
- Kwon, B.K.; Streijger, F.; Fallah, N.; Noonan, V.K.; Belanger, L.M.; Ritchie, L.; Paquette, S.J.; Ailon, T.; Boyd, M.C.; Street, J.; et al. Cerebrospinal Fluid Biomarkers To Stratify Injury Severity and Predict Outcome in Human Traumatic Spinal Cord Injury. J. Neurotrauma 2017, 34, 567–580. [Google Scholar] [CrossRef]
- Ogurcov, S.; Shulman, I.; Garanina, E.; Sabirov, D.; Baichurina, I.; Kuznetcov, M.; Masgutova, G.; Kostennikov, A.; Rizvanov, A.; James, V.; et al. Blood Serum Cytokines in Patients with Subacute Spinal Cord Injury: A Pilot Study to Search for Biomarkers of Injury Severity. Brain Sci. 2021, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Yunta, M.; Nieto-Diaz, M.; Esteban, F.J.; Caballero-Lopez, M.; Navarro-Ruiz, R.; Reigada, D.; Pita-Thomas, D.W.; del Aguila, A.; Munoz-Galdeano, T.; Maza, R.M. MicroRNA Dysregulation in the Spinal Cord following Traumatic Injury. PLoS ONE 2012, 7, e34534. [Google Scholar] [CrossRef] [PubMed]
- Morganti, S.; Tarantino, P.; Ferraro, E.; D’Amico, P.; Viale, G.; Trapani, D.; Duso, B.A.; Curigliano, G. Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit. Rev. Oncol. Hematol. 2019, 133, 171–182. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinchi, E.; Frati, A.; Cantatore, S.; D’Errico, S.; La Russa, R.; Maiese, A.; Palmieri, M.; Pesce, A.; Viola, R.V.; Frati, P.; et al. Acute Spinal Cord Injury: A Systematic Review Investigating miRNA Families Involved. Int. J. Mol. Sci. 2019, 20, 1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, M. Cardio-miRNAs and onco-miRNAs: Circulating miRNA-based diagnostics for non-cancerous and cancerous diseases. Front. Cell Dev. Biol. 2014, 2, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.K.; Wang, X.F.; Lu, Q.B.; Xu, X.M. Altered microRNA expression following traumatic spinal cord injury. Exp. Neurol. 2009, 219, 424–429. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Tigchelaar, S.; Gupta, R.; Shannon, C.P.; Streijger, F.; Sinha, S.; Flibotte, S.; Rizzuto, M.A.; Street, J.; Paquette, S.; Ailon, T.; et al. MicroRNA Biomarkers in Cerebrospinal Fluid and Serum Reflect Injury Severity in Human Acute Traumatic Spinal Cord Injury. J. Neurotrauma 2019, 36, 2358–2371. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.Q.; Yin, H.F.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Carlsen, A.L.; Schetter, A.J.; Nielsen, C.T.; Lood, C.; Knudsen, S.; Voss, A.; Harris, C.C.; Hellmark, T.; Segelmark, M.; Jacobsen, S.; et al. Circulating MicroRNA Expression Profiles Associated With Systemic Lupus Erythematosus. Arthritis Rheum. 2013, 65, 1324–1334. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z.F.; Le, W.D. Tiny But Mighty: Promising Roles of MicroRNAs in the Diagnosis and Treatment of Parkinson’s Disease. Neurosci. Bull. 2017, 33, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef]
- Wang, W.T.; Chen, Y.Q. Circulating miRNAs in cancer: From detection to therapy. J. Hematol. Oncol. 2014, 7, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachisuka, S.; Kamei, N.; Ujigo, S.; Miyaki, S.; Yasunaga, Y.; Ochi, M. Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal Cord 2014, 52, 596–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.Q.; Chen, J.; Wang, S.N.; Duan, F.X.; Chen, Y.Q.; Shi, Y.J.; Hu, J.G.; Lu, H.Z. Highlight article: Identification of serum exosomal microRNAs in acute spinal cord injured rats. Exp. Biol. Med. 2019, 244, 1149–1161. [Google Scholar] [CrossRef]
