pCTX-M3—Structure, Function, and Evolution of a Multi-Resistance Conjugative Plasmid of a Broad Recipient Range
Abstract
1. Introduction
2. Isolation of pCTX-M3
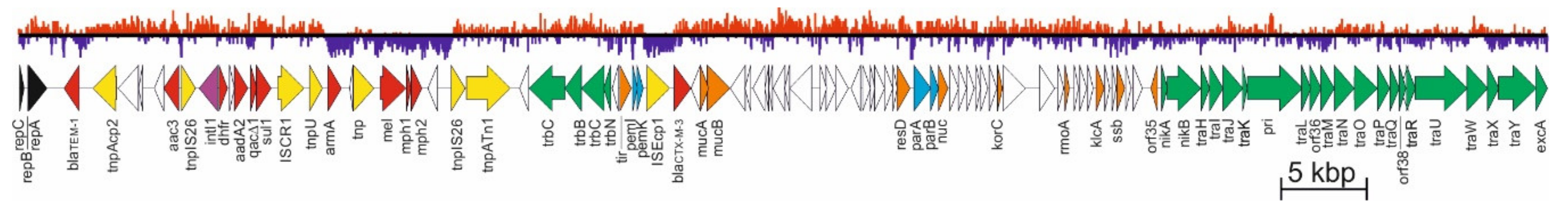
3. Analysis of pCTX-M3 Nucleotide Sequence
3.1. Replicon
3.2. Stable Maintenance Systems
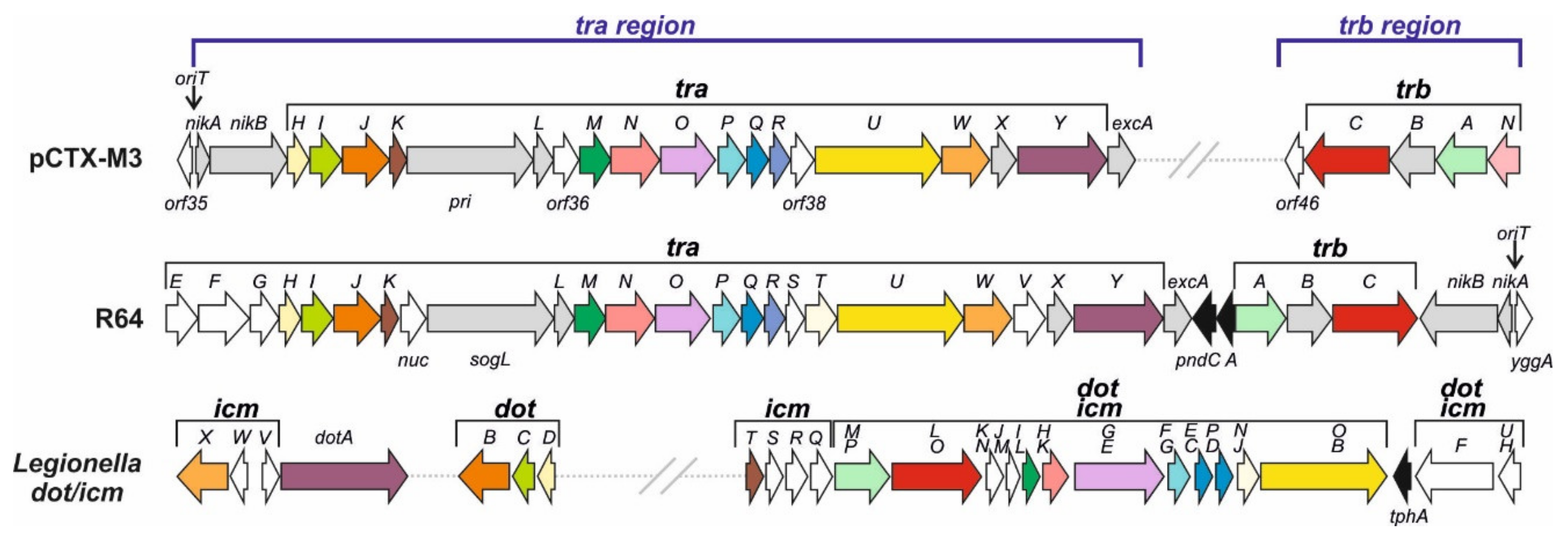
3.2.1. pCTX-M3 Conjugative Transfer System
- The transmembrane core subcomplex. Its OMC comprises TraN, TraI, and TraH, the homologues of Legionella DotH, DotC, and DotD, respectively. These Legionella proteins localize to the outer membrane forming the pore, similar to that formed by the VirB7–VirB9 proteins; by combining with the inner membrane proteins DotF and DotG they form the transmembrane core subcomplex [40]. In pCTX-M3, the DotF and DotG homologues are TraP and TraO; the latter is also a distant homologue of VirB10 [21,41];
- The pilus—the TraR and TraQ proteins are distant homologues of VirB2, the major pilus subunit;
- The VirB11 and VirB4 ATPases—TraJ is a homologue of VirB11 and TraU is a distant homologue of Legionella DotO and Agrobacterium VirB4, the only component common to all T4SSs of Gram-negative and Gram-positive bacteria, and archaea [39];
- The coupling protein—the putative CP is TrbC. The CP subcomplex may also comprise TrbA, a DotM homologue;
- The entry exclusion system—TraY and ExcA of pCTX-M3 are close homologues of respective R64 proteins [42].
