YABBY Genes in the Development and Evolution of Land Plants
Abstract
1. Introduction
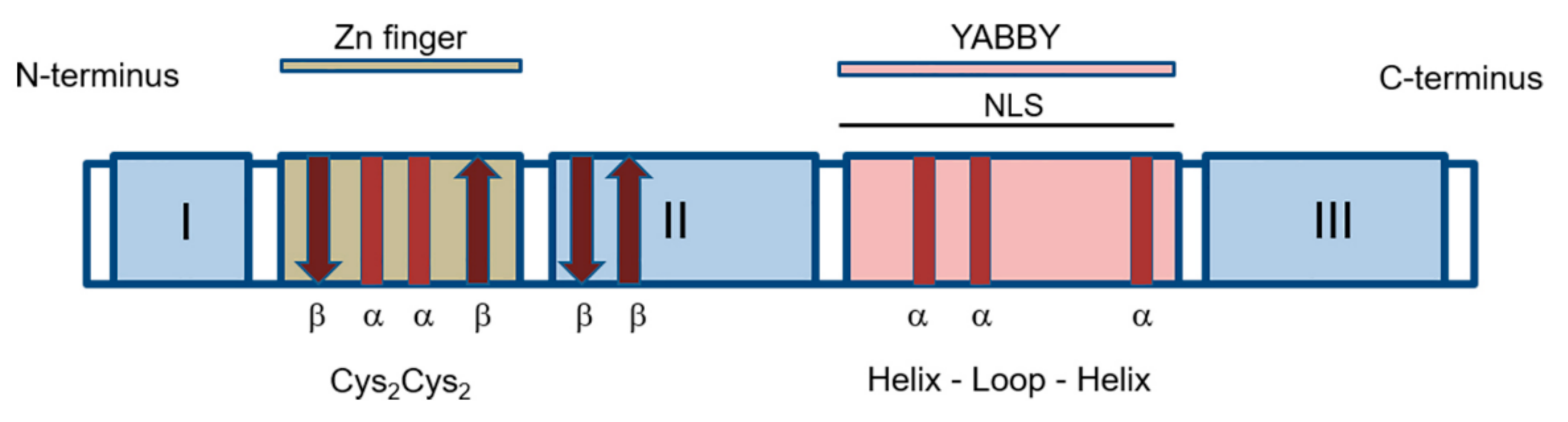
2. Molecular Characterization of YABBY Proteins
3. Phylogeny of YABBY Genes
4. Expression Patterns and Putative Functions of Members of the YABBY Gene Family
4.1. “Vegetative” YABBY Genes
4.2. The “Reproductive” YABBY Genes
4.2.1. CRC Gene Subfamily
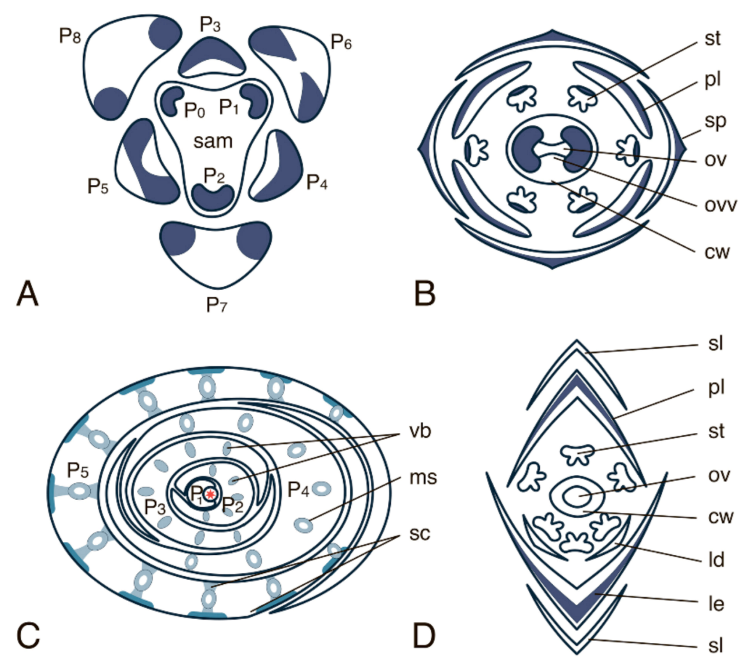
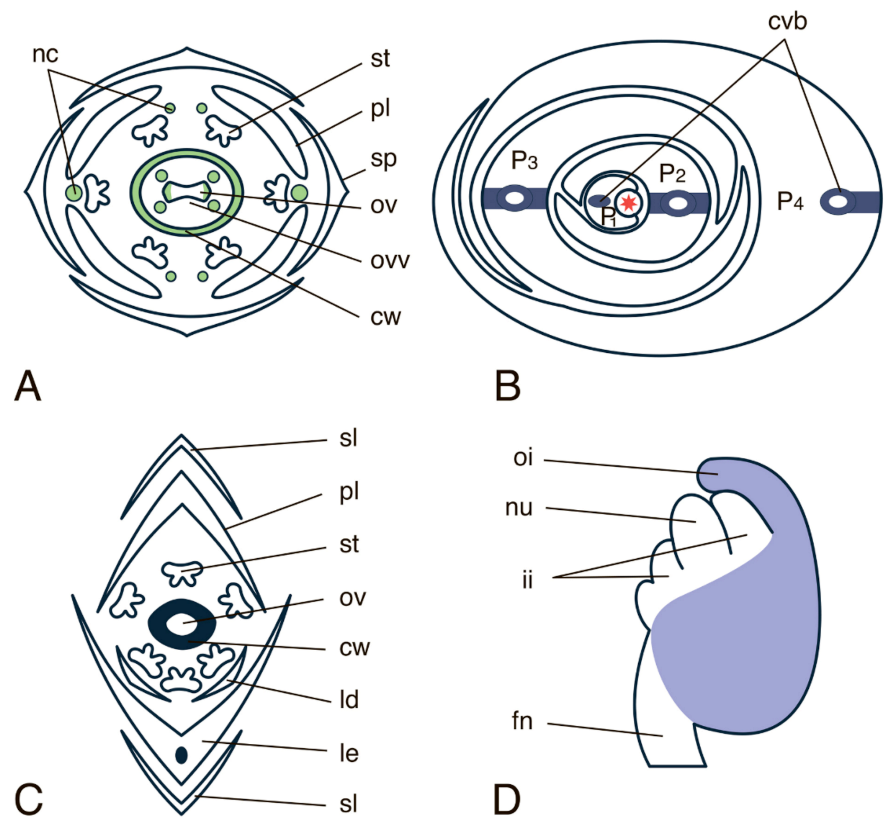
4.2.2. INO (INNER NO OUTER) Gene Subfamily
4.3. Comparison of YABBY Genes in Different Groups of Plants
4.3.1. YABBY Genes in Angiosperms
4.3.2. YABBY Genes in Gymnosperms
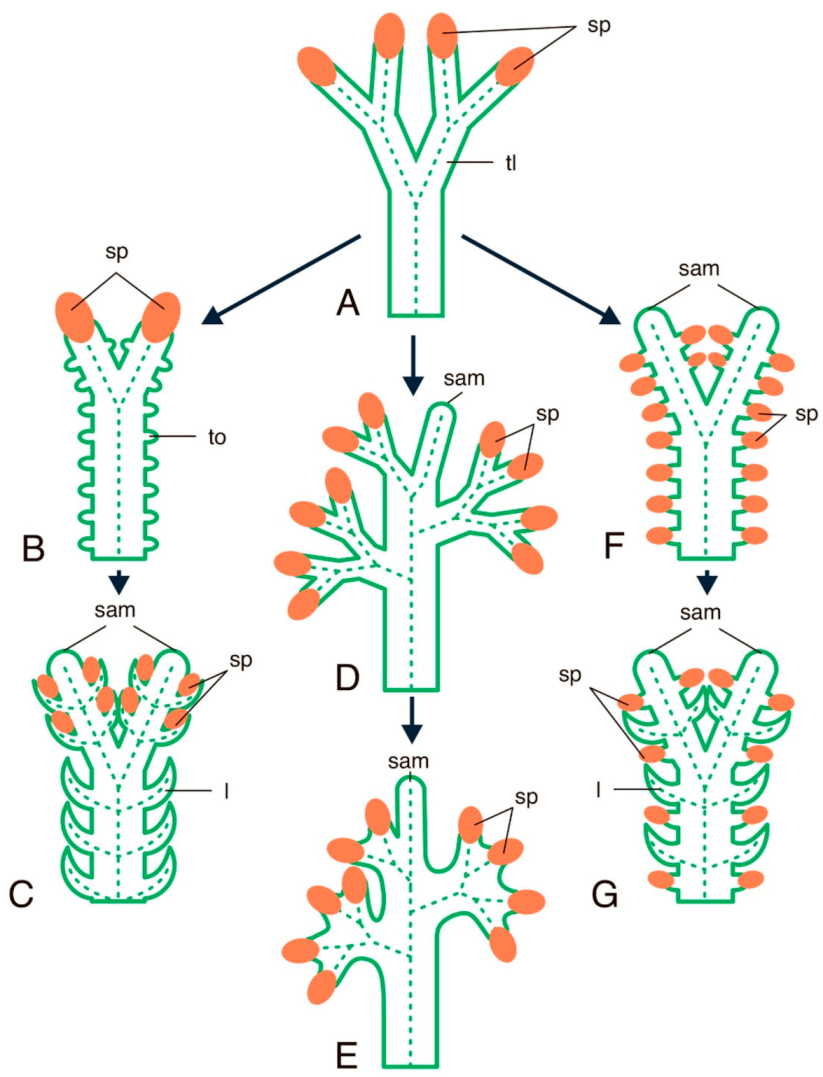
4.3.3. YABBY Genes in Seedless Plants
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gifford, E.M.; Foster, A.S. Morphology and Evolution of Vascular Plants, 3rd ed.; W. H. Freeman and Co.: New York, NY, USA, 1989; ISBN 0716719460. [Google Scholar]
- Kenrick, P.; Crane, P.R. The origin and early evolution of plants on land. Nature 1997, 389, 33–39. [Google Scholar] [CrossRef]
- Zimmerman, W. Main results of the telome theory. Paleobotanist 1952, 1, 456–470. [Google Scholar]
- Zimmermann, W. Die Phylogenie de Pflanzen: Ein Uberblick über TATSACHEN und Probleme; G. Fischer: Jena, Germany, 1930; p. 452. [Google Scholar]
- Tomescu, A.M.F. Megaphylls, microphylls and the evolution of leaf development. Trends Plant Sci. 2009, 14, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Tomescu, A.M.F.; Wyatt, S.E.; Hasebe, M.; Rothwell, G.W. Early evolution of the vascular plant body plan—The missing mechanisms. Curr. Opin. Plant Biol. 2014, 17, 126–136. [Google Scholar] [CrossRef]
- Vasco, A.; Ambrose, B.A. Simple and divided leaves in ferns: Exploring the genetic basis for leaf morphology differences in the genus Elaphoglossum (Dryopteridaceae). Int. J. Mol. Sci. 2020, 21, 5180. [Google Scholar] [CrossRef]
- Vasco, A.; Smalls, T.L.; Graham, S.W.; Cooper, E.D.; Wong, G.K.; Stevenson, D.W.; Moran, R.C.; Ambrose, B.A. Challenging the paradigms of leaf evolution: Class III HD-Zips in ferns and lycophytes. New Phytol. 2016, 212, 745–758. [Google Scholar] [CrossRef]
- Cronk, Q. Plant evolution and development in a post-genomic context. Nat. Rev. Genet. 2001, 2, 607–619. [Google Scholar] [CrossRef]
- Friedman, W.E.; Moore, R.C.; Purugganan, M.D. The evolution of plant development. Am. J. Bot. 2004, 91, 1726–1741. [Google Scholar] [CrossRef]
- Du, H.; Ran, J.-H.; Feng, Y.-Y.; Wang, X.-Q. The flattened and needlelike leaves of the pine family (Pinaceae) share a conserved genetic network for adaxial-abaxial polarity but have diverged for photosynthetic adaptation. BMC Evol. Biol. 2020, 20, 131. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix- loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar]
- Sawa, S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A.A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef]
- Itoh, J.-I.; Hibara, K.-I.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef]
- Caggiano, M.P.; Yu, X.; Bhatia, N.; Larsson, A.; Ram, H.; Ohno, C.K.; Sappl, P.; Meyerowitz, E.M.; Jönsson, H.; Heisler, M.G. Cell type boundaries organize plant development. eLife 2017, 6, e27421. [Google Scholar] [CrossRef]
