Novel SCN5A p.Val1667Asp Missense Variant Segregation and Characterization in a Family with Severe Brugada Syndrome and Multiple Sudden Deaths
Abstract
1. Introduction
2. Material and Methods
2.1. Subjects
2.2. DNA Extraction and Genetic Analysis
2.3. Plasmid Generation
2.4. Cell Culture and Transfection
2.5. Functional Analysis
2.6. Statistical Analysis
3. Results
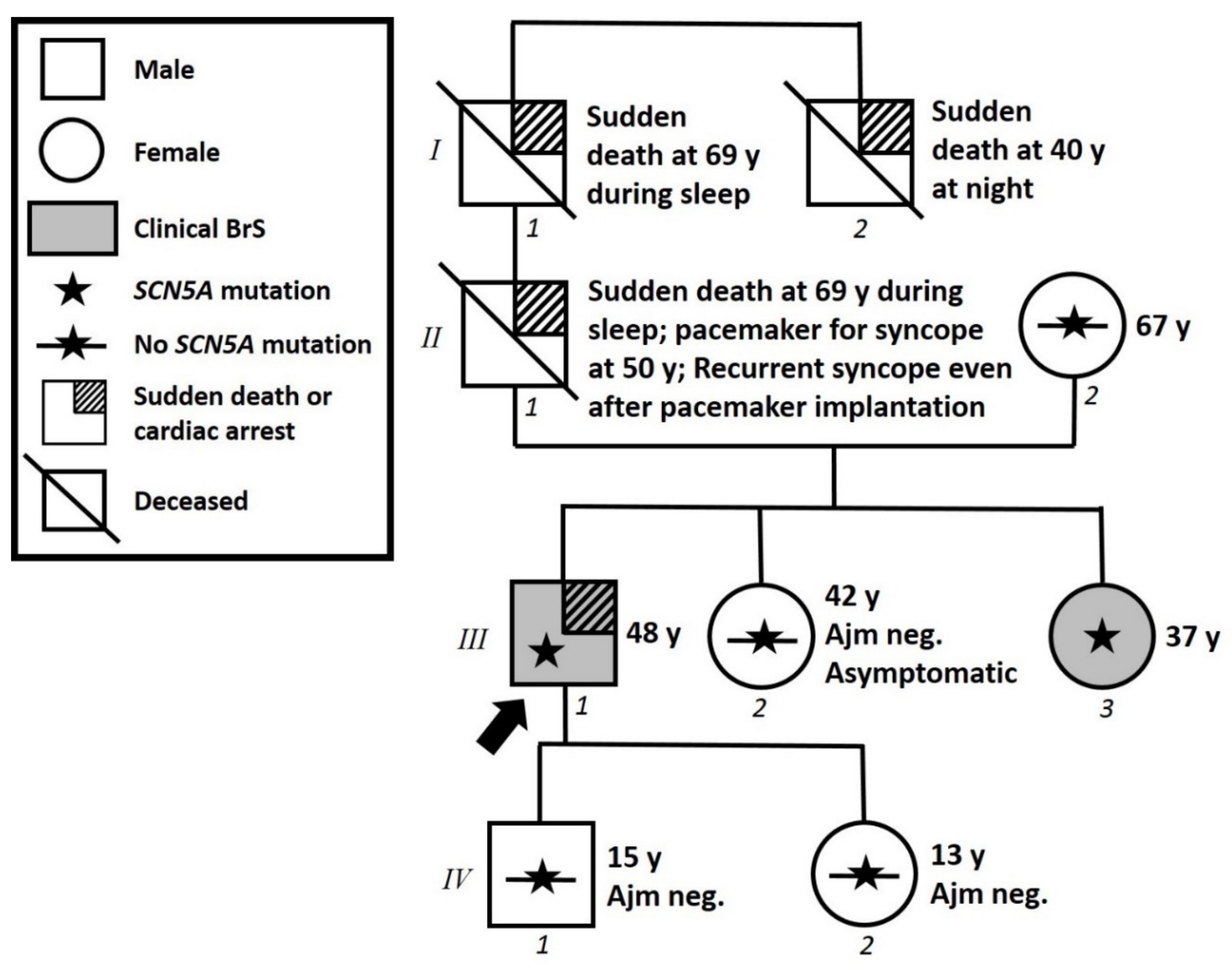
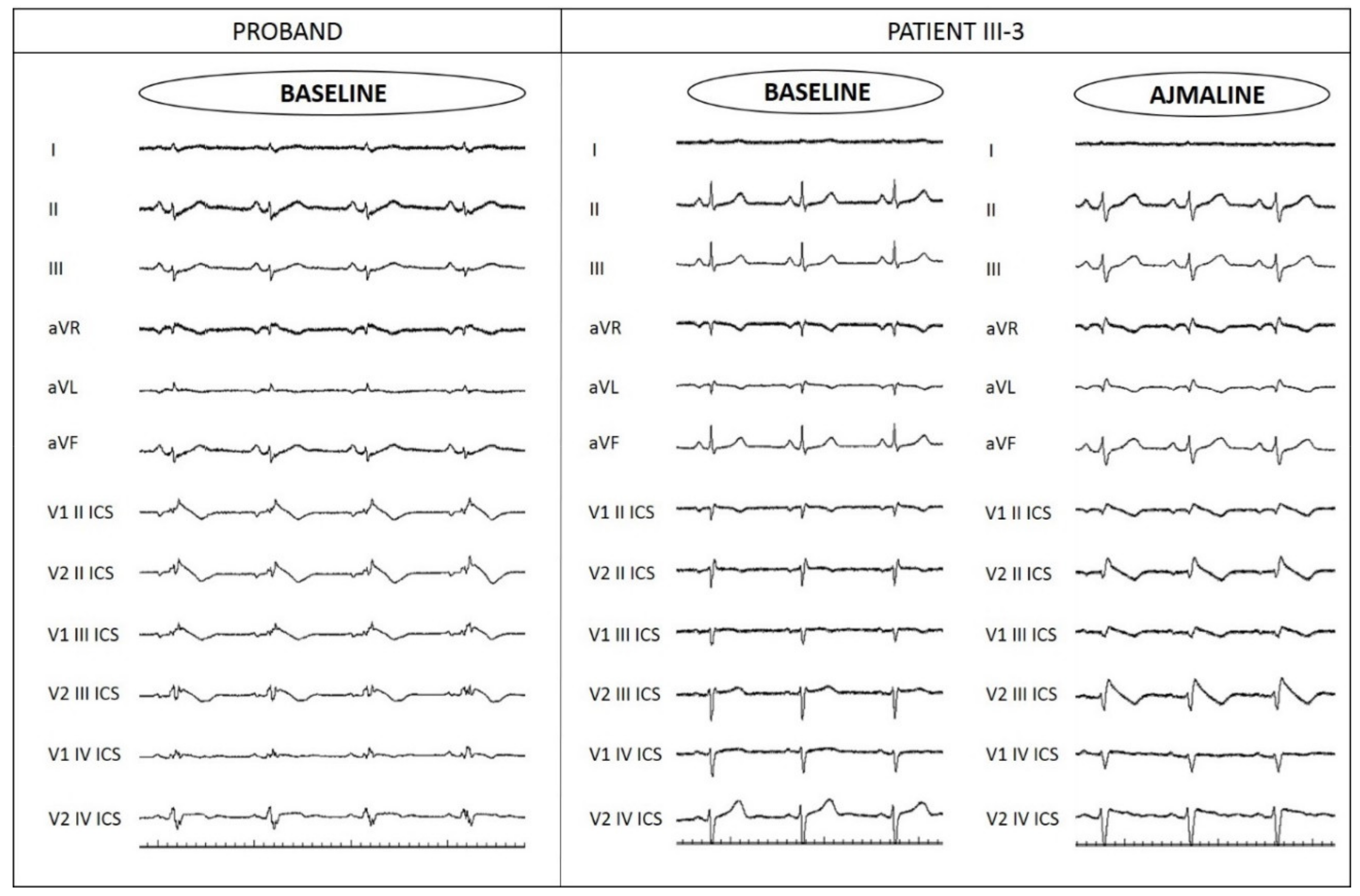
3.1. Proband Patient Characteristics and Family History
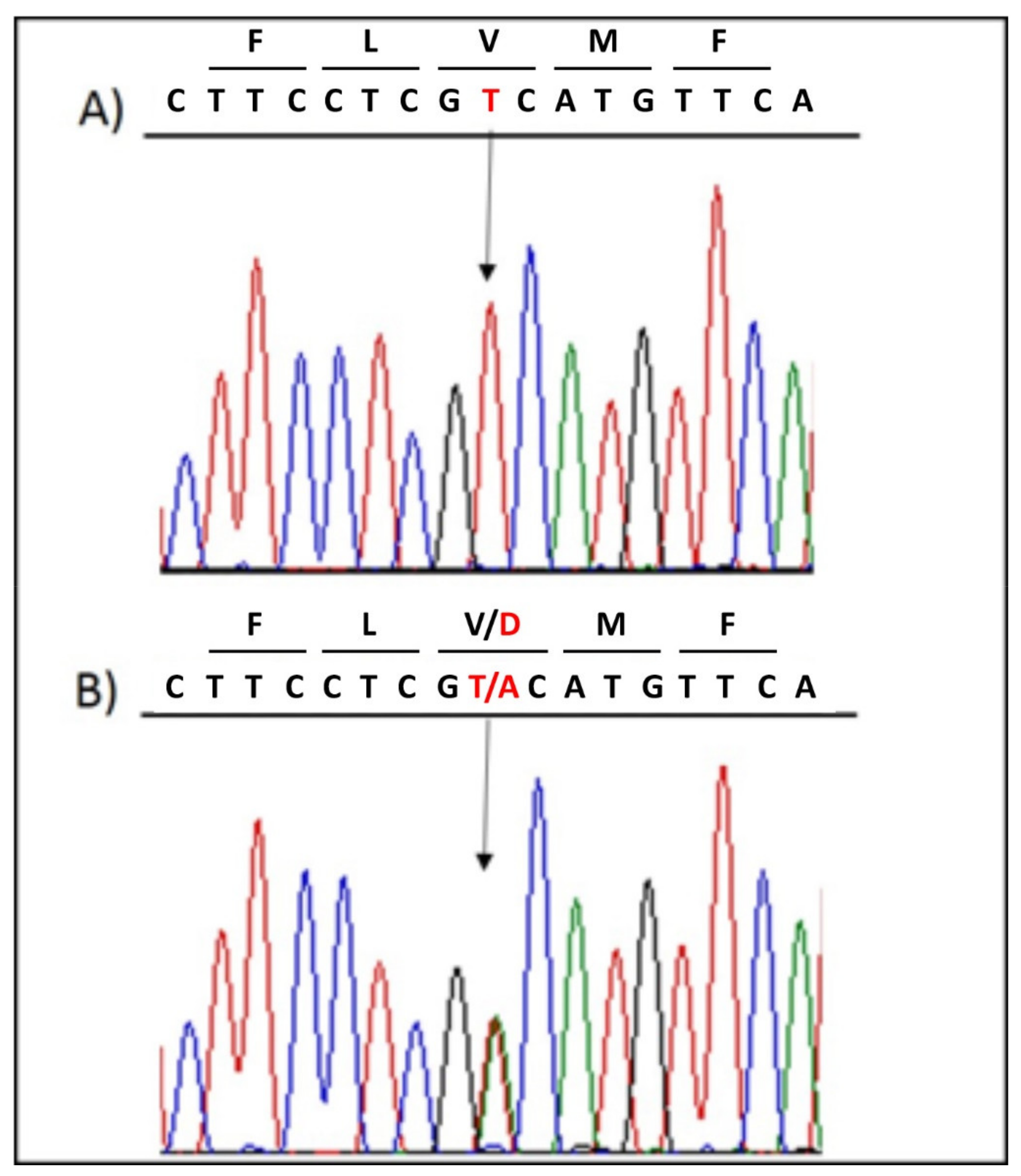
3.2. Genetic Testing Results and In Silico Prediction
- PM1, Moderate: UniProt protein SCN5A_HUMAN trans-membrane region ‘Helical’ has 4 non-VUS missense/in-frame/non-synonymous, variants (4 pathogenic and 0 benign), pathogenicity = 100.0% which is more than threshold 50.0%.