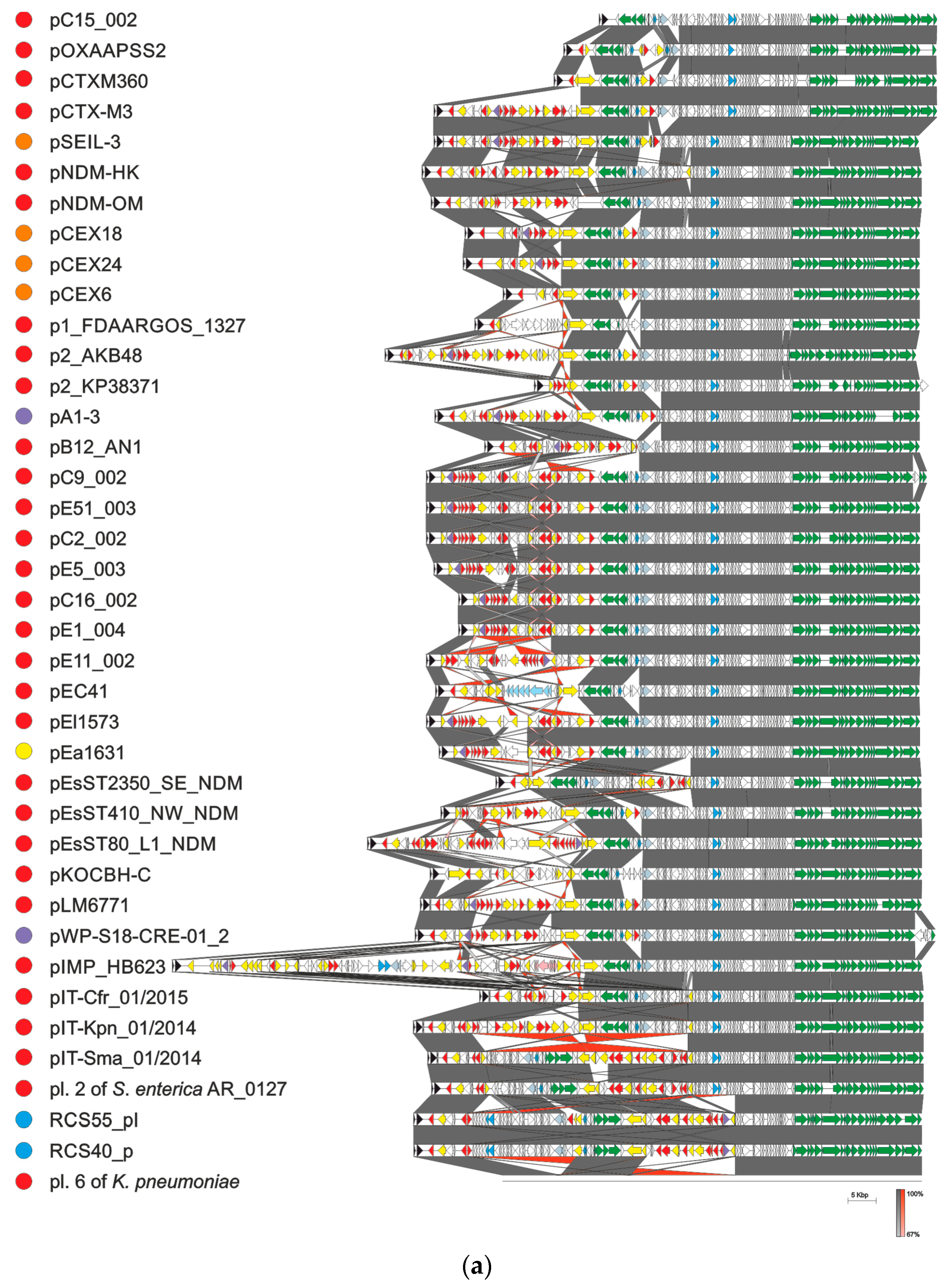
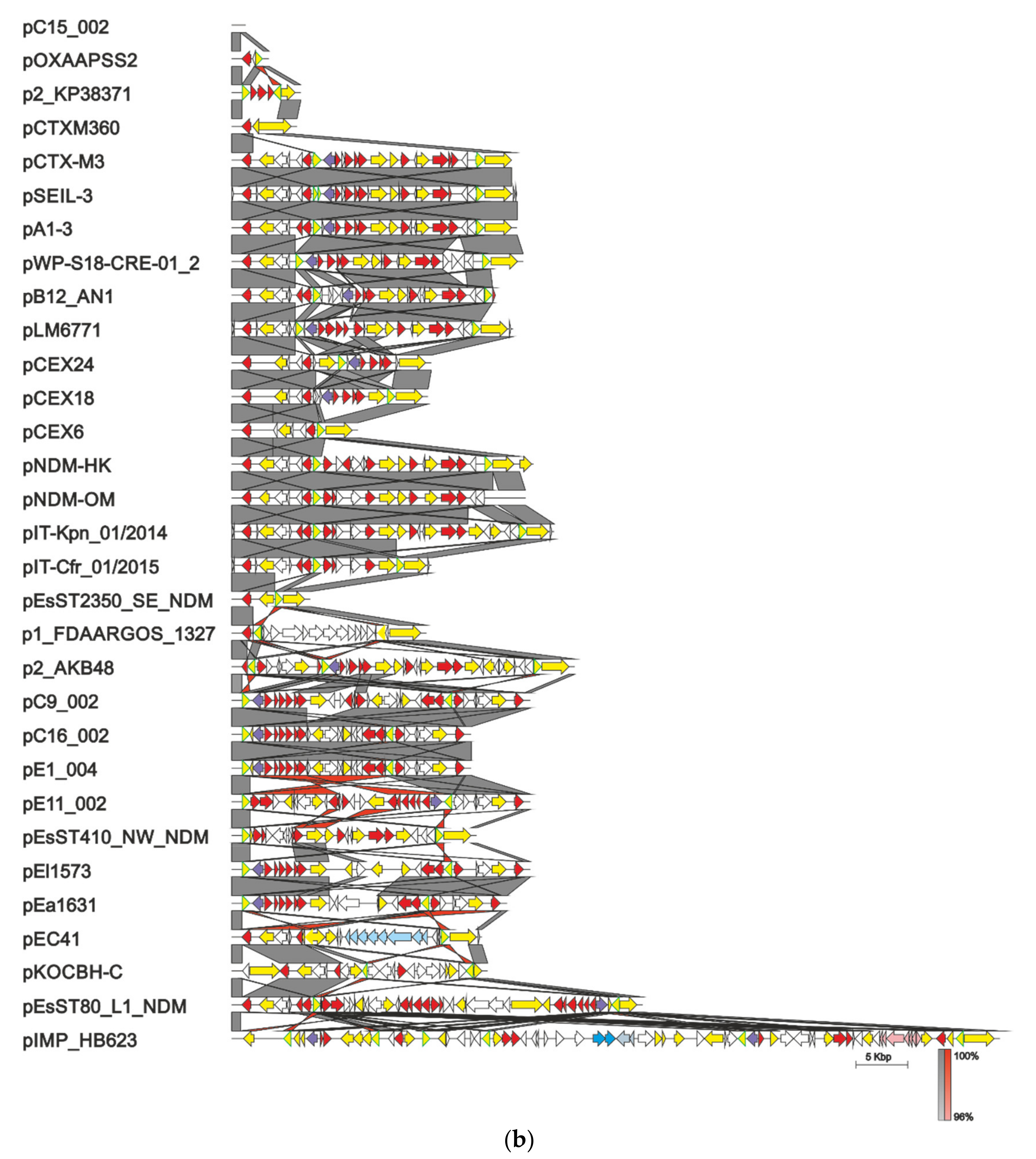
4. Evolution of pCTX-M3-Related IncM Plasmids
- β-lactam antibiotics—blaTEM-1, usually located in proximity of the rep region, blaIMP-4 located within the integron, and also blaNDM-1, blaDHA-1, blaOXA-16, blaOXA-48, and blaIMP-34;
- aminoglycosides—armA (nbrB) imported into the plasmid with ISCR1 as a part of a composite transposon flanked by two IS26 elements, aac(6’)-Ib4 related to IS26 and aac(3)-IId located within an integron, also aac(6’)-Ib-cr, aadA1, aadA16, aadA2, and strAB;
- macrolides—mel, mph1, and mph2, frequently ISCR1 related;
- chloramphenicol—catB3 located within the integron;
- quinolones—qnrB2 and aac(6’)-Ib-cr;
- sulphonamides and trimethoprim—sul1 and dfrA12, located within an integron, and also dfrA14 or dfrA27;
- quaternary ammonium compounds—qacE and qacG2;
- mercury—the mer operon;
- fosfomycin—fosC2;
- bleomycin—bleMBL.