- Xue, Z.; Liu, L.; Zhang, C. Regulation of Shoot apical meristem and axillary meristem development in plants. Int. J. Mol. Sci. 2020, 21, 2917. [Google Scholar] [CrossRef]
- Bowman, J.L.; Eshed, Y.; Baum, S.F. Establishment of polarity in angiosperm lateral organs. Trends Genet. 2002, 18, 134–141. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Nukazuka, A.; Tsukaya, H. Leaf adaxial–abaxial polarity specification and lamina outgrowth: Evolution and development. Plant Cell Physiol. 2012, 53, 1180–1194. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, F.; Jackson, D. All together now, a magical mystery tour of the maize shoot meristem. Curr. Opin. Plant Biol. 2018, 45, 26–35. [Google Scholar] [CrossRef]
- Finet, C.; Floyd, S.K.; Conway, S.J.; Zhong, B.; Scutt, C.P.; Bowman, J.L. Evolution of the YABBY gene family in seed plants. Evol. Dev. 2016, 18, 116–126. [Google Scholar] [CrossRef]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; de Pamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr. Biol. 2006, 16, 1911–1917. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.K.; Bowman, J.L. The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 2007, 168, 1–35. [Google Scholar] [CrossRef]
- Floyd, S.K.; Zalewski, C.S.; Bowman, J.L. Evolution of Class III homeodomain-leucine zipper genes in streptophytes. Genetics 2006, 173, 373–388. [Google Scholar] [CrossRef]
- Harrison, C.J.; Corley, S.B.; Moylan, E.C.; Alexander, D.L.; Scotland, R.W.; Langdale, J.A. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 2005, 434, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Prigge, M.; Clark, S. Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 2006, 8, 350–361. [Google Scholar] [CrossRef]
- Sano, R.; Juárez, C.M.; Hass, B.; Sakakibara, K.; Ito, M.; Banks, J.A.; Hasebe, M. KNOX homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems. Evol. Dev. 2005, 7, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zumajo-Cardona, C.; Vasco, A.; Ambrose, B.A. The evolution of the KANADI gene family and leaf development in lycophytes and ferns. Plants 2019, 8, 313. [Google Scholar] [CrossRef]
- Maksimova, A.I.; Berke, L.; Saldago, M.; Klimova, E.; Pawlowski, K.; Romanova, M.; Voitsekhovskaja, O. What can the phylogeny of class I KNOX genes and their expression patterns in land plants tell us about the evolution of shoot development? Bot. J. Linn. Soc. 2021, 195, 254–280. [Google Scholar] [CrossRef]
- Hernández-Hernández, B.; Tapia-López, R.; Ambrose, B.A.; Vasco, A. R2R3-MYB gene evolution in plants, incorporating ferns into the story. Int. J. Plant Sci. 2021, 182, 1–8. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. Gene expression patterns in seed plant shoot meristems and leaves: Homoplasy or homology? J. Plant Res. 2010, 123, 43–55. [Google Scholar] [CrossRef]
- Evkaikina, A.I.; Berke, L.; Romanova, M.A.; Proux-Wéra, E.; Ivanova, A.N.; Rydin, C.; Pawlowski, K.; Voitsekhovskaja, O.V. The Huperzia selago shoot tip transcriptome sheds new light on the evolution of leaves. Genome Biol. Evol. 2017, 9, 2444–2460. [Google Scholar] [CrossRef]
- Yang, M.; You, W.; Wu, S.; Fan, Z.; Xu, B.; Zhu, M.; Li, X.; Xiao, Y. Global transcriptome analysis of Huperzia serrata and identification of critical genes involved in the biosynthesis of huperzine A. BMC Genom. 2017, 18, 245. [Google Scholar] [CrossRef]
