Evidence for a Negative Correlation between Human Reactive Enamine-Imine Intermediate Deaminase A (RIDA) Activity and Cell Proliferation Rate: Role of Lysine Succinylation of RIDA
Abstract
1. Introduction
2. Results
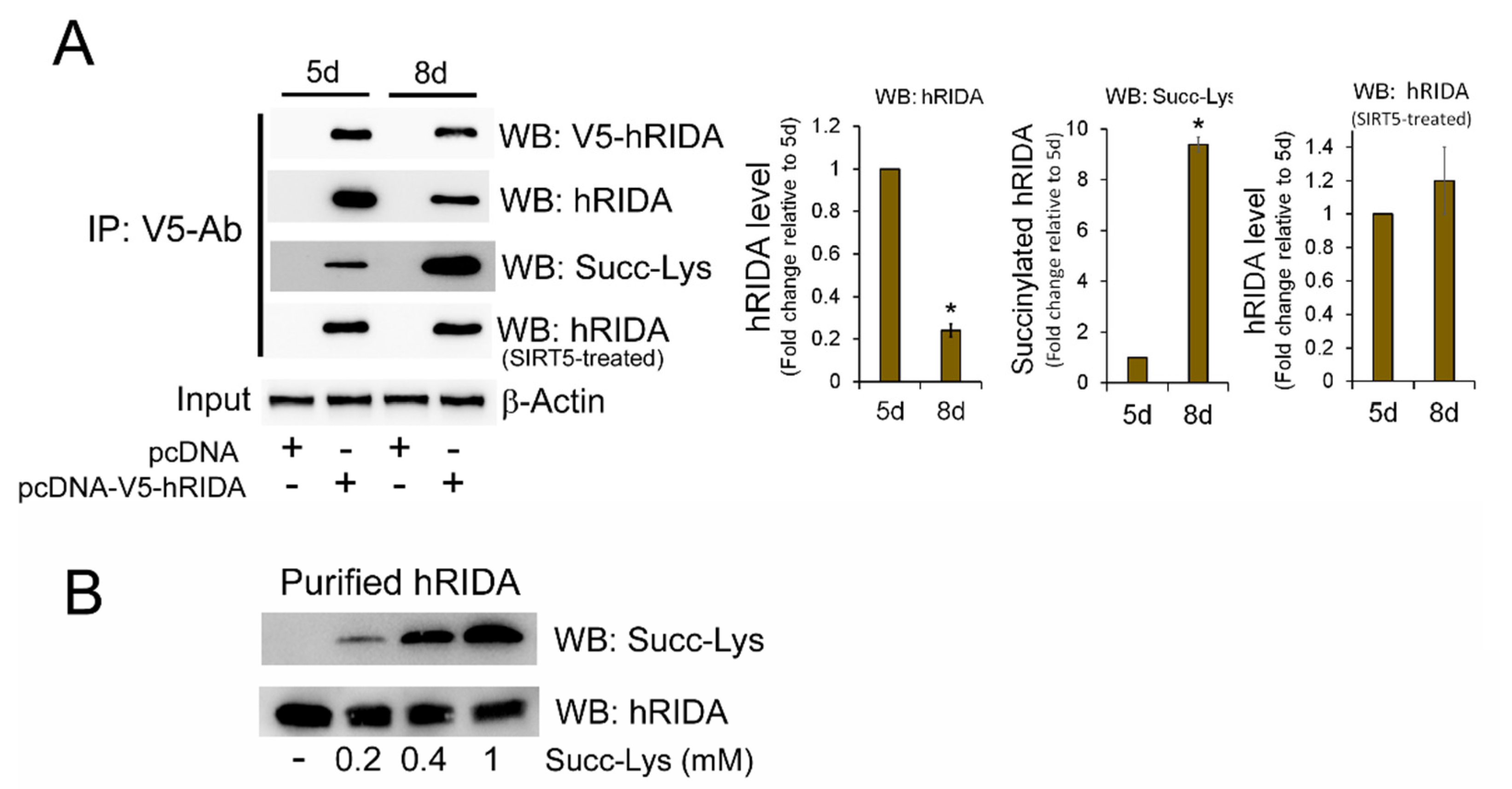
2.1. Analysis of hRIDA Expression in Different Human Cell Lines and Post-Translational Modification
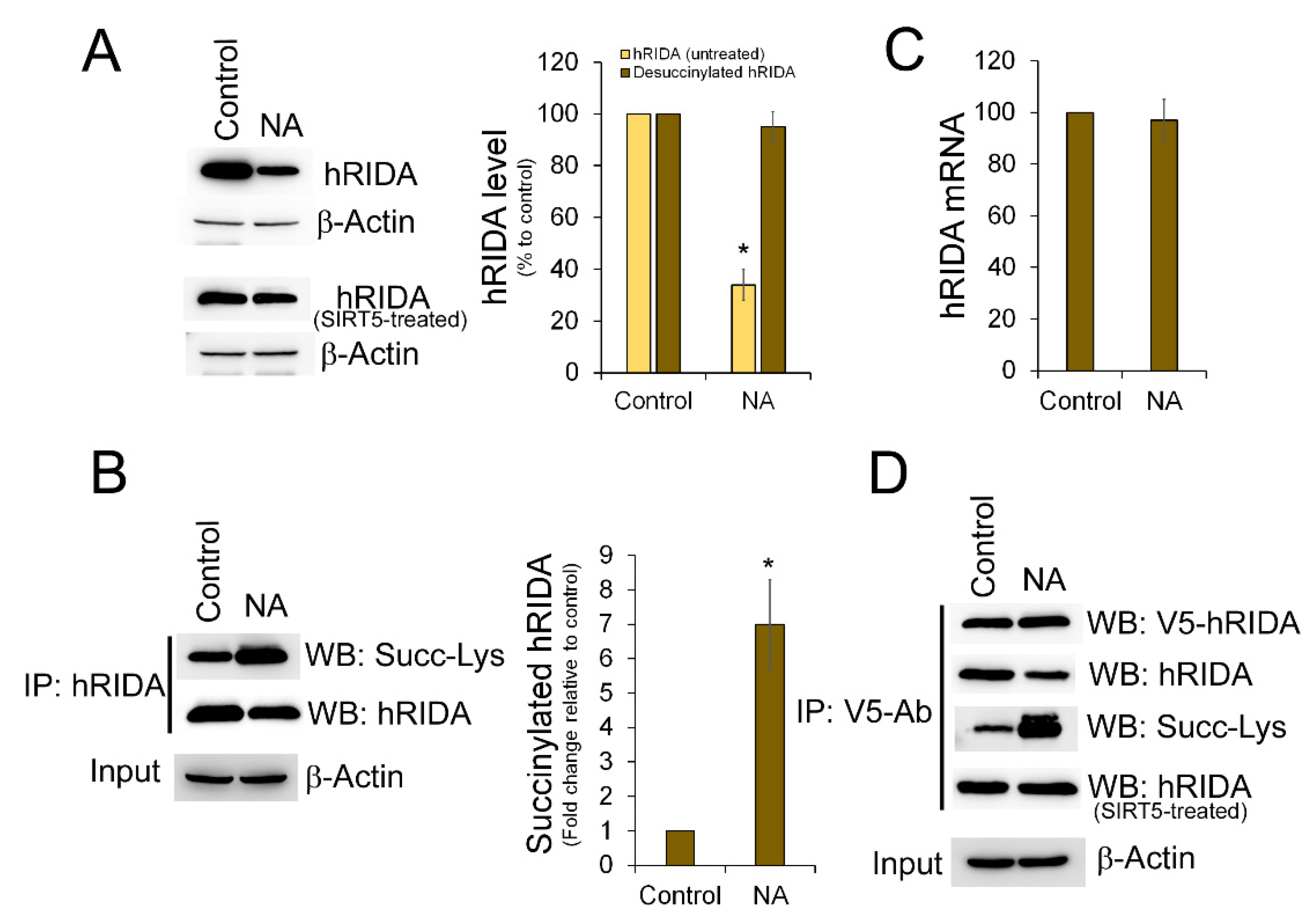
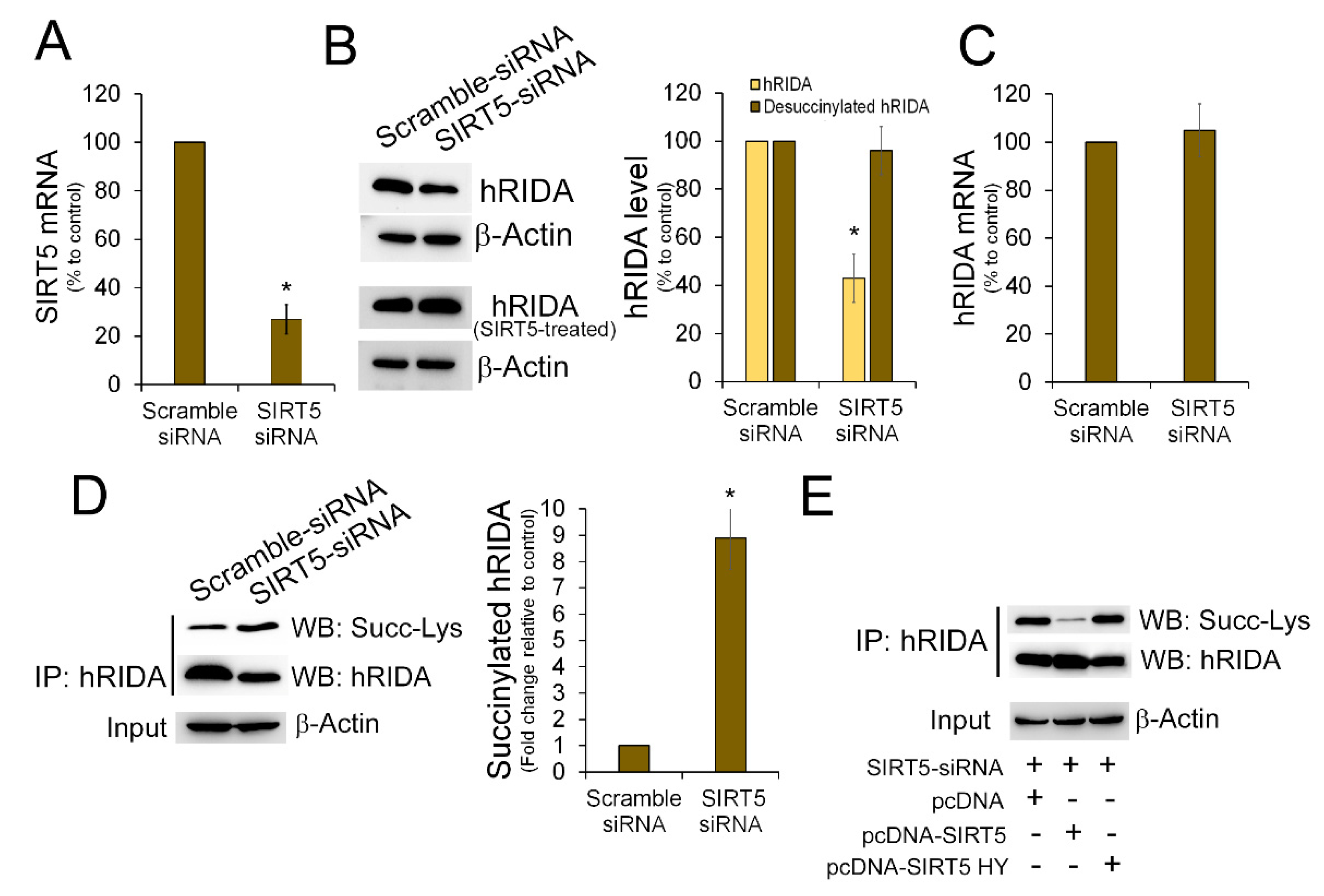
2.2. K-Succinylation of hRIDA Was Under the Control of SIRT5
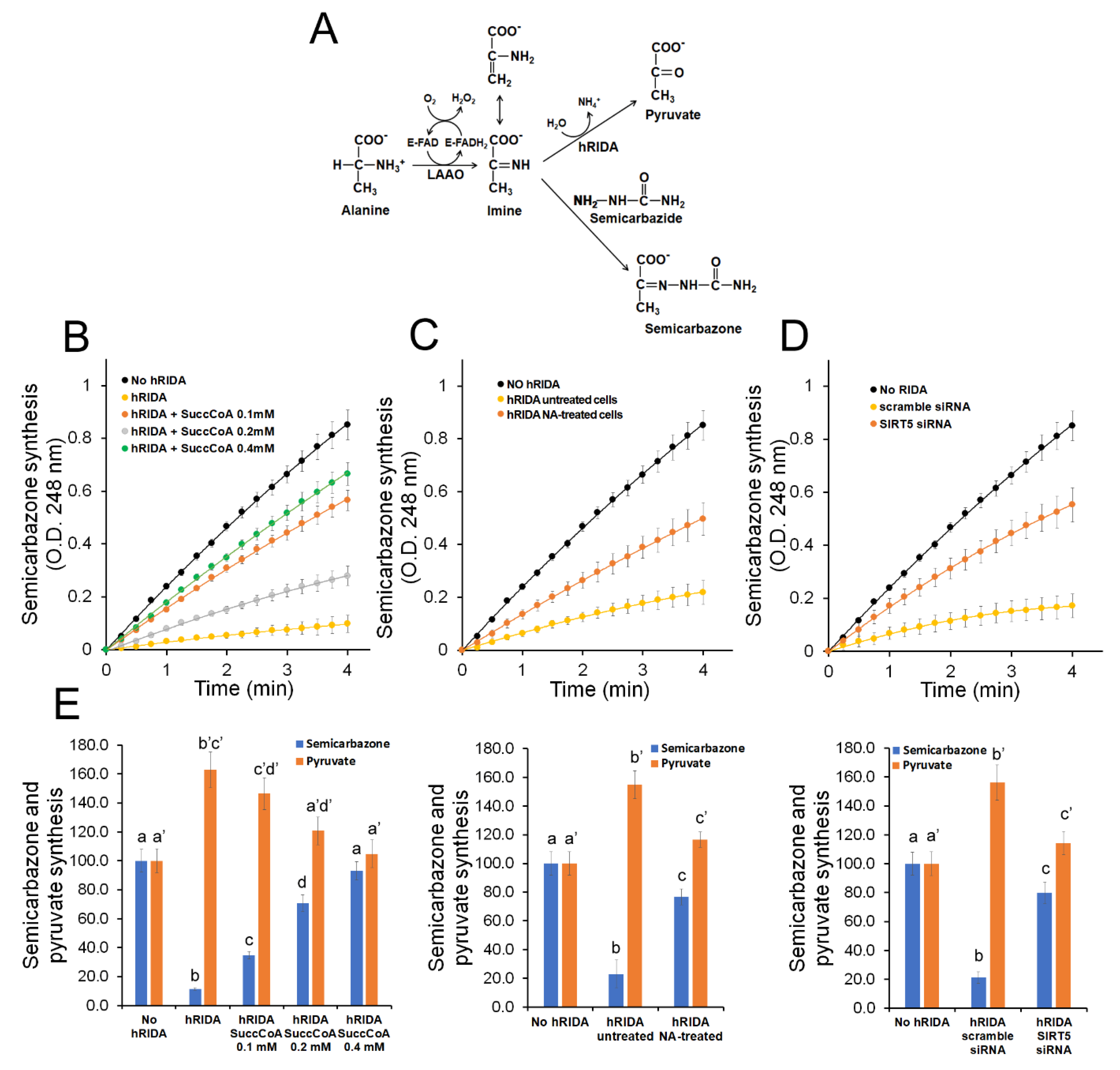
2.3. K-Succinylation Negatively Affects Enzymatic Activity of hRIDA
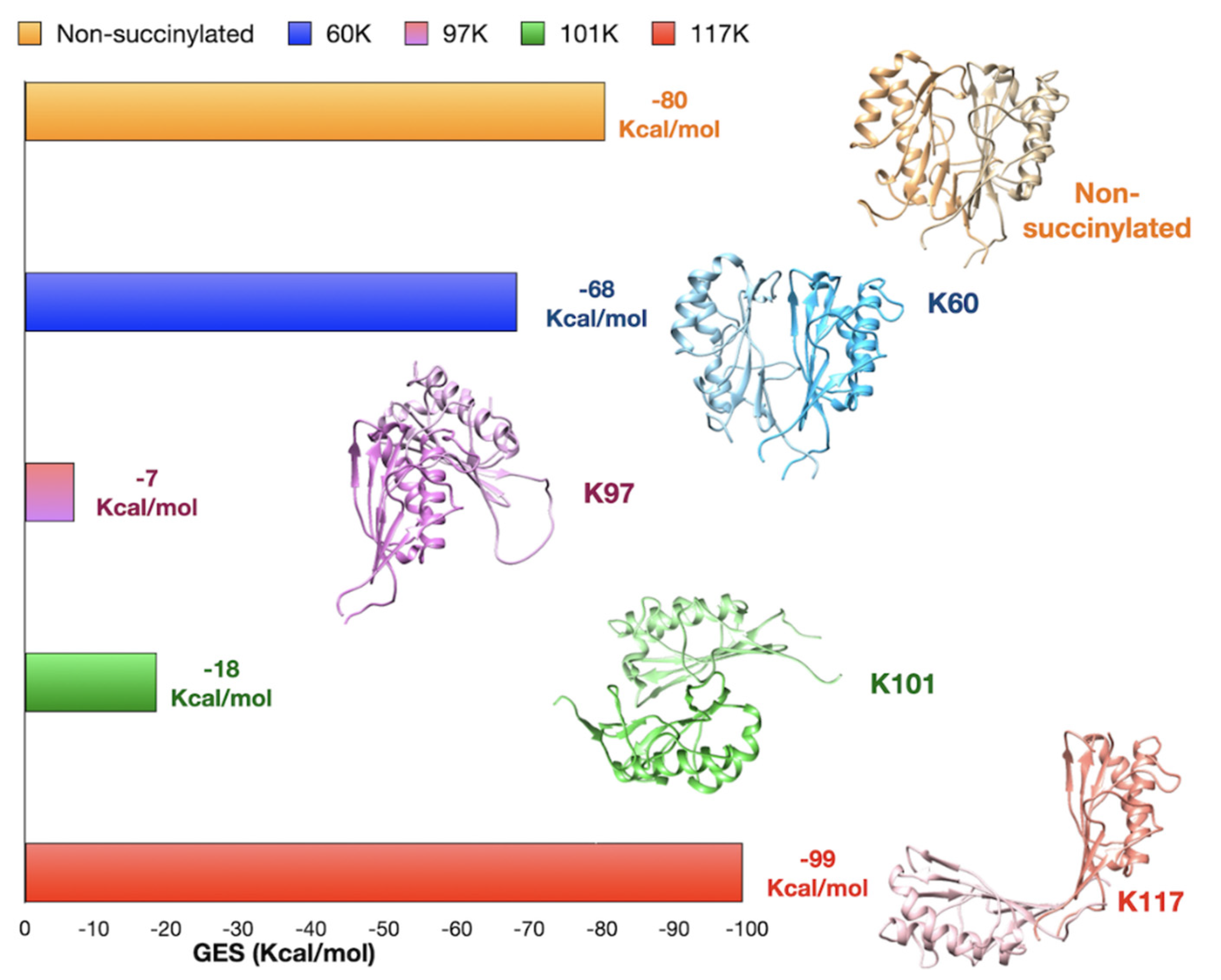
2.4. In Silico Analysis of K-Succinylation Influence on hRIDA Structures
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Growth Curve Analysis
4.2. Isolation of RNA from Cultured Cells and Real-Time qPCR Analysis
4.3. Western Blotting Analysis and Immunoprecipitation
4.4. Plasmids and Recombinant Proteins Synthesis
4.5. RNA Interference Analysis
4.6. In Vitro hRIDA Desuccinylation and Succinylation Assays
4.7. hRIDA Enzymatic Assay
4.8. 3D Modeling and Docking Simulations
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Downs, D.M.; Ernst, D.C. From microbiology to cancer biology: The Rid protein family prevents cellular damage caused by endogenously generated reactive nitrogen species. Mol. Microbiol. 