5. Taxonomy of IncL/M Plasmids
6. Conclusions
7. Patents
Funding
Acknowledgments
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morgane. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef]
- Cornaglia, G.; Garau, J.; Livermore, D.M. Living with ESBLs. Clin. Microbiol. Infect. 2007, 14, 1–2. [Google Scholar] [CrossRef]
- Livermore, D.M.; Canton, R.; Gniadkowski, M.; Nordmann, P.; Rossolini, G.M.; Arlet, G.; Ayala, J.; Coque, T.M.; Kern-Zdanowicz, I.; Luzzaro, F.; et al. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2007, 59, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, A.; Schweighart, S.; Grimm, H. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 1990, 18, 294–298. [Google Scholar] [CrossRef]
- Rodríguez, M.M.; Power, P.; Radice, M.; Vay, C.; Famiglietti, A.; Galleni, M.; Ayala, J.A.; Gutkind, G. Chromosome-Encoded CTX-M-3 from Kluyvera ascorbata: A Possible Origin of Plasmid-Borne CTX-M-1-Derived Cefotaximases. Antimicrob. Agents Chemother. 2004, 48, 4895–4897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Olson, A.B.; Silverman, M.; Boyd, D.A.; Mcgeer, A.; Willey, B.M.; Daneman, N.; Mulvey, M.R. Identification of a Progenitor of the CTX-M-9 Group of Extended-Spectrum beta-Lactamases from Kluyvera georgiana Isolated in Guyana. Antimicrob. Agents Chemother. 2005, 49, 2112–2115. [Google Scholar] [CrossRef]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Woodford, N. Successful, multiresistant bacterial clones. J. Antimicrob. Chemother. 2008, 61, 233–234. [Google Scholar] [CrossRef]
- Gniadkowski, M.; Schneider, I.; Pałucha, A.; Jungwirth, R.; Mikiewicz, B.; Bauernfeind, A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: Identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 1998, 42, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, A.; Fiett, J.; Sulikowska, A.; Hryniewicz, W.; Gniadkowski, M. Countrywide Spread of CTX-M-3 Extended-Spectrum β-Lactamase-Producing Microorganisms of the Family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 2002, 46, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Gołębiewski, M.; Kern-Zdanowicz, I.; Zienkiewicz, M.; Adamczyk, M.; Żylinska, J.; Baraniak, A.; Gniadkowski, M.; Bardowski, J.; Cegłowski, P. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 2007, 51, 3789–3795. [Google Scholar] [CrossRef]
- Granier, S.A.; Hidalgo, L.; San Millan, A.; Escudero, J.A.; Gutierrez, B.; Brisabois, A.; Gonzalez-Zorn, B. ArmA Methyltransferase in a Monophasic Salmonella enterica Isolate from Food. Antimicrob. Agents Chemother. 2011, 55, 5262–5266. [Google Scholar] [CrossRef] [PubMed]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. ISCR elements: Novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 2006, 70, 296–316. [Google Scholar] [CrossRef]
- Blackwell, G.A.; Holt, K.E.; Bentley, S.D.; Hsu, L.Y.; Hall, R.M. Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 2017, 72, dkw542. [Google Scholar] [CrossRef]
- Athanasopoulos, V.; Praszkier, J.; Pittard, A.J. The replication of an IncL/M plasmid is subject to antisense control. J. Bacteriol. 1995, 177, 4730–4741. [Google Scholar] [CrossRef][Green Version]
- Athanasopoulos, V.; Praszkier, J.; Pittard, A.J. Analysis of elements involved in pseudoknot-dependent expression and regulation of the repA gene of an IncL/M plasmid. J. Bacteriol. 1999, 181, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Mierzejewska, J.; Kulińska, A.; Jagura-Burdzy, G. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 2007, 57, 95–107. [Google Scholar] [CrossRef]
- Carattoli, A.; Seiffert, S.N.; Schwendener, S.; Perreten, V.; Endimiani, A. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS ONE 2015, 10, e0123063. [Google Scholar] [CrossRef]