- Li, F.W.; Nishiyama, T.; Waller, M.; Frangedakis, E.; Keller, J.; Li, Z.; Fernandez-Pozo, N.; Barker, M.S.; Bennett, T.; Blazquez, M.A.; et al. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants 2020, 6, 259–272. [Google Scholar] [CrossRef]
- Worden, A.Z.; Lee, J.H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef]
- Gross, T.; Broholm, S.; Becker, A. CRABS CLAW acts as a bifunctional transcription factor in flower development. Front. Plant Sci. 2018, 9, 835. [Google Scholar] [CrossRef]
- Bowman, J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000, 3, 17–22. [Google Scholar] [CrossRef]
- Bustin, M.; Reeves, R. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 35–100. [Google Scholar] [CrossRef]
- Yamada, T.; Yokota, S.; Hirayama, Y.; Imaichi, R.; Kato, M.; Gasser, C.S. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 2011, 67, 26–36. [Google Scholar] [CrossRef]
- Toriba, T.; Harada, K.; Takamura, A.; Nakamura, H.; Ichikawa, H.; Suzaki, T.; Hirano, H.-Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 2007, 277, 457–468. [Google Scholar] [CrossRef]
- Morioka, K.; Yockteng, R.; Almeida, A.M.R.; Specht, C.D. Loss of YABBY2-like gene expression may underlie the evolution of the laminar style in Canna and contribute to floral morphological diversity in the Zingiberales. Front. Plant Sci. 2005, 6, 1106. [Google Scholar] [CrossRef]
- Bartholmes, C.; Hidalgo, O.; Gleissberg, S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol. 2012, 14, 11–23. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Baum, S.F.; Alvarez, J.; Patel, A.; Chitwood, D.H.; Bowman, J.L. Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 2005, 17, 25–36. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Baum, S.F.; Oh, S.H.; Jiang, C.Z.; Chen, J.C.; Bowman, J.L. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 2005, 132, 5021–5032. [Google Scholar] [CrossRef]
- Yamada, T.; Ito, M.; Kato, M. YABBY2 homologue expression in lateral organs of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 2004, 165, 917–924. [Google Scholar] [CrossRef]
- Byng, J.W.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Judd, W.S.; Mabberley, D.J.; Sennikov, A.N.; Soltis, D.E.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Yang, Z.; Gong, Q.; Wang, L.; Jin, Y.; Xi, J.; Li, Z.; Qin, W.; Yang, Z.; Lu, L.; Chen, Q.; et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Front. Genet. 2018, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Shchennikova, A.V.; Slugina, M.A.; Beletsky, A.V.; Filyushin, M.A.; Mardanov, A.A.; Shulga, O.A.; Kochieva, E.Z.; Ravin, N.V.; Skryabin, K.G. The YABBY genes of leaf and leaf-like organ polarity in leafless plant Monotropa hypopitys. Int. J. Genom. 2018, 2018, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, A.B.; Yang, Y.; Sharif, R.; Wu, S.N.; Xie, Y.; Wang, C. Genome wide identification, characterization, and expression analysis of YABBY-gene family in wheat (Triticum aestivum L.). Agronomy 2020, 10, 1189. [Google Scholar] [CrossRef]
- Huang, Z.; Houten, J.V.; Gonzalez, G.; Xiao, H.; van der Knaap, E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013, 288, 111–129. [Google Scholar] [CrossRef]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef]
