Characterization of the mbsA Gene Encoding a Putative APSES Transcription Factor in Aspergillus fumigatus
Abstract
1. Introduction
2. Results
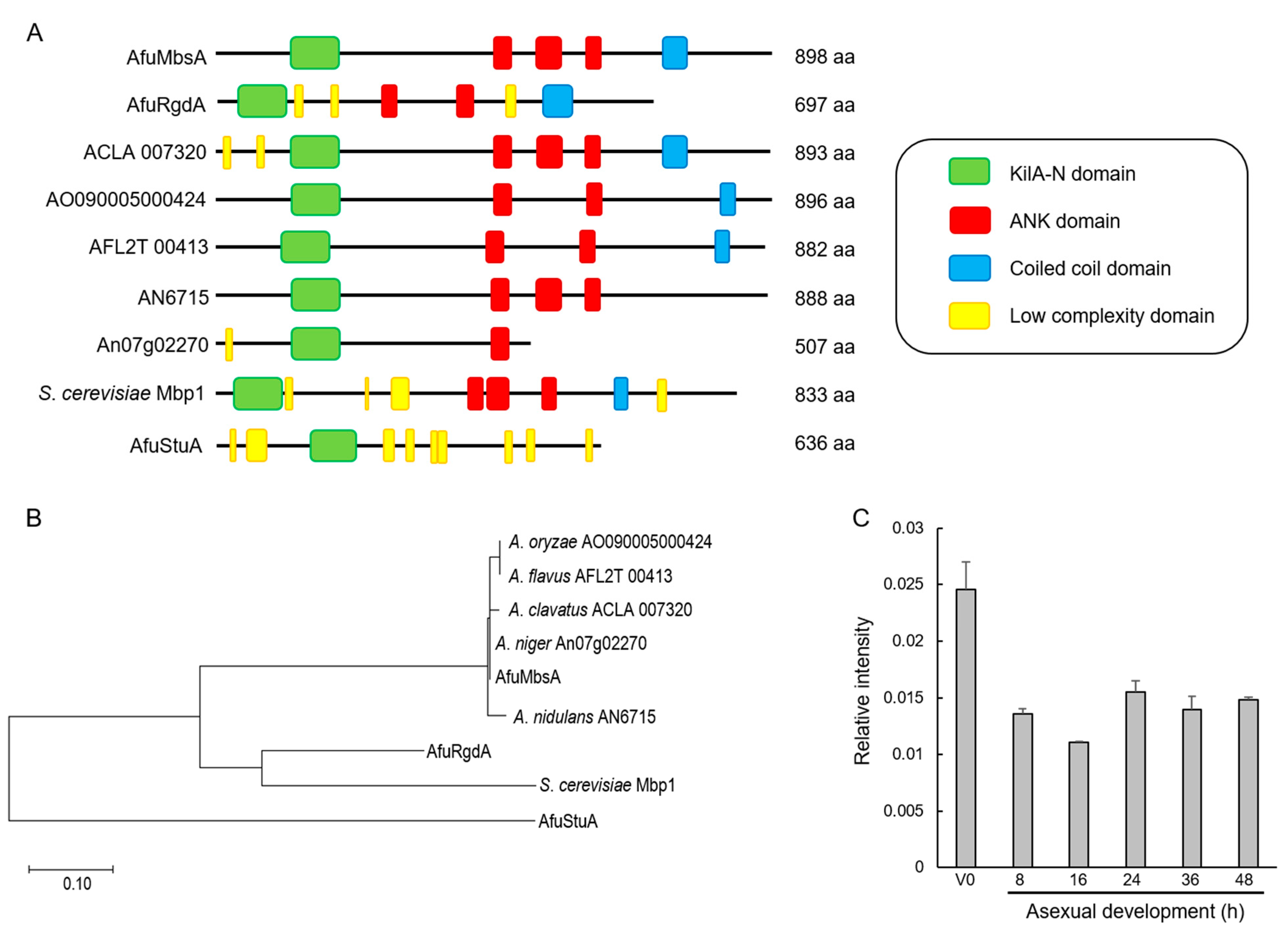
2.1. Summary of A. fumigatus MbsA
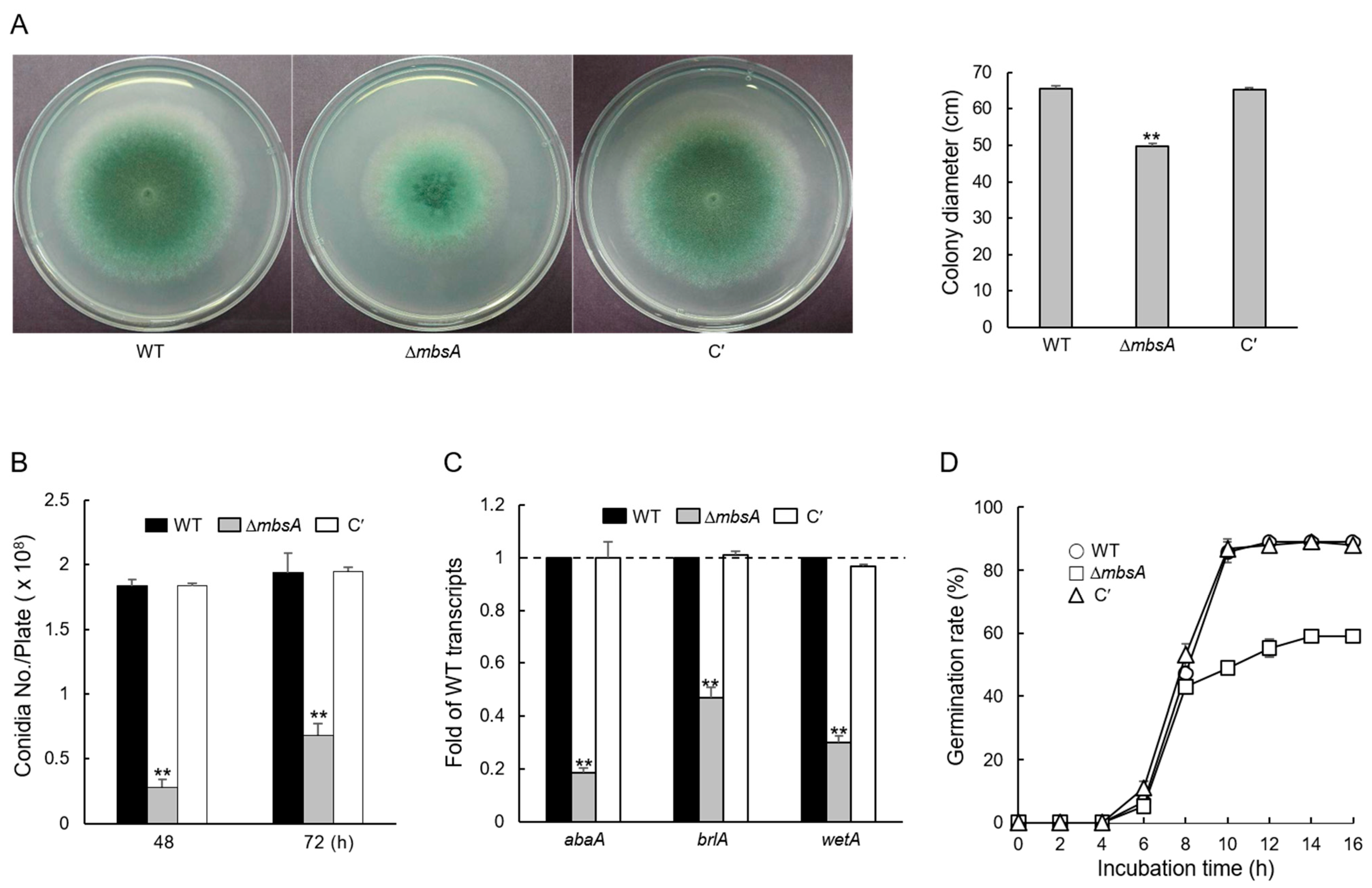