- Nieto-Diaz, M.; Esteban, F.J.; Reigada, D.; Munoz-Galdeano, T.; Yunta, M.; Caballero-Loopez, M.; Navarro-Ruiz, R.; del Aguila, A.; Maza, R.M. MicroRNA dysregulation in spinal cord injury: Causes, consequences, and therapeutics. Front. Cell. Neurosci. 2014, 8, 53. [Google Scholar] [CrossRef]
- Strickland, E.R.; Hook, M.A.; Balaraman, S.; Huie, J.R.; Grau, J.W.; Miranda, R.C. Microrna dysregulation following spinal cord contusion: Implications for neural plasticity and repair. Neuroscience 2011, 186, 146–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jee, M.K.; Jung, J.S.; Choi, J.I.; Jang, J.A.; Kang, K.S.; Im, Y.B.; Kang, S.K. MicroRNA 486 is a potentially novel target for the treatment of spinal cord injury. Brain 2012, 135, 1237–1252. [Google Scholar] [CrossRef] [Green Version]
- Jee, M.K.; Jung, J.S.; Im, Y.B.; Jung, S.J.; Kang, S.K. Silencing of miR20a Is Crucial for Ngn1-Mediated Neuroprotection in Injured Spinal Cord. Hum. Gene Ther. 2012, 23, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Detloff, M.R.; Miller, K.N.; Santi, L.; Houlé, J.D. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Exp. Neurol. 2012, 233, 447–456. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.Z.; Huang, J.H.; Zeng, L.; Wang, G.; Cao, M.; Lu, H.B. Anti-Apoptotic Effect of MicroRNA-21 after Contusion Spinal Cord Injury in Rats. J. Neurotrauma 2013, 30, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.K.; Zhang, Y.P.; Titsworth, W.L.; Jiang, X.Y.; Han, S.; Lu, P.H.; Shields, C.B.; Xu, X.M. A novel role of phospholipase A(2) in mediating spinal cord secondary injury. Ann. Neurol. 2006, 59, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.L.; Gomes, M.; Medeiros, R. EGFR signaling pathway and related-miRNAs in age-relateddiseases: The example of miR-221 and miR-222. Front. Genet. 2012, 3, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tigchelaar, S.; Streijger, F.; Sinha, S.; Flibotte, S.; Manouchehri, N.; So, K.; Shortt, K.; Okon, E.; Rizzuto, M.A.; Malenica, I.; et al. Serum MicroRNAs Reflect Injury Severity in a Large Animal Model of Thoracic Spinal Cord Injury. Sci. Rep. 2017, 7, 1376. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.J.; Zhou, H.X.; Lu, L.; Li, X.Y.; Fu, Z.; Liu, J.; Kang, Y.; Wei, Z.J.; Pan, B.; Liu, L.; et al. The roles of microRNAs in spinal cord injury. Int. J. Neurosci. 2017, 127, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Barbato, C.; Ruberti, F.; Cogoni, C. Searching for MIND: microRNAs in neurodegenerative diseases. J. Biomed. Biotechnol. 2009, 2009, 871313. [Google Scholar] [CrossRef] [Green Version]
- Denk, J.; Boelmans, K.; Siegismund, C.; Lassner, D.; Arlt, S.; Jahn, H. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimers disease. PLoS ONE 2015, 10, e0126423. [Google Scholar] [CrossRef] [Green Version]
- Denk, J.; Oberhauser, F.; Kornhuber, J.; Wiltfang, J.; Fassbender, K.; Schroeter, M.L.; Volk, A.E.; Diehl-Schmid, J.; Prudlo, J.; Danek, A.; et al. Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal dementia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE 2018, 13, e0197329. [Google Scholar] [CrossRef]
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of Extracellular miRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer’s and Parkinson’s Diseases Correlate with Disease Status and Features of Pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef]