- Sawa, S.; Ito, T.; Shimura, Y.; Okada, K. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 1999, 11, 69–86. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar]
- Stahle, M.I.; Kuehlich, J.; Staron, L.; von Arnim, A.G.; Golz, J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 2009, 21, 3105–3118. [Google Scholar] [CrossRef]
- Watanabe, K.; Okada, K. Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 2003, 15, 2592–2602. [Google Scholar] [CrossRef]
- Villanueva, J.M.; Broadhvest, J.; Hauser, B.A.; Meister, R.J.; Schneitz, K.; Gasser, C.S. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 1999, 13, 3160–3169. [Google Scholar] [CrossRef]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef]
- Nurani, A.M.; Ozawa, Y.; Furuya, T.; Sakamoto, Y.; Ebine, K.; Matsunaga, S.; Ueda, T.; Fukuda, H.; Kondo, Y. Deep imaging analysis in VISUAL reveals the role of YABBY genes in vascular stem cell fate determination. Plant Cell Physiol. 2020, 61, 255–264. [Google Scholar] [CrossRef]
- Jang, S.; Hur, J.; Kim, S.J.; Han, M.J.; Kim, S.R.; An, G. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol. Biol. 2004, 56, 133–143. [Google Scholar] [CrossRef]
- Strable, J.; Wallace, J.G.; Unger-Wallace, E.; Briggs, S.; Bradbury, P.J.; Buckler, E.S.; Vollbrecht, E. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture. Plant Cell 2017, 29, 1622–1641. [Google Scholar] [CrossRef]
- Soundararajan, P.; Won, S.Y.; Park, D.S.; Lee, Y.H.; Kim, J.S. Comparative analysis of the YABBY gene family of Bienertia sinuspersici, a single-cell C4 plant. Plants 2019, 8, 536. [Google Scholar] [CrossRef]
- Shao, H.X.; Chen, H.F.; Zhang, D.; Wu, H.Q.; Zhao, C.P.; Han, M.Y. Identification, evolution and expression analysis of the YABBY gene family in apple (Malus × domestica Borkh.). Acta Agric. Zhejiangensis 2017, 29, 1129–1138. [Google Scholar]
- Li, J.; Zhang, X.; Lu, Y.; Feng, D.; Gu, A.; Wang, S.; Wu, F.; Su, X.; Chen, X.; Li, X.; et al. Characterization of non-heading mutation in heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci 2019, 10, 112. [Google Scholar] [CrossRef]
- Liu, X.F.; Ning, K.; Che, G.; Yan, S.S.; Han, L.J.; Gu, R.; Li, Z.; Weng, Y.Q.; Zhang, X.L. CsSPL functions as an adaptor between HD-ZIP III and CsWUS transcription factors regulating anther and ovule development in Cucumis sativus (cucumber). Plant J. 2018, 94, 535–547. [Google Scholar] [CrossRef]
- Golz, J.F.; Roccaro, M.; Kuzoff, R.; Hudson, A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 2004, 131, 3661–3670. [Google Scholar] [CrossRef]
- Navarro, C.; Efremova, N.; Golz, J.F.; Rubiera, R.; Schwarz-Sommer, Z. Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 2004, 131, 3649–3659. [Google Scholar] [CrossRef]
- Xiang, J.; Liu, R.Q.; Li, T.M.; Han, L.J.; Zou, Y.; Xu, T.F.; Wei, J.Y.; Wang, Y.J.; Xu, Y. Isolation and characterization of two VpYABBY genes from wild Chinese Vitis pseudoreticulata. Protoplasma 2013, 250, 1315–1325. [Google Scholar] [CrossRef]
- Fourquin, C.A.; Vinauger-Douard, M.; Fogliani, B.; Dumas, C.; Scutt, C.P. Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. USA 2005, 102, 4649–4654. [Google Scholar] [CrossRef]