2.2. MbsA Affects Vegetative Growth and Conidial Development
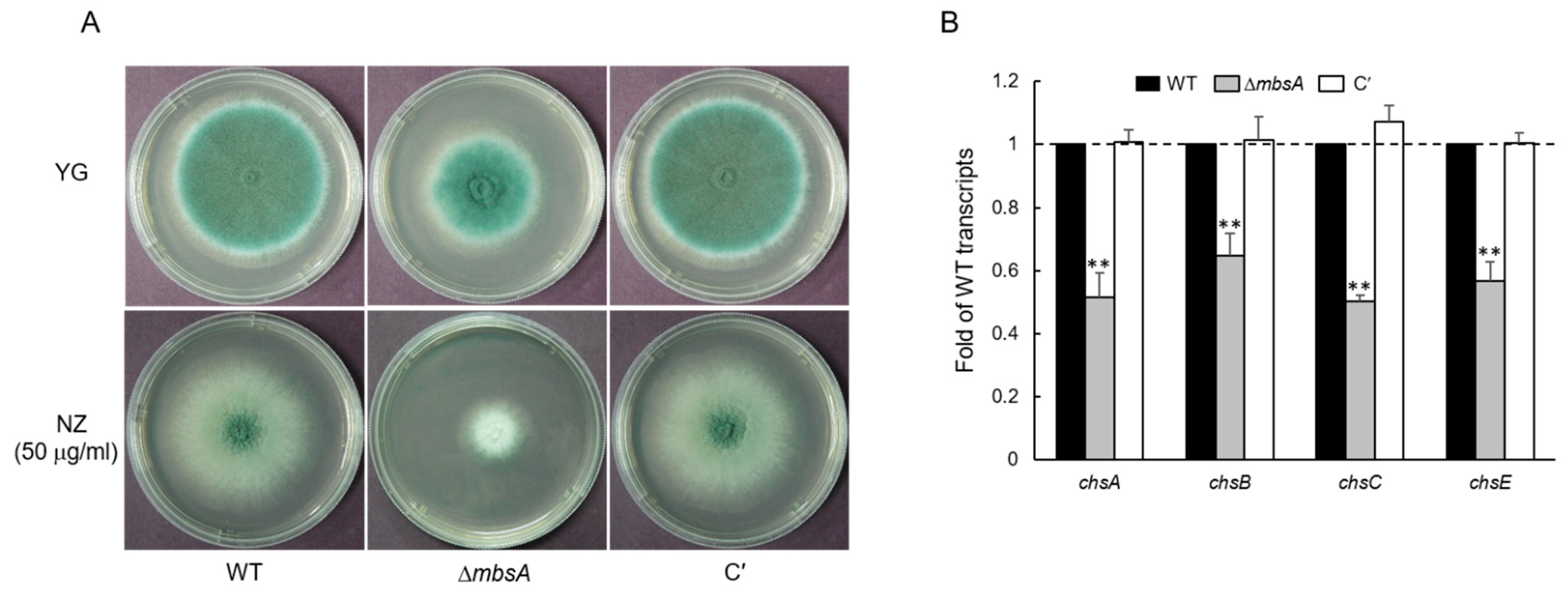