- Wang, W.X.; Huang, Q.W.; Hu, Y.L.; Stromberg, A.J.; Nelson, P.T. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: White matter versus gray matter. Acta Neuropathol. 2011, 121, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Van Harten, A.C.; Mulders, J.; Scheltens, P.; van der Flier, W.M.; Oudejans, C.B.M. Differential Expression of microRNA in Cerebrospinal Fluid as a Potential Novel Biomarker for Alzheimer’s Disease. J. Alzheimers Dis. 2015, 47, 243–252. [Google Scholar] [CrossRef]
- Dangla-Valls, A.; Molinuevo, J.L.; Altirriba, J.; Sanchez-Valle, R.; Alcolea, D.; Fortea, J.; Rami, L.; Balasa, M.; Munoz-Garcia, C.; Ezquerra, M.; et al. CSF microRNA Profiling in Alzheimer’s Disease: A Screening and Validation Study. Mol. Neurobiol. 2017, 54, 6647–6654. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. Micro RNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef] [Green Version]
- Pogue, A.I.; Cui, J.G.; Li, Y.Y.; Zhao, Y.; Culicchia, F.; Lukiw, W.J. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci. Lett. 2010, 476, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Marchegiani, F.; Matacchione, G.; Ramini, D.; Marcheselli, F.; Recchioni, R.; Casoli, T.; Mercuri, E.; Lazzarini, M.; Giorgetti, B.; Cameriere, V.; et al. Diagnostic performance of new and classic CSF biomarkers in age-related dementias. Aging 2019, 11, 2420–2429. [Google Scholar] [CrossRef] [PubMed]
- Riancho, J.; Vazquez-Higuera, J.L.; Pozueta, A.; Lage, C.; Kazimierczak, M.; Bravo, M.; Calero, M.; Gonzalez, A.; Rodriguez, E.; Lleo, A.; et al. MicroRNA Profile in Patients with Alzheimer’s Disease: Analysis of miR-9-5p and miR-598 in Raw and Exosome Enriched Cerebrospinal Fluid Samples. J. Alzheimer’s Dis. 2017, 57, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Kiko, T.; Nakagawa, K.; Tsuduki, T.; Furukawa, K.; Arai, H.; Miyazawa, T. MicroRNAs in Plasma and Cerebrospinal Fluid as Potential Markers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 39, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolaı, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunez-Iglesias, J.; Liu, C.C.; Morgan, T.E.; Finch, C.E.; Zhou, X.J. Joint genomewide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS ONE 2010, 5, e8898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, M.C.T.; Barreto-Sanz, M.A.; Correia, B.R.S.; Bell, R.; Widnall, C.; Perez, L.T.; Berteau, C.; Schulte, C.; Scheller, D.; Berg, D.; et al. miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget 2018, 9, 17455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, Y.X.; Liu, H.; Zhang, L.S.; Lv, W.; Hu, X.Y. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [Green Version]
- Starhof, C.; Hejl, A.M.; Heegaard, N.H.H.; Carlsen, A.L.; Burton, M.; Lilje, B.; Winge, K. The biomarker potential of cell-free microRNA from cerebrospinal fluid in Parkinsonian Syndromes. Mov. Disord. 2019, 34, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Wanet, A.; Tacheny, A.; Arnould, T.; Renard, P. miR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res. 2012, 40, 4742–4753. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.J.; Liu, G.; Jin, S.M.; Parisiadou, L.; Xie, C.; Yu, J.; Sun, L.; Ma, B.; Ding, J.; Vancraenenbroeck, R.; et al. MicroRNA-205 regulates the expression of Parkinson’s diseaserelated leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2012, 22, 608–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basak, I.; Patil, K.S.; Alves, G.; Larsen, J.P.; Moller, S.G. microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell. Mol. Life Sci. 2016, 73, 811–827. [Google Scholar] [CrossRef] [PubMed]