2015, 96, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Borchert, A.J.; Ernst, D.C.; Downs, D.M. Reactive Enamines and Imines In Vivo: Lessons from the RidA Paradigm. Trends Biochem. Sci. 2019, 44, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, T.D.; Gerdes, S.; Hodge-Hanson, K.; Zhukov, A.; Cooper, A.J.; ElBadawi-Sidhu, M.; Fiehn, O.; Downs, D.M.; Hanson, A.D. Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genom. 2015, 16, 382. [Google Scholar] [CrossRef] [PubMed]
- Hodge-Hanson, K.M.; Downs, D.M. Members of the Rid protein family have broad imine deaminase activity and can accelerate the Pseudomonas aeruginosa D-arginine dehydrogenase (DauA) reaction in vitro. PLoS ONE 2017, 12, e0185544. [Google Scholar] [CrossRef] [PubMed]
- Degani, G.; Barbiroli, A.; Regazzoni, R.; Popolo, L.; Vanoni, M.A. Imine Deaminase Activity and Conformational Stability of UK114, the Mammalian Member of the Rid Protein Family Active in Amino Acid Metabolism. Int. J. Mol. Sci. 2018, 19, 945. [Google Scholar] [CrossRef]
- Irons, J.L.; Hodge-Hanson, K.; Downs, D.M. RidA Proteins Protect against Metabolic Damage by Reactive Intermediates. Microbiol. Mol. Biol. Rev. 2020, 84, e00024-20. [Google Scholar] [CrossRef]