- PM2, Moderate: Variant not found in gnomAD exomes (good gnomAD exomes coverage = 99.6). Variant not found in gnomAD genomes (good gnomAD genomes coverage = 31.9).
- PM5, Moderate: A nearby variant chr3:38592864 G>A (Val1649Ile) is classified Pathogenic by UniProt Variants (and confirmed using ACMG).
- PP2, Supporting: The gnomAD missense Z-Score = 2.75 is greater than 0.647.
- PP3, Supporting: Pathogenic computational verdict based on 11 pathogenic predictions from BayesDel_addAF, DEOGEN2, EIGEN, FATHMM-MKL, LIST-S2, M-CAP, MVP, MutationAssessor, MutationTaster, PrimateAI, and SIFT vs. no benign predictions (1 uncertain prediction from DANN).
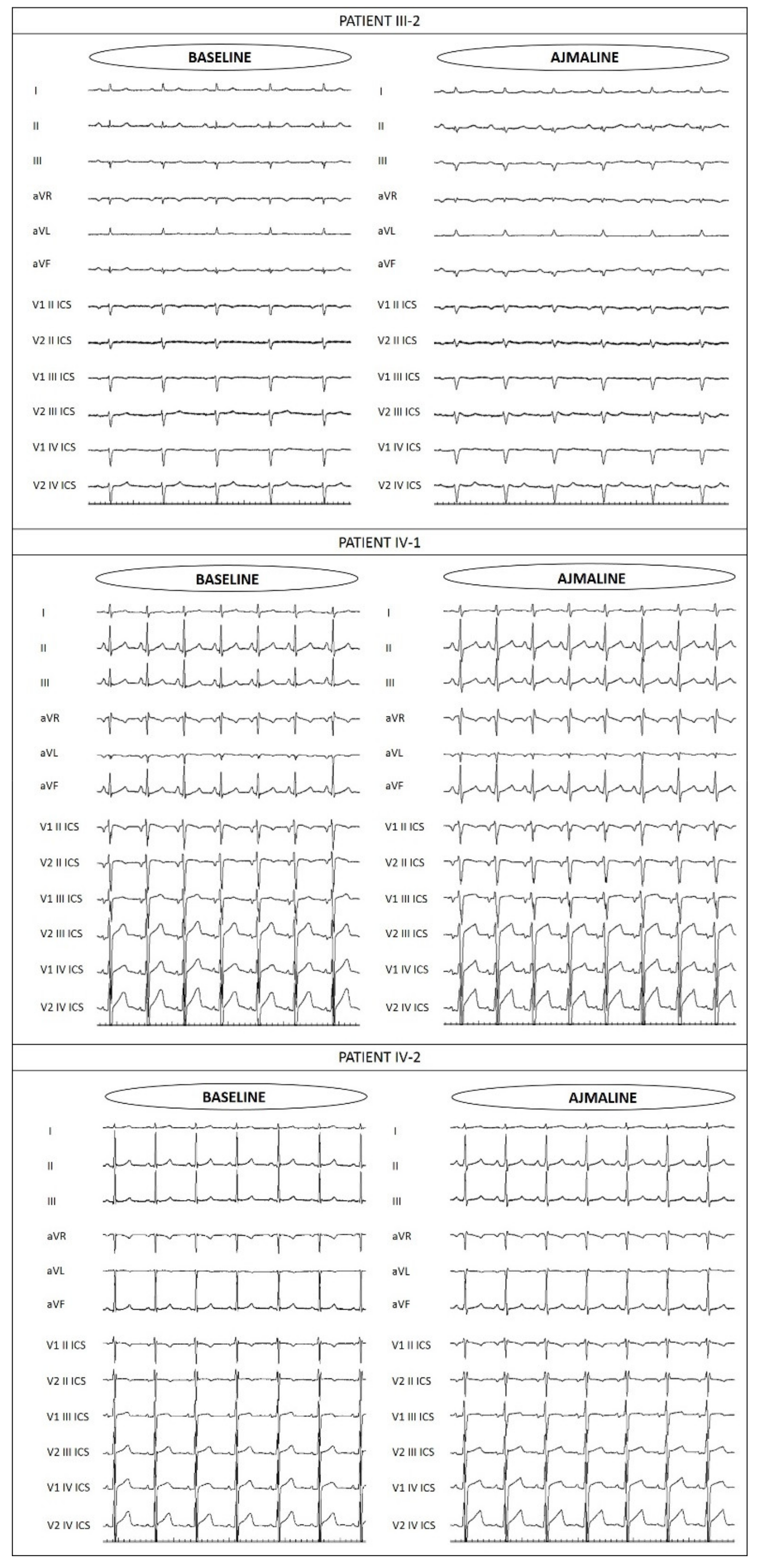
3.3. Assessment of Family Members
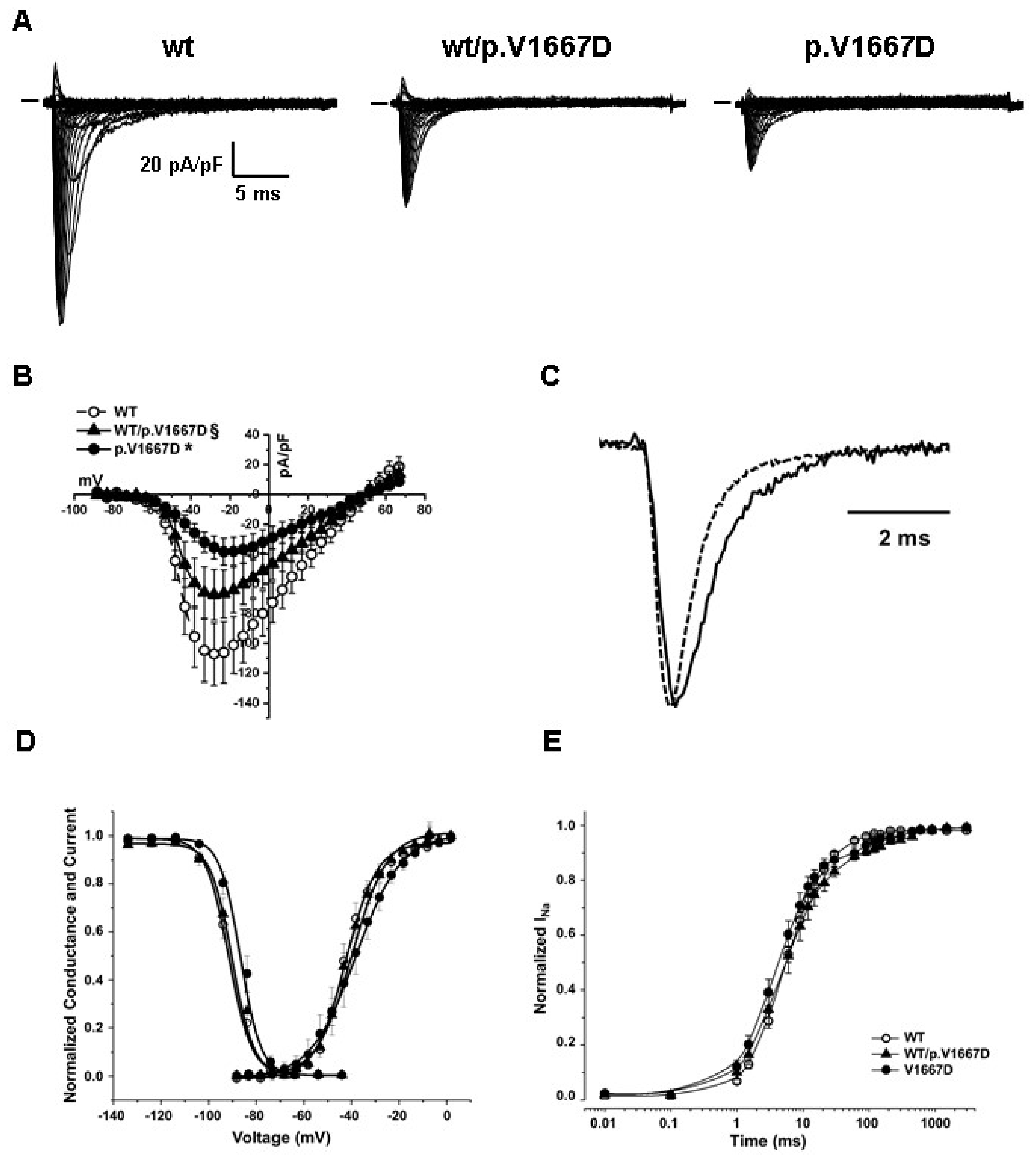
3.4. Functional Analysis of p.V1667D NaV1.5 Channels