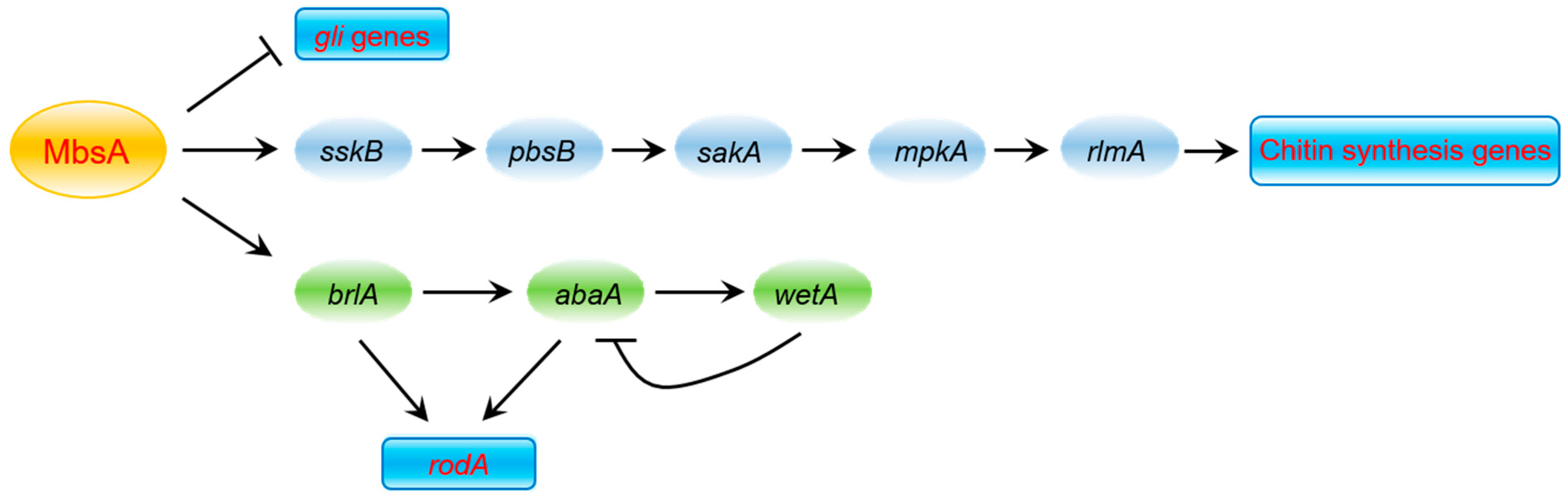
2.3. MbsA Is Involved in Chtin Synthesis in A. fumigatus
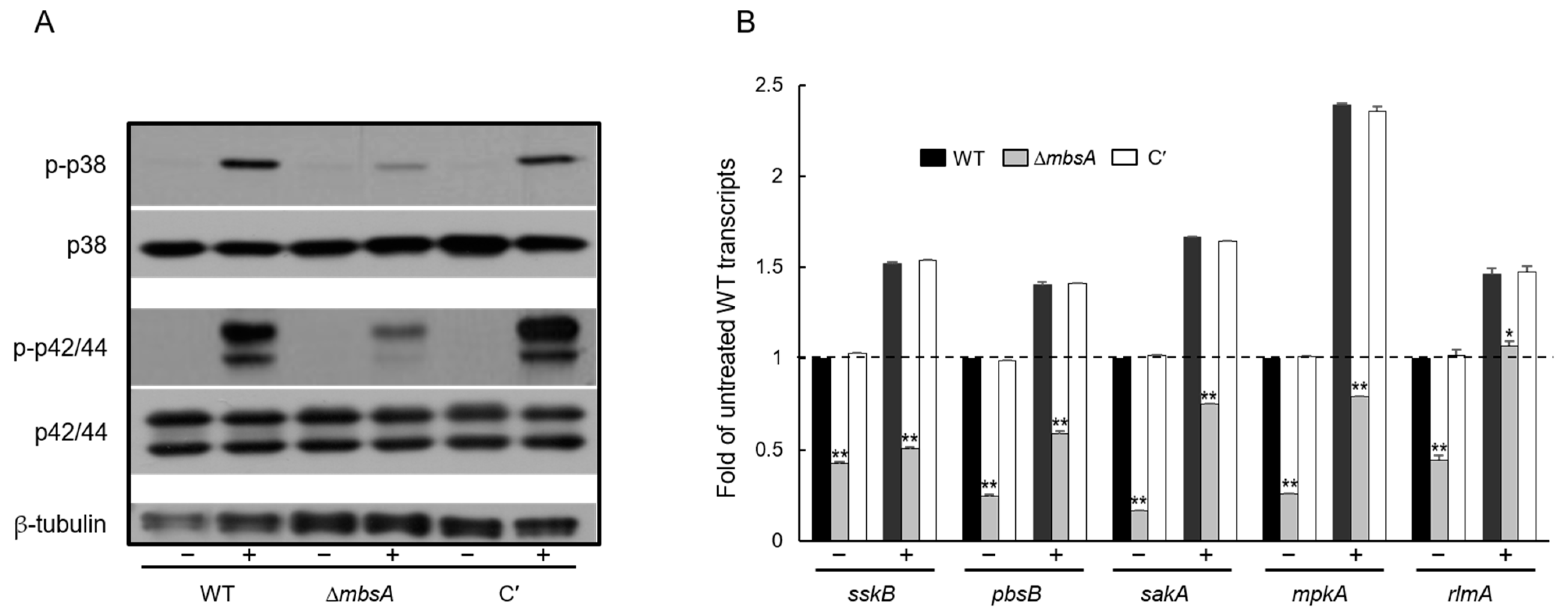