- Colombo, I.; Ceciliani, F.; Ronchi, S.; Bartorelli, A.; Berra, B. cDNA cloning and Escherichia coli expression of UK114 tumor antigen. Biochim. Biophys. Acta 1998, 1442, 49–59. [Google Scholar] [CrossRef]
- Schmiedeknecht, G.; Kerkhoff, C.; Orsó, E.; Stöhr, J.; Aslanidis, C.; Nagy, G.M.; Knuechel, R.; Schmitz, G. Isolation and characterization of a 14.5-kDa trichloroacetic-acid-soluble translational inhibitor protein from human monocytes that is upregulated upon cellular differentiation. Eur. J. Biochem. 1996, 242, 339–351. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteomics 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Antonenkov, V.D.; Ohlmeier, S.; Sormunen, R.T.; Hiltunen, J.K. UK114, a YjgF/Yer057p/UK114 family protein highly conserved from bacteria to mammals, is localized in rat liver peroxisomes. Biochem. Biophys. Res. Commun. 2007, 357, 252–257. [Google Scholar] [CrossRef]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef]
- Funaro, A.; Horenstein, A.L.; Ghisolfi, G.; Bussolati, B.; Bartorelli, A.; Bussolati, G. Identification of a 220-kDa membrane tumor-associated antigen by human anti-UK114 monoclonal antibodies selected from the immunoglobulin repertoire of a cancer patient. Exp. Cell Res. 1999, 247, 441–450. [Google Scholar] [CrossRef]
- Bartorelli, A.; Bussolati, B.; Millesimo, M.; Gugliotta, P.; Bussolati, G. Antibody-dependent cytotoxic activity on human cancer cells expressing UK 114 tumor membrane antigen. Int. J. Oncol. 1996, 8, 543–548. [Google Scholar] [CrossRef]