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapplinger, J.D.; Tester, D.J.; Alders, M.; Benito, B.; Berthet, M.; Brugada, J.; Brugada, P.; Fressart, V.; Guerchicoff, A.; Harris-Kerr, C.; et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 2010, 7, 33–46. [Google Scholar] [CrossRef]
- Probst, V.; Veltmann, C.; Eckardt, L.; Meregalli, P.G.; Gaita, F.; Tan, H.L.; Babuty, D.; Sacher, F.; Giustetto, C.; Schulze-Bahr, E.; et al. Long-term prognosis of patients diagnosed with Brugada syndrome: Results from the FINGER Brugada Syndrome Registry. Circulation 2010, 121, 635–643. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Kim, R.; Udupa, S.; Costain, G.; Jobling, R.; Liston, E.; Jamal, S.M.; Szybowska, M.; Morel, C.F.; Bowdin, S.; et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death. Circulation 2018, 138, 1195–1205. [Google Scholar] [CrossRef]
- Monasky, M.M.; Pappone, C.; Piccoli, M.; Ghiroldi, A.; Micaglio, E.; Anastasia, L. Calcium in Brugada Syndrome: Questions for Future Research. Front. Physiol. 2018, 9, 1088. [Google Scholar] [CrossRef]
- Denham, N.C.; Pearman, C.M.; Ding, W.Y.; Waktare, J.; Gupta, D.; Snowdon, R.; Hall, M.; Cooper, R.; Modi, S.; Todd, D.; et al. Systematic re-evaluation of SCN5A variants associated with Brugada syndrome. J. Cardiovasc. Electrophysiol. 2019, 30, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Rivolta, I.; Abriel, H.; Tateyama, M.; Liu, H.; Memmi, M.; Vardas, P.; Napolitano, C.; Priori, S.G.; Kass, R.S. Inherited Brugada and long QT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J. Biol. Chem. 2001, 276, 30623–30630. [Google Scholar] [CrossRef] [PubMed]
- Bugiardini, E.; Rivolta, I.; Binda, A.; Soriano Caminero, A.; Cirillo, F.; Cinti, A.; Giovannoni, R.; Botta, A.; Cardani, R.; Wicklund, M.P.; et al. SCN4A mutation as modifying factor of myotonic dystrophy type 2 phenotype. Neuromuscul. Disord. 2015, 25, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- VarSome: The Human Genomics Community. Available online: https://varsome.com/ (accessed on 22 March 2021).