- Osborn, A.M.; Fernanda, M.; Tatley, S.; Steyn, L.M.; Pickup, R.W.; Saunders, J.R. Mosaic plasmids and mosaic replicons: Evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 2000, 146, 2267–2275. [Google Scholar] [CrossRef]
- Dmowski, M.; Gołębiewski, M.; Kern-Zdanowicz, I. Characteristics of the conjugative transfer system of the IncM plasmid pCTX-M3 and identification of its putative regulators. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Salje, J.; Gayathri, P.; Löwe, J. The ParMRC system: Molecular mechanisms of plasmid segregation by actin-like filaments. Nat. Rev. Microbiol. 2010, 8, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Komano, T.; Yoshida, T.; Narahara, K.; Furuya, N. The transfer region of Incl1 plasmid R64: Similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 2000, 35, 1348–1359. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Zhu, L.; Suzuki, M.; Inouye, M. Interference of mRNA Function by Sequence-specific Endoribonuclease PemK*. J. Biol. Chem. 2004, 279. [Google Scholar] [CrossRef]
- Dmowski, M.; Kern-Zdanowicz, I. A novel mobilizing tool based on the conjugative transfer system of the IncM Plasmid pCTX-M3. Appl. Environ. Microbiol. 2020, 86, 1–18. [Google Scholar] [CrossRef]
- Yoshida, T.; Furuya, N.; Ishikura, M.; Isobe, T.; Haino-fukushima, K.; Ogawa, T.; Komano, T. Purification and Characterization of Thin Pili of IncI1 Plasmids ColIb-P9 and R64: Formation of PilV-Specific Cell Aggregates by Type IV Pili. J. Bacteriol. 1998, 180, 2842–2848. [Google Scholar] [CrossRef]
- Bradley, D.E. Determination of pili by conjugative bacterial drug resistance plasmids of incompatibility groups B, C, H, J, K, M, V, and X. J. Bacteriol. 1980, 141, 828–837. [Google Scholar] [CrossRef]
- Llosa, M.; Gomis-Rüth, F.X.; Coll, M.; de la Cruz, F. Bacterial conjugation: A two-step mechanism for DNA transport. Mol. Microbiol. 2002, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zechner, E.L.; Lang, S.; Schildbach, J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1073–1087. [Google Scholar] [CrossRef]
- Koraimann, G.; Wagner, M.A. Social behavior and decision making in bacterial conjugation. Front. Cell. Infect. Microbiol. 2014, 4, 54. [Google Scholar] [CrossRef] [PubMed]
- Wallden, K.; Rivera-Calzada, A.; Waksman, G. Type IV secretion systems: Versatility and diversity in function. Cell. Microbiol. 2010, 12, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Hirt, H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010, 29, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Fronzes, R.; Schäfer, E.; Wang, L.; Saibil, H.R.; Orlova, E.V.; Waksman, G. Structure of a Type IV Secretion System Core Complex. Science 2009, 323, 266–268. [Google Scholar] [CrossRef]
- Christie, P.J. The mosaic type IV secretion systems. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Bhatty, M.; Laverde Gomez, J.A.; Christie, P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013, 164, 620–639. [Google Scholar] [CrossRef]
- Grohmann, E.; Christie, P.J.; Waksman, G.; Backert, S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 2018, 107, 455–471. [Google Scholar] [CrossRef]
- Segal, G.; Feldman, M.; Zusman, T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2005, 29, 65–81. [Google Scholar] [CrossRef]
- Nagai, H.; Kubori, T. Type IVB secretion systems of Legionella and other gram-negative bacteria. Front. Microbiol. 2011, 2, 1–12. [Google Scholar] [CrossRef]
- Guglielmini, J.; Bertrand, N.; Abby, S.S.; Garcillan-Barcia, M.P.; de la Cruz, F.; Rocha, E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or. Nucleic Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.D.; Friedman, J.R.; Jeong, K.C.; Sutherland, M.C.; Vogel, J.P. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 2012, 85, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Quintais, L.; Garcillán-Barcia, M.P.; de la Cruz, F.; Rocha, E.P.C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011, 7, e1002222. [Google Scholar] [CrossRef]