- Chong, C.L.; Huang, S.F.; Hu, C.P.; Chen, Y.L.; Chou, H.Y.; Chau, G.Y.; Shew, J.Y.; Tsai, Y.L.; Chen, C.T.; Chang, C.; et al. Decreased expression of UK114 is related to the differentiation status of human hepatocellular carcinoma. Cancer Epidemiol. Biomark. Prev. 2008, 17, 535–542. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Asagi, K.; Oka, T.; Arao, K.; Suzuki, I.; Thakur, M.K.; Izumi, K.; Natori, Y. Purification, characterization and differentiation-dependent expression of a perchloric acid soluble protein from rat kidney. Nephron 1998, 79, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Kanouchi, H.; Oka, T.; Asagi, K.; Tachibana, H.; Yamada, K. Expression and cellular distribution of perchloric acid-soluble protein is dependent on the cell proliferating states of NRK-52E. Cell. Mol. Life Sci. 2000, 57, 1103–1108. [Google Scholar] [CrossRef]
- Kanouchi, H.; Taga, M.; Okamoto, T.; Yamasaki, M.; Oka, T.; Yamada, K.; Tone, S.; Minatogawa, Y. Reduced expression of perchloric acid-soluble protein after partial hepatectomy in rats. Biosci. Biotechnol. Biochem. 2006, 70, 290–292. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef]