2.4. MbsA Positively Regulates the SakA MAP Kinase Pathway
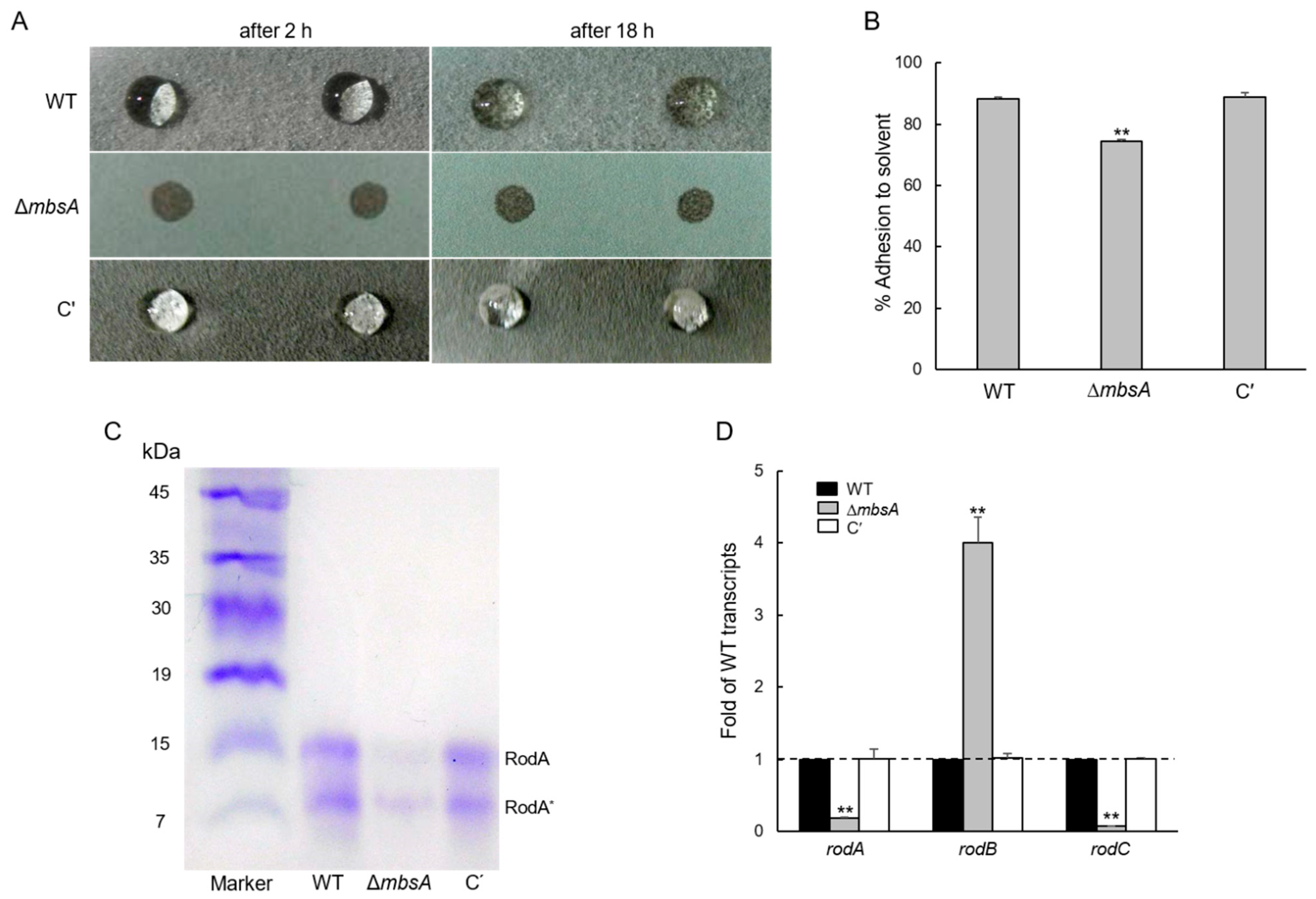
2.5. MbsA Is Necessary