- de Almeida, A.M.R.; Yockteng, R.; Schnable, J.; Alvarez-Buylla, E.R.; Freeling, M.; Specht, C.D. Co-option of the polarity gene network shapes filament morphology in angiosperms. Sci. Rep. 2014, 4, 6194. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Hsiao, Y.Y.; Chang, S.B.; Zhang, D.; Lan, S.R.; Liu, Z.J.; Tsai, W.C. Genome-wide identification of YABBY genes in Orchidaceae and their expression patterns in Phalaenopsis orchid. Genes 2020, 11, 955. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, Y.; Yamada, T.; Oya, Y.; Ito, M.; Kato, M.; Imaichi, R. Expression patterns of class I KNOX and YABBY genes in Ruscus aculeatus (Asparagaceae) with implications for phylloclade homology. Dev. Genes Evol. 2007, 217, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Tang, J.; Li, D.; Chen, C.; Zhao, X.; Zhu, L. Isolation and functional analysis of LiYAB1, a YABBY family gene, from lily (Lilium longiflorum). J. Plant Physiol. 2009, 166, 988–995. [Google Scholar] [CrossRef]
- Kumaran, M.K.; Bowman, J.L.; Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 2002, 14, 2761–2770. [Google Scholar] [CrossRef]
- Sarvepalli, K.; Das Gupta, M.; Challa, K.R.; Nath, U. Molecular cartography of leaf development—Role of transcription factors. Curr. Opin. Plant Biol. 2019, 47, 22–31. [Google Scholar] [CrossRef]
- Nole-Wilson, S.; Krizek, B.A. AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 2006, 141, 977–987. [Google Scholar] [CrossRef]
- Chung, Y.; Zhu, Y.; Wu, M.F.; Simonini, S.; Kuhn, A.; Armenta-Medina, A.; Jin, R.; Østergaard, L.; Gillmor, C.S.; Wagner, D. Auxin Response Factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat. Commun. 2019, 10, 886. [Google Scholar] [CrossRef]
- Bonaccorso, O.; Lee, J.E.; Puah, L.; Scutt, C.P.; Golz, J.F. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 2012, 12, 176. [Google Scholar] [CrossRef]
- la Rota, C.; Chopard, J.; Das, P.; Paindavoine, S.; Rozier, F.; Farcot, E.; Godin, C.; Traas, J.; Moneger, F. A data-driven integrative model of sepal primordium polarity in Arabidopsis. Plant Cell 2011, 23, 4318–4333. [Google Scholar] [CrossRef]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef]
- Iwasaki, M.; Takahashi, H.; Iwakawa, H.; Nakagawa, A.; Ishikawa, T.; Tanaka, H.; Matsumura, Y.; Pekker, I.; Eshed, Y.; Vial-Pradel, S.; et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 2013, 140, 1958–1969. [Google Scholar] [CrossRef]
- Xu, B.; Li, Z.; Zhu, Y.; Wang, H.; Ma, H.; Dong, A.; Huang, H. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary specifying genes to promote sepal and petal development. Plant Physiol. 2008, 146, 566–575. [Google Scholar] [CrossRef]
- Nakata, M.; Matsumoto, N.; Tsugeki, R.; Rikirsch, E.; Laux, T.; Okada, K. Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 2012, 24, 519–535. [Google Scholar] [CrossRef]
- Kuchen, E.E.; Fox, S.; de Reuille, P.B.; Kennaway, R.; Bensmihen, S.; Avondo, J.; Calder, G.M.; Southam, P.; Bangham, A.; Coen, E. Generation of leaf shape through early patterns of growth and tissue polarity. Science 2012, 335, 1092–1096. [Google Scholar] [CrossRef]
- Goldshmidt, A.; Alvarez, J.P.; Bowman, J.L.; Eshed, Y. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 2008, 20, 1217–1230. [Google Scholar] [CrossRef]
- Boter, M.; Golz, J.F.; Giménez-Ibañez, S.; Fernandez-Barbero, G.; Franco-Zorrilla, J.M.; Solano, R. FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 2015, 27, 3160–3174. [Google Scholar] [CrossRef]