- Bringman-Rodenbarger, L.R.; Guo, A.H.; Lyssiotis, C.A.; Lombard, D.B. Emerging Roles for SIRT5 in Metabolism and Cancer. Antioxid. Redox Signal. 2018, 28, 677–690. [Google Scholar] [CrossRef]
- Fischer, F.; Gertz, M.; Suenkel, B.; Lakshminarasimhan, M.; Schutkowski, M.; Steegborn, C. Sirt5 deacylation activities show differential sensitivities to nicotinamide inhibition. PLoS ONE 2012, 7, e45098. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Z.; Li, X.; Liu, B.; Liu, M.; Liu, L.; Chen, S.; Ren, M.; Wang, Y.; Yu, M.; et al. SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res. 2018, 78, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Roy, A.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; Park, T.; Heo, L.; Park, C.; Seok, C. GalaxyHomomer: A web server for protein homo-oligomer structure prediction from a monomer sequence or structure. Nucleic Acids Res. 2017, 45, W320–W324. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E.; et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013, 18, 920–933. [Google Scholar] [CrossRef]
- Matsushita, N.; Yonashiro, R.; Ogata, Y.; Sugiura, A.; Nagashima, S.; Fukuda, T.; Inatome, R.; Yanagi, S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells 2011, 16, 190–202. [Google Scholar] [CrossRef]