- Belhassen, B. Management of Brugada Syndrome 2016: Should All High Risk Patients Receive an ICD? Alternatives to Implantable Cardiac Defibrillator Therapy for Brugada Syndrome. Circ. Arrhythm. Electrophysiol. 2016, 9, e004185. [Google Scholar] [CrossRef]
- Brodehl, A.; Ebbinghaus, H.; Deutsch, M.A.; Gummert, J.; Gartner, A.; Ratnavadivel, S.; Milting, H. Human Induced Pluripotent Stem-Cell-Derived Cardiomyocytes as Models for Genetic Cardiomyopathies. Int. J. Mol. Sci. 2019, 20, 4381. [Google Scholar] [CrossRef]
- Hu, J.; Han, J.; Li, H.; Zhang, X.; Liu, L.L.; Chen, F.; Zeng, B. Human Embryonic Kidney 293 Cells: A Vehicle for Biopharmaceutical Manufacturing, Structural Biology, and Electrophysiology. Cells Tiss. Organs 2018, 205, 1–8. [Google Scholar] [CrossRef]
- Walsh, R.; Tadros, R.; Bezzina, C.R. When genetic burden reaches threshold. Eur. Heart J. 2020, 41, 3849–3855. [Google Scholar] [CrossRef] [PubMed]
- Monasky, M.M.; Micaglio, E.; Ciconte, G.; Pappone, C. Brugada Syndrome: Oligogenic or Mendelian Disease? Int. J. Mol. Sci. 2020, 21, 1687. [Google Scholar] [CrossRef]
- Piippo, K.; Holmstrom, S.; Swan, H.; Viitasalo, M.; Raatikka, M.; Toivonen, L.; Kontula, K. Effect of the antimalarial drug halofantrine in the long QT syndrome due to a mutation of the cardiac sodium channel gene SCN5A. Am. J. Cardiol. 2001, 87, 909–911. [Google Scholar] [CrossRef]
- Nakajima, T.; Dharmawan, T.; Kawabata-Iwakawa, R.; Tamura, S.; Hasegawa, H.; Kobari, T.; Kaneko, Y.; Nishiyama, M.; Kurabayashi, M. Biophysical defects of an SCN5A V1667I mutation associated with epinephrine-induced marked QT prolongation. J. Cardiovasc. Electrophysiol. 2020, 31, 2107–2115. [Google Scholar] [CrossRef]
- Chen, C.; Tan, Z.; Zhu, W.; Fu, L.; Kong, Q.; Xiong, Q.; Yu, J.; Hong, K. Brugada syndrome with SCN5A mutations exhibits more pronounced electrophysiological defects and more severe prognosis: A meta-analysis. Clin. Genet. 2020, 97, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K.; Horie, M.; Aiba, T.; Ogawa, S.; Aizawa, Y.; Ohe, T.; Yamagishi, M.; Makita, N.; Sakurada, H.; Tanaka, T.; et al. Genotype-Phenotype Correlation of SCN5A Mutation for the Clinical and Electrocardiographic Characteristics of Probands With Brugada Syndrome: A Japanese Multicenter Registry. Circulation 2017, 135, 2255–2270. [Google Scholar] [CrossRef]
- Priori, S.G.; Napolitano, C.; Gasparini, M.; Pappone, C.; Della Bella, P.; Giordano, U.; Bloise, R.; Giustetto, C.; De Nardis, R.; Grillo, M.; et al. Natural history of Brugada syndrome: Insights for risk stratification and management. Circulation 2002, 105, 1342–1347. [Google Scholar] [CrossRef]
- Antzelevitch, C.; Yan, G.X.; Ackerman, M.J.; Borggrefe, M.; Corrado, D.; Guo, J.; Gussak, I.; Hasdemir, C.; Horie, M.; Huikuri, H.; et al. J-Wave syndromes expert consensus conference report: Emerging concepts and gaps in knowledge. J. Arrhythm. 2016, 32, 315–339. [Google Scholar] [CrossRef] [PubMed]
- Antzelevitch, C.; Yan, G.X.; Ackerman, M.J.; Borggrefe, M.; Corrado, D.; Guo, J.; Gussak, I.; Hasdemir, C.; Horie, M.; Huikuri, H.; et al. J-Wave syndromes expert consensus conference report: Emerging concepts and gaps in knowledge. Heart Rhythm 2016, 13, e295–e324. [Google Scholar] [CrossRef] [PubMed]