for the Conidia Hydrophobicity and Plays a Role in rodA Expression
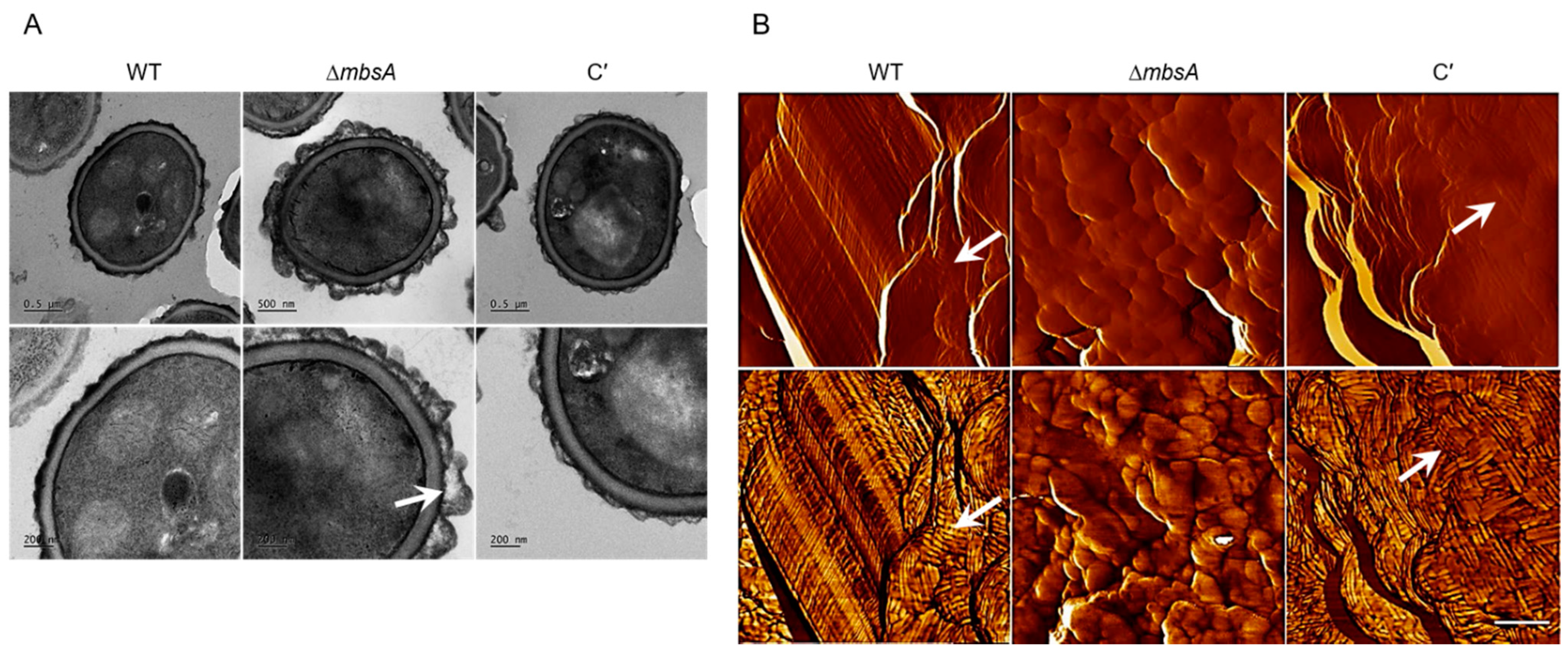