- Manjasetty, B.A.; Delbrück, H.; Pham, D.T.; Mueller, U.; Fieber-Erdmann, M.; Scheich, C.; Sievert, V.; Büssow, K.; Niesen, F.H.; Weihofen, W.; et al. Crystal structure of Homo sapiens protein hp14.5. Proteins 2004, 54, 797–800. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tapias, V.; Acosta, D.; Xu, H.; Chen, H.; Bhawal, R.; Anderson, E.; Ivanova, E.; Lin, H.; Sagdullaev, B.T.; et al. Succinylation Links Metabolic Reductions to Amyloid and Tau Pathology. bioRxiv 2019. [Google Scholar] [CrossRef]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. EMBO J. 2017, 36, 1302–1315. [Google Scholar] [CrossRef] [PubMed]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [PubMed]
- Damiano, F.; Gnoni, G.V.; Siculella, L. Citrate carrier promoter is target of peroxisome proliferator-activated receptor alpha and gamma in hepatocytes and adipocytes. Int. J. Biochem. Cell Biol. 2012, 44, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Siculella, L.; Giannotti, L.; Testini, M.; Gnoni, G.V.; Damiano, F. In steatotic cells, ATP-citrate lyase mRNA is efficiently translated through a cap-independent mechanism, contributing to the stimulation of de novo lipogenesis. Int. J. Mol. Sci. 2020, 21, 1206. [Google Scholar] [CrossRef]
- Damiano, F.; Tocci, R.; Gnoni, G.V.; Siculella, L. Expression of citrate carrier gene is activated by ER stress effectors XBP1 and ATF6α, binding to an UPRE in its promoter. Biochim. Biophys. Acta 2015, 1849, 23–31. [Google Scholar] [CrossRef]