- Sakuma, T.; Tazumi, S.; Furuya, N.; Komano, T. ExcA proteins of IncI1 plasmid R64 and IncIγ plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid 2013, 69, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, M.; Kern-Zdanowicz, I. Attempts to overproduce the product of the orf36 gene of the pCTX-M3 plasmid of Enterobacteriaceae; Institute of Biochemistry and Biophysics, Polish Academy of Sciences: Warsaw, Poland, 2017. [Google Scholar]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Tietze, E.; Tschäpe, H. Temperature-dependent expression of conjugation pili by IncM plasmid-harbouring bacteria: Identification of plasmid-encoded regulatory functions. J. Basic Microbiol. 1994, 34, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Foster, G.C.; McGhee, G.C.; Jones, A.L.; Sundin, G.W. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 2004, 70, 7539–7544. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, W.; Luo, L.; Wang, J.; Zhuang, X.; Zhong, L.; Liao, K.; Zeng, Y.; Lu, Y. Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 2009, 53, 5291–5293. [Google Scholar] [CrossRef]
- Ho, P.L.; Lo, W.U.; Yeung, M.K.; Lin, C.H.; Chow, K.H.; Ang, I.; Tong, A.H.Y.; Bao, J.Y.-J.; Lok, S.; Lo, J.Y.C. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS ONE 2011, 6, e17989. [Google Scholar] [CrossRef][Green Version]
- Bonnin, R.A.; Nordmann, P.; Carattoli, A.; Poirel, L. Comparative Genomics of IncL/M-Type Plasmids: Evolution by Acquisition of Resistance Genes and Insertion Sequences. Antimicrob. Agents Chemother. 2013, 57, 674–676. [Google Scholar] [CrossRef]
- Wang, Y.; Lo, W.-U.; Lai, E.L.; Chow, K.-H.; Ho, P.-L. Complete Sequence of the Multidrug-Resistant IncL/M Plasmid pIMP-HB623 Cocarrying blaIMP-34 and fosC2 in an Enterobacter cloacae Strain Associated with Medical Travel to China. Antimicrob. Agents Chemother. 2015, 59. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Thomas, L.C.; Ginn, A.N.; Wiklendt, A.M.; Kyme, P.; Iredell, J.R. A novel gene cassette, aacA43, in a plasmid-borne class 1 integron. Antimicrob. Agents Chemother. 2011, 55, 2979–2982. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hall, R.M. IS26-Mediated Formation of Transposons Carrying Antibiotic Resistance Genes. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, G.A.; Doughty, E.L.; Moran, R.A. Evolution and dissemination of L and M plasmid lineages carrying antibiotic resistance genes in diverse Gram-negative bacteria. Plasmid 2021, 113, 102528. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kern-Zdanowicz, I. pCTX-M3—Structure, Function, and Evolution of a Multi-Resistance Conjugative Plasmid of a Broad Recipient Range. Int. J. Mol. Sci. 2021, 22, 4606. https://doi.org/10.3390/ijms22094606
Kern-Zdanowicz I. pCTX-M3—Structure, Function, and Evolution of a Multi-Resistance Conjugative Plasmid of a Broad Recipient Range. International Journal of Molecular Sciences. 2021; 22(9):4606. https://doi.org/10.3390/ijms22094606
Chicago/Turabian StyleKern-Zdanowicz, Izabela. 2021. "pCTX-M3—Structure, Function, and Evolution of a Multi-Resistance Conjugative Plasmid of a Broad Recipient Range" International Journal of Molecular Sciences 22, no. 9: 4606. https://doi.org/10.3390/ijms22094606
APA StyleKern-Zdanowicz, I. (2021). pCTX-M3—Structure, Function, and Evolution of a Multi-Resistance Conjugative Plasmid of a Broad Recipient Range. International Journal of Molecular Sciences, 22(9), 4606. https://doi.org/10.3390/ijms22094606