- Li, Z.; Li, G.; Cai, M.; Priyadarshani, S.V.G.N.; Aslam, M.; Zhou, Q.; Huang, X.; Wang, X.; Liu, Y.; Qin, Y. Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int. J. Mol. Sci. 2019, 20, 5863. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Sun, X.; Li, Y.; Yao, J.; van Nocker, S.; Wang, X. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Front. Plant Sci. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Zhao, S.P.; Lu, D.; Yu, T.F.; Ji, Y.J.; Zheng, W.J.; Zhang, S.X.; Chai, S.C.; Chen, Z.Y.; Cui, X.Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017, 119, 132–146. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, C.; Ge, D.; Yan, M.; Ren, Y.; Huang, X.; Yuan, Z. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene 2020, 752, 144784. [Google Scholar] [CrossRef]
- Yang, H.; Shi, G.; Li, X.; Hu, D.; Cui, Y.; Hou, J.; Yu, D.; Huang, F. Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 234. [Google Scholar] [CrossRef]
- Hou, H.; Lin, Y.; Hou, X. Ectopic expression of a Pak-choi YABBY gene, BcYAB3, causes leaf curvature and flowering stage delay in Arabidopsis thaliana. Genes 2020, 11, 370. [Google Scholar] [CrossRef]
- Kidner, C.; Timmermans, M. Signaling sides: Adaxial-abaxial patterning in leaves. Curr. Top. Dev. Biol. 2010, 91, 141–168. [Google Scholar] [CrossRef]
- Ohmori, Y.; Toriba, T.; Nakamura, H.; Ichikawa, H.; Hirano, H.Y. Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. Plant J. 2011, 65, 77–86. [Google Scholar] [CrossRef]
- Skinner, D.J.; Brown, R.H.; Kuzoff, R.K.; Gasser, C.S. Conservation of the role of INNER NO OUTER in development of unitegmic ovules of the Solanaceae despite a divergence in protein function. BMC Plant Biol. 2016, 16, 143. [Google Scholar] [CrossRef]
- Prunet, N.; Morel, P.; Thierry, A.M.; Eshed, Y.; Bowman, J.L.; Negrutiu, I.; Trehin, C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 2008, 20, 901–919. [Google Scholar] [CrossRef][Green Version]
- Sun, W.; Huang, W.; Li, Z.; Lv, H.; Huang, H.; Wang, Y. Characterization of a Crabs Claw gene in basal eudicot species Epimedium sagittatum (Berberidaceae). Int. J. Mol. Sci. 2013, 14, 119–1131. [Google Scholar] [CrossRef]
- Orashakova, S.; Lange, M.; Lange, S.; Wege, S.; Becker, A. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 2009, 58, 682–693. [Google Scholar] [CrossRef]
- Ishikawa, M.; Ohmori, Y.; Tanaka, W.; Hirabayashi, C.; Murai, K.; Ogihara, Y.; Yamaguchi, T.; Hirano, H.Y. The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes Genet. Syst. 2009, 84, 137–146. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, N.; Miyoshi, M.; Sano, Y.; Satoh, H.; Hirano, H.-Y.; Sakai, H.; Nagato, Y. SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 2003, 130, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Dukowic-Schulze, S.; Harris, A.; Li, J.; Sundararajan, A.; Mudge, J.; Retzel, E.F.; Pawlowski, W.P.; Chen, C. Comparative transcriptomics of early meiosis in Arabidopsis and maize. J. Genet. Genom 2014, 41, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Fourquin, C.A.; Primo, A.; Martínez-Fernández, I.; Huet-Trujillo, E.; Ferrándiz, C. The CRC orthologue from Pisum sativum shows conserved functions in carpel morphogenesis and vascular development. Ann. Bot. 2014, 114, 1535–1544. [Google Scholar] [CrossRef]