- Kumar, S.; Lombard, D.B. Generation and Purification of Catalytically Active Recombinant Sirtuin5 (SIRT5) Protein. Methods Mol. Biol. 2016, 1436, 241–257. [Google Scholar] [CrossRef]
- Talà, A.; Damiano, F.; Gallo, G.; Pinatel, E.; Calcagnile, M.; Testini, M.; Fico, D.; Rizzo, D.; Sutera, A.; Renzone, G.; et al. Pirin: A novel redox-sensitive modulator of primary and secondary metabolism in Streptomyces. Metab. Eng. 2018, 48, 254–268. [Google Scholar] [CrossRef]
- Siculella, L.; Tocci, R.; Rochira, A.; Testini, M.; Gnoni, A.; Damiano, F. Lipid accumulation stimulates the cap-independent translation of SREBP-1a mRNA by promoting hnRNP A1 binding to its 5′-UTR in a cellular model of hepatic steatosis. Biochim. Biophys. Acta 2016, 1861, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Okanishi, H.; Kim, K.; Fukui, K.; Yano, T.; Kuramitsu, S.; Masui, R. Proteome-wide identification of lysine succinylation in thermophilic and mesophilic bacteria. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 1–17. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siculella, L.; Giannotti, L.; Di Chiara Stanca, B.; Calcagnile, M.; Rochira, A.; Stanca, E.; Alifano, P.; Damiano, F. Evidence for a Negative Correlation between Human Reactive Enamine-Imine Intermediate Deaminase A (RIDA) Activity and Cell Proliferation Rate: Role of Lysine Succinylation of RIDA. Int. J. Mol. Sci. 2021, 22, 3804. https://doi.org/10.3390/ijms22083804