- Doxakis, E. Post-transcriptional Regulation of alpha-Synuclein Expression by mir-7 and mir-153. J. Biol. Chem. 2010, 285, 12726–12734. [Google Scholar] [CrossRef] [Green Version]
- Burgos, K.L.; Javaherian, A.; Bomprezzi, R.; Ghaffari, L.; Rhodes, S.; Courtright, A.; Tembe, W.; Kim, S.; Metpally, R.; Van Keuren-Jensen, K. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 2013, 19, 712–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAlexander, M.A.; Phillips, M.J.; Witwer, K.W. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front. Genet. 2013, 4, 83. [Google Scholar] [CrossRef] [Green Version]
- Bache, S.; Rasmussen, R.; Rossing, M.; Laigaard, F.P.; Nielsen, F.C.; Moller, K. MicroRNA Changes in Cerebrospinal Fluid After Subarachnoid Hemorrhage. Stroke 2017, 48, 2391. [Google Scholar] [CrossRef]
- Akers, J.C.; Hua, W.; Li, H.Y.; Ramakrishnan, V.; Yang, Z.X.; Quan, K.; Zhu, W.; Li, J.; Figueroa, J.; Hirshman, B.R.; et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget 2017, 8, 68769–68779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopkova, A.; Sana, J.; Fadrus, P.; Slaby, O. Cerebrospinal fluid microRNAs as diagnostic biomarkers in brain tumors. Clin. Chem. Lab. Med. 2018, 56, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Saugstad, J.A.; Lusardi, T.A.; Van Keuren-Jensen, K.R.; Phillips, J.I.; Lind, B.; Harrington, C.A.; McFarland, T.J.; Courtright, A.L.; Reiman, R.A.; Yeri, A.S.; et al. Analysis of extracellular RNA in cerebrospinal fluid. J. Extracell. Vesicles 2017, 6, 1317577. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, S.S.; Nygaard, A.B.; Carlsen, A.L.; Heegaard, N.H.H.; Bak, M.; Christensen, T. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Transl. Stroke Res. 2017, 5, 24. [Google Scholar] [CrossRef]
- Sorensen, S.S.; Nygaard, A.B.; Nielsen, M.Y.; Jensen, K.; Christensen, T. miRNA Expression Profiles in Cerebrospinal Fluid and Blood of Patients with Acute Ischemic Stroke. Transl. Stroke Res. 2014, 5, 711–718. [Google Scholar] [CrossRef]
- Masotti, A.; Preckel, T. Analysis of small RNAs with the Agilent 2100 Bioanalyzer. Nat. Methods 2006, 3, 658. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Kodani, M.; Yang, G.Y.; Conklin, L.M.; Travis, T.C.; Whitney, C.G.; Anderson, L.J.; Schrag, S.J.; Taylor, T.H.; Beall, B.W.; Breiman, R.F.; et al. Application of TaqMan Low-Density Arrays for Simultaneous Detection of Multiple Respiratory Pathogens. J. Clin. Microbiol. 2011, 49, 2175–2182. [Google Scholar] [CrossRef] [Green Version]
- Kopkova, A.; Sana, J.; Fadrus, P.; Machackova, T.; Vecera, M.; Vybihal, V.; Juracek, J.; Vychytilova-Faltejskova, P.; Smrcka, M.; Slaby, O. MicroRNA isolation and quantification in cerebrospinal fluid: A comparative methodical study. PLoS ONE 2018, 13, e0208580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalaby, T.; Grotzer, M.A. Tumor-Associated CSF MicroRNAs for the Prediction and Evaluation of CNS Malignancies. Int. J. Mol. Sci. 2015, 16, 29103–29119. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Maghnouj, A.; Zollner, H.; Schmiegel, W.; Hahn, S.; Schroers, R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012, 14, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Chomiak, M.; Ahle, G.; Gress, T.; Buchholz, M.; Turewicz, M.; Eisenacher, M.; Margold, M.; Schlegel, U.; Schmiegel, W.; et al. MicroRNA-30c as a novel diagnostic biomarker for primary and secondary B-cell lymphoma of the CNS. J. Neurooncol. 2018, 137, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Fardo, D.W.; Jicha, G.A.; Nelson, P.T. A Customized Quantitative PCR MicroRNA Panel Provides a Technically Robust Context for Studying Neurodegenerative Disease Biomarkers and Indicates a High Correlation Between Cerebrospinal Fluid and Choroid Plexus MicroRNA Expression. Mol. Neurobiol. 2017, 54, 8191–8202. [Google Scholar] [CrossRef] [PubMed]