- Lora, J.; Hormaza, J.I.; Herrero, M.; Gasser, C.S. Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. USA 2011, 108, 5461–5465. [Google Scholar] [CrossRef]
- Simon, M.K.; Williams, L.A.; Brady-Passerini, K.; Brown, R.H.; Gasser, C.S. Positive- and negative-acting regulatory elements contribute to the tissue-specific expression of INNER NO OUTER, a YABBY-type transcription factor gene in Arabidopsis. BMC Plant Biol. 2012, 12, 214. [Google Scholar] [CrossRef]
- Simon, M.K.; Skinner, D.J.; Gallagher, T.L.; Gasser, C.S. Integument development in Arabidopsis depends on interaction of YABBY protein INNER NO OUTER with coactivators and corepressors. Genetics 2017, 207, 1489–1500. [Google Scholar] [CrossRef]
- Yamada, T.; Ito, M.; Kato, M. Expression pattern of INNER NO OUTER homologue in Nymphaea (water lily family, Nymphaeaceae). Dev. Genes Evol. 2003, 213, 510–513. [Google Scholar] [CrossRef]
- McAbee, J.M.; Kuzoff, R.K.; Gasser, C.S. Mechanisms of derived unitegmy among Impatiens species. Plant Cell 2005, 17, 1674–1684. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Schneitz, K. NOZZLE regulates proximal–distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development 2000, 127, 4227–4238. [Google Scholar]
- Meister, R.J.; Kotow, L.M.; Gasser, C.S. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 2002, 129, 4281–4289. [Google Scholar]
- Franks, R.G.; Liu, Z.; Fischer, R.L. SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals petals. Planta 2006, 224, 801–811. [Google Scholar] [CrossRef]
- Kelley, D.R.; Skinner, D.J.; Gasser, C.S. Roles of polarity determinants in ovule development. Plant J. 2009, 57, 1054–1064. [Google Scholar] [CrossRef]
- Du, F.; Guan, C.M.; Jiao, Y.L. Molecular mechanisms of leaf morphogenesis. Mol. Plant 2018, 11, 1117–1134. [Google Scholar] [CrossRef]
- Nishiyama, T.; Fujita, T.; Shin-I, T.; Seki, M.; Nishide, H.; Uchiyama, I.; Kamiya, A.; Carninci, P.; Hayashizaki, Y.; Shinozaki, K.; et al. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: Implication for land plant evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 8007–8012. [Google Scholar] [CrossRef]
- Li, F.W.; Brouwer, P.; Carretero-Paulet, L.; Cheng, S.; de Vries, J.; Delaux, P.-M.; Eily, A.; Koppers, N.; Kuo, L.-Y.; Li, Z.; et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 2018, 4, 460–472. [Google Scholar] [CrossRef]
- Bowman, J.L.; Briginshaw, L.N.; Florent, S.N. Evolution and co-option of developmental regulatory networks in early land plants. Curr. Top. Devel. Biol. 2019, 131, 35–54. [Google Scholar] [CrossRef]
- Nakata, M.; Okada, K. The leaf adaxial-abaxial boundary and lamina growth. Plants 2013, 2, 174–202. [Google Scholar] [CrossRef]
- Harrison, C.J.; Rezvani, M.; Langdale, J.A. Growth from two transient apical initials in the meristem of Selaginella kraussiana. Development 2007, 134, 881–889. [Google Scholar] [CrossRef]
- Romanova, M.A.; Naumenko, A.N.; Evkaikina, A.I. Special features of apical morphogenesis in various taxa of seedless plants (in Russian). Vestn. St.-Petersbg. State Uni. 2010, 3, 29–36. Available online: https://cyberleninka.ru/article/n/osobennosti-apikalnogo-morfogeneza-v-raznyh-taksonah-nesemennyh-rasteniy. (accessed on 13 April 2021). (In Russian).