Siculella L, Giannotti L, Di Chiara Stanca B, Calcagnile M, Rochira A, Stanca E, Alifano P, Damiano F. Evidence for a Negative Correlation between Human Reactive Enamine-Imine Intermediate Deaminase A (RIDA) Activity and Cell Proliferation Rate: Role of Lysine Succinylation of RIDA. International Journal of Molecular Sciences. 2021; 22(8):3804. https://doi.org/10.3390/ijms22083804
Chicago/Turabian StyleSiculella, Luisa, Laura Giannotti, Benedetta Di Chiara Stanca, Matteo Calcagnile, Alessio Rochira, Eleonora Stanca, Pietro Alifano, and Fabrizio Damiano. 2021. "Evidence for a Negative Correlation between Human Reactive Enamine-Imine Intermediate Deaminase A (RIDA) Activity and Cell Proliferation Rate: Role of Lysine Succinylation of RIDA" International Journal of Molecular Sciences 22, no. 8: 3804. https://doi.org/10.3390/ijms22083804
APA StyleSiculella, L., Giannotti, L., Di Chiara Stanca, B., Calcagnile, M., Rochira, A., Stanca, E., Alifano, P., & Damiano, F. (2021). Evidence for a Negative Correlation between Human Reactive Enamine-Imine Intermediate Deaminase A (RIDA) Activity and Cell Proliferation Rate: Role of Lysine Succinylation of RIDA. International Journal of Molecular Sciences, 22(8), 3804. https://doi.org/10.3390/ijms22083804