- Tsang, H.F.; Xue, V.W.; Koh, S.P.; Chiu, Y.M.; Ng, L.P.W.; Wong, S.C.C. NanoString, a novel digital color-coded barcode technology: Current and future applications in molecular diagnostics. Expert Rev. Mol. Diagn. 2017, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Horbinski, C.; Wu, H.; Liu, Y.X.; Sheng, S.Y.; Liu, J.P.; Weiss, H.; Stromberg, A.J.; Wang, C. NanoStringDiff: A novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic Acids Res. 2016, 44, e151. [Google Scholar] [CrossRef] [Green Version]
- Hong, L.Z.; Zhou, L.; Zou, R.; Khoo, C.M.; San Chew, A.L.; Chin, C.L.; Shih, S.J. Systematic evaluation of multiple qPCR platforms, NanoString and miRNA-Seq for microRNA biomarker discovery in human biofluids. Sci. Rep. 2021, 11, 4435. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Umansky, S.R. Circulating cell-free microRNA as biomarkers for screening, diagnosis, and monitoring of neurodegenerative diseases and other neurologic pathologies. Front. Cell. Neurosci. 2013, 7, 150. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Vigneron, N.; Meryet-Figuiere, M.; Guttin, A.; Issartel, J.P.; Lambert, B.; Briand, M.; Louis, M.H.; Vernon, M.; Lebailly, P.; Lecluse, Y.; et al. Towards a new standardized method for circulating miRNAs profiling in clinical studies: Interest of the exogenous normalization to improve miRNA signature accuracy. Mol. Oncol. 2016, 10, 981–992. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncol. 2012, 14, 689–700. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. miR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): A Platform for Glioblastoma Biomarker Development. PLoS ONE 2013, 8, e78115. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baichurina, I.; Valiullin, V.; James, V.; Rizvanov, A.; Mukhamedshina, Y. The Study of Cerebrospinal Fluid microRNAs in Spinal Cord Injury and Neurodegenerative Diseases: Methodological Problems and Possible Solutions. Int. J. Mol. Sci. 2022, 23, 114. https://doi.org/10.3390/ijms23010114
Baichurina I, Valiullin V, James V, Rizvanov A, Mukhamedshina Y. The Study of Cerebrospinal Fluid microRNAs in Spinal Cord Injury and Neurodegenerative Diseases: Methodological Problems and Possible Solutions. International Journal of Molecular Sciences. 2022; 23(1):114. https://doi.org/10.3390/ijms23010114
Chicago/Turabian StyleBaichurina, Irina, Victor Valiullin, Victoria James, Albert Rizvanov, and Yana Mukhamedshina. 2022. "The Study of Cerebrospinal Fluid microRNAs in Spinal Cord Injury and Neurodegenerative Diseases: Methodological Problems and Possible Solutions" International Journal of Molecular Sciences 23, no. 1: 114. https://doi.org/10.3390/ijms23010114
APA StyleBaichurina, I., Valiullin, V., James, V., Rizvanov, A., & Mukhamedshina, Y. (2022). The Study of Cerebrospinal Fluid microRNAs in Spinal Cord Injury and Neurodegenerative Diseases: Methodological Problems and Possible Solutions. International Journal of Molecular Sciences, 23(1), 114. https://doi.org/10.3390/ijms23010114