2.6. MbsA Is Required for Proper Spore Wall Formation
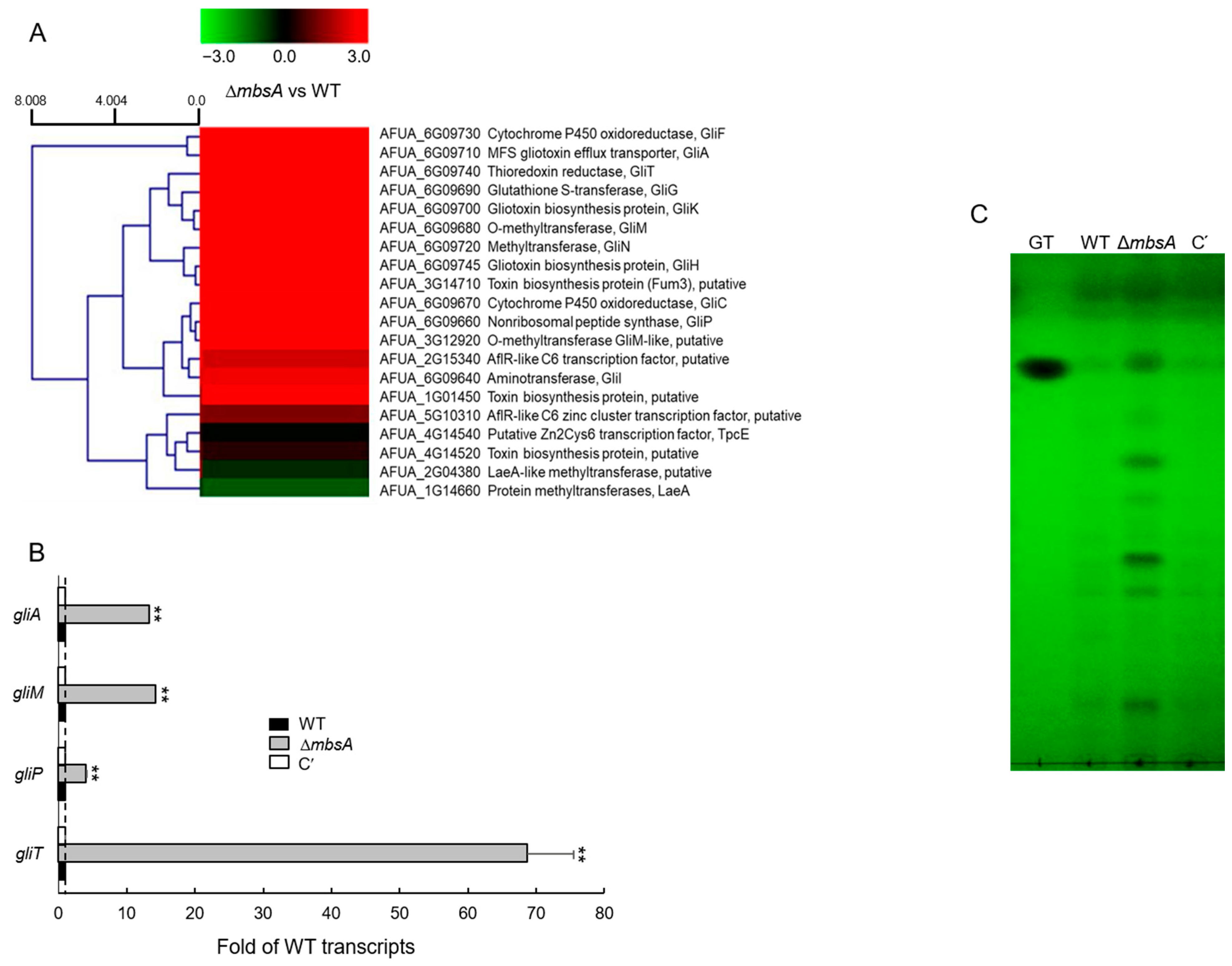
2.7. MbsA Down-Regulates Gliotoxin Biosynthesis