- Antzelevitch, C.; Yan, G.X.; Ackerman, M.J.; Borggrefe, M.; Corrado, D.; Guo, J.; Gussak, I.; Hasdemir, C.; Horie, M.; Huikuri, H.; et al. J-Wave syndromes expert consensus conference report: Emerging concepts and gaps in knowledge. Europace 2017, 19, 665–694. [Google Scholar] [PubMed]
| WT | WT/p.V1667D | p.V1667D | ||
|---|---|---|---|---|
| Current density @-28 mV (pA/pF) | −107.11 ± 21 (n = 16) | −67.30 ± 17 * (n = 16) | −35.58 ± 7 * (n = 13) | |
| Time to peak (@-28 mV, msec) | 0.58 ± 0.02 | 0.70 ± 0.03 * | 0.69 ± 0.04* | |
| Kinetic of fast inactivation (@-28 mV) | τ (ms) | 0.75 ± 0.02 (n = 17) | 1.02 ± 0.1 * (n = 10) | 0.94 ± 0.04 * (n = 10) |
| Steady state of activation | V1/2 (mV) | −42.6 ± 1.8 (n = 16) | −41.6 ± 1.5 (n = 16) | −38.9 ± 3.5 (n=13) |
| k | 4.5 ± 0.3 | 4.8 ± 0.1 | 5.3 ± 0.5 * | |
| Availability curve | V1/2 (mV) | −90.9 ± 1.3 (n = 20) | −89.3 ± 1.9 (n = 13) | −85.8 ± 1.6 * (n = 12) |
| k | 4.8 ± 0.2 | 4.8 ± 0.3 | 4.6 ± 0.3 | |
| Recovery from inactivation | t1 (ms) | 7.0 ± 0.5 (n = 18) | 6.0 ± 1 (n = 8) | 5.4 ± 0.5 (n = 11) |
| t2 (ms) | 67.30 ± 14 | 193.3 ± 31 * | 142.5 ± 22 * | |
| Development of intermediate inactivation | t1 (ms) | 485 ± 114 (n = 9) | 492 ± 253 (n = 9) | 469 ± 178 (n = 6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monasky, M.M.; Micaglio, E.; Ciconte, G.; Rivolta, I.; Borrelli, V.; Ghiroldi, A.; D’Imperio, S.; Binda, A.; Melgari, D.; Benedetti, S.; et al. Novel SCN5A p.Val1667Asp Missense Variant Segregation and Characterization in a Family with Severe Brugada Syndrome and Multiple Sudden Deaths. Int. J. Mol. Sci. 2021, 22, 4700. https://doi.org/10.3390/ijms22094700
Monasky MM, Micaglio E, Ciconte G, Rivolta I, Borrelli V, Ghiroldi A, D’Imperio S, Binda A, Melgari D, Benedetti S, et al. Novel SCN5A p.Val1667Asp Missense Variant Segregation and Characterization in a Family with Severe Brugada Syndrome and Multiple Sudden Deaths. International Journal of Molecular Sciences. 2021; 22(9):4700. https://doi.org/10.3390/ijms22094700
Chicago/Turabian StyleMonasky, Michelle M., Emanuele Micaglio, Giuseppe Ciconte, Ilaria Rivolta, Valeria Borrelli, Andrea Ghiroldi, Sara D’Imperio, Anna Binda, Dario Melgari, Sara Benedetti, and et al. 2021. "Novel SCN5A p.Val1667Asp Missense Variant Segregation and Characterization in a Family with Severe Brugada Syndrome and Multiple Sudden Deaths" International Journal of Molecular Sciences 22, no. 9: 4700. https://doi.org/10.3390/ijms22094700
APA StyleMonasky, M. M., Micaglio, E., Ciconte, G., Rivolta, I., Borrelli, V., Ghiroldi, A., D’Imperio, S., Binda, A., Melgari, D., Benedetti, S., Mitrovic, P., Anastasia, L., Mecarocci, V., Ćalović, Ž., Casari, G., & Pappone, C. (2021). Novel SCN5A p.Val1667Asp Missense Variant Segregation and Characterization in a Family with Severe Brugada Syndrome and Multiple Sudden Deaths. International Journal of Molecular Sciences, 22(9), 4700. https://doi.org/10.3390/ijms22094700