- Imaichi, R.; Hiratsuka, R. Evolution of shoot apical meristem structures in vascular plants with respect to plasmodesmatal network. Am. J. Bot. 2007, 94, 1911–1921. [Google Scholar] [CrossRef]
- Frank, M.H.; Edwards, M.B.; Schultz, E.R.; McKain, M.R.; Fei, Z.; Sørensen, I.; Rose, J.K.C.; Scanlon, M.J. Dissecting the molecular signatures of apical cell-type shoot meristems from two ancient land plant lineages. New Phytol. 2015, 207, 893–904. [Google Scholar] [CrossRef]
- Kidston, R.; Lang, W.H. On Old Red Sandstone plants showing structure, from the Rhynie Chert Bed, Aberdeenshire. Part III. Asteroxylon mackie, Kidston and Lang. Trans. R. Soc. Edinb. 1920, 52, 643–680. [Google Scholar] [CrossRef]
- Cooke, T.D.; Tilney, M.S.; Tilney, L. Plasmodesmatal networks in apical meristems and mature structures: Geometric evidence for both primary and secondary formation of plasmodesmata. In Membranes: Specialized Functions in Plants; Smallwood, M., Knox, J.P., Bowles, D.J., Eds.; BIOS Scientific: Cambridge, UK, 1996; pp. 471–488. ISBN 1-85996-200-9. [Google Scholar]
- Szovenyi, P.; Waller, M.; Kirbis, A. Evolution of the plant body plan. In Plant Development and Evolution, 1st ed.; Grossniklaus, U., Ed.; Academic Press: Amsterdam, The Netherlands; Elsevier: New York, NY, USA, 2019; Volume 131, pp. 1–34. ISBN 978-0-12-809804-2. [Google Scholar]
- Bowman, J.M.; Sakakibara, K.; Furumizu, C.; Dierschke, T. The evolution of the cycle of life. Annu. Rev. Genet. 2016, 50, 133–154. [Google Scholar] [CrossRef]
- Harrison, C.J.; Morris, J.L. The origin and early evolution of vascular plant shoots and leaves. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 373, 20160496. [Google Scholar] [CrossRef]
- Sakakibara, K.; Ando, S.; Yip, H.K.; Tamada, Y.; Hiwatashi, Y.; Murata, T.; Deguchi, H.; Hasebe, M.; Bowman, J.L. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 2013, 339, 1067–1070. [Google Scholar] [CrossRef]
- Whitewoods, C.D.; Cammarata, J.; Venza, Z.N.; Sang, S.; Crook, A.D.; Aoyama, T.; Wang, X.Y.; Waller, M.; Kamisugi, Y.; Cuming, A.C.; et al. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr. Biol. 2018, 28, 2365–2376. [Google Scholar] [CrossRef]
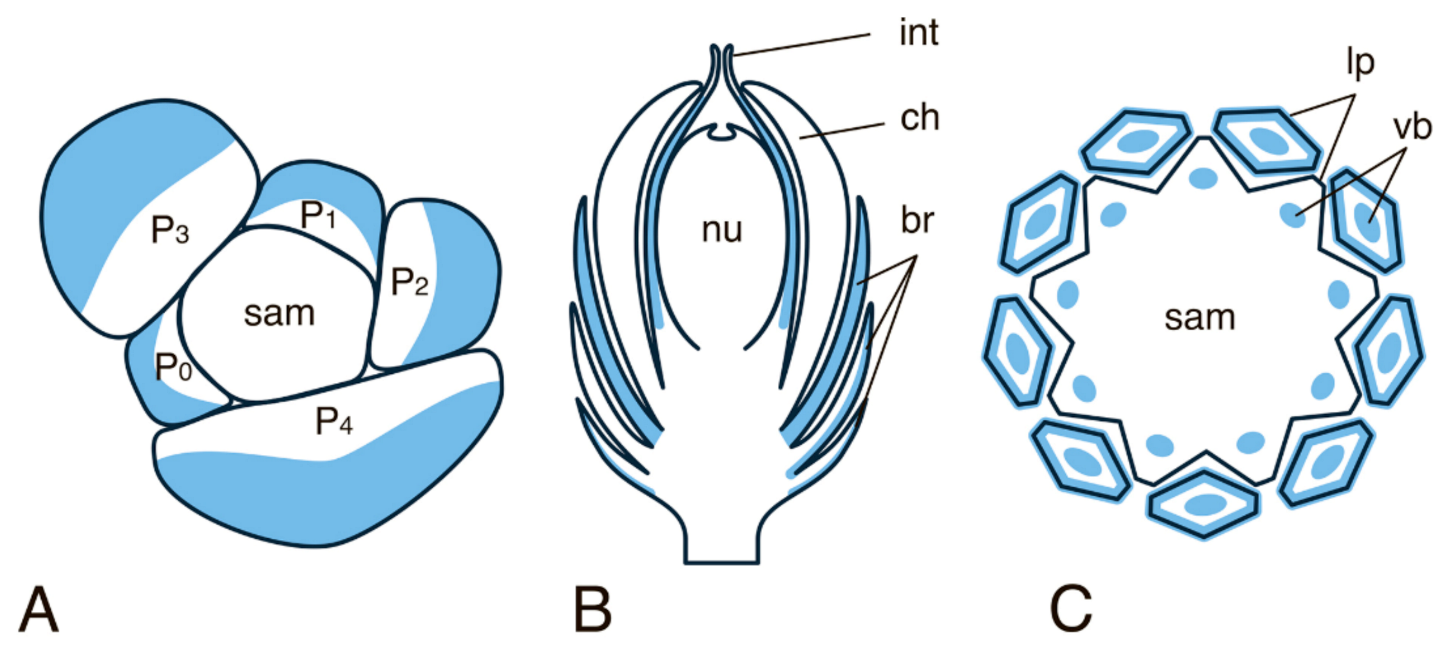
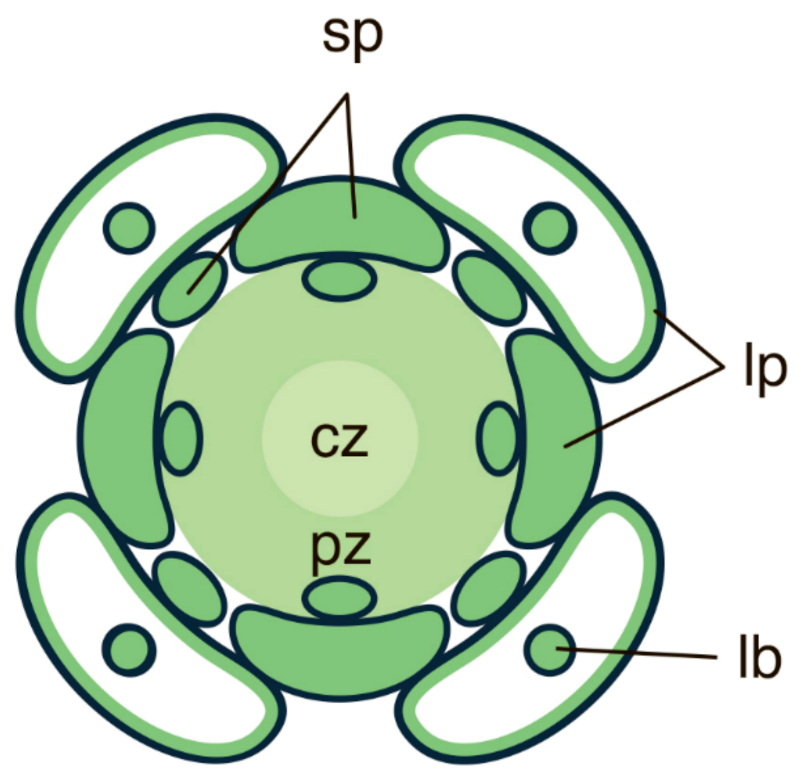
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romanova, M.A.; Maksimova, A.I.; Pawlowski, K.; Voitsekhovskaja, O.V. YABBY Genes in the Development and Evolution of Land Plants. Int. J. Mol. Sci. 2021, 22, 4139. https://doi.org/10.3390/ijms22084139
Romanova MA, Maksimova AI, Pawlowski K, Voitsekhovskaja OV. YABBY Genes in the Development and Evolution of Land Plants. International Journal of Molecular Sciences. 2021; 22(8):4139. https://doi.org/10.3390/ijms22084139
Chicago/Turabian StyleRomanova, Marina A., Anastasiia I. Maksimova, Katharina Pawlowski, and Olga V. Voitsekhovskaja. 2021. "YABBY Genes in the Development and Evolution of Land Plants" International Journal of Molecular Sciences 22, no. 8: 4139. https://doi.org/10.3390/ijms22084139
APA StyleRomanova, M. A., Maksimova, A. I., Pawlowski, K., & Voitsekhovskaja, O. V. (2021). YABBY Genes in the Development and Evolution of Land Plants. International Journal of Molecular Sciences, 22(8), 4139. https://doi.org/10.3390/ijms22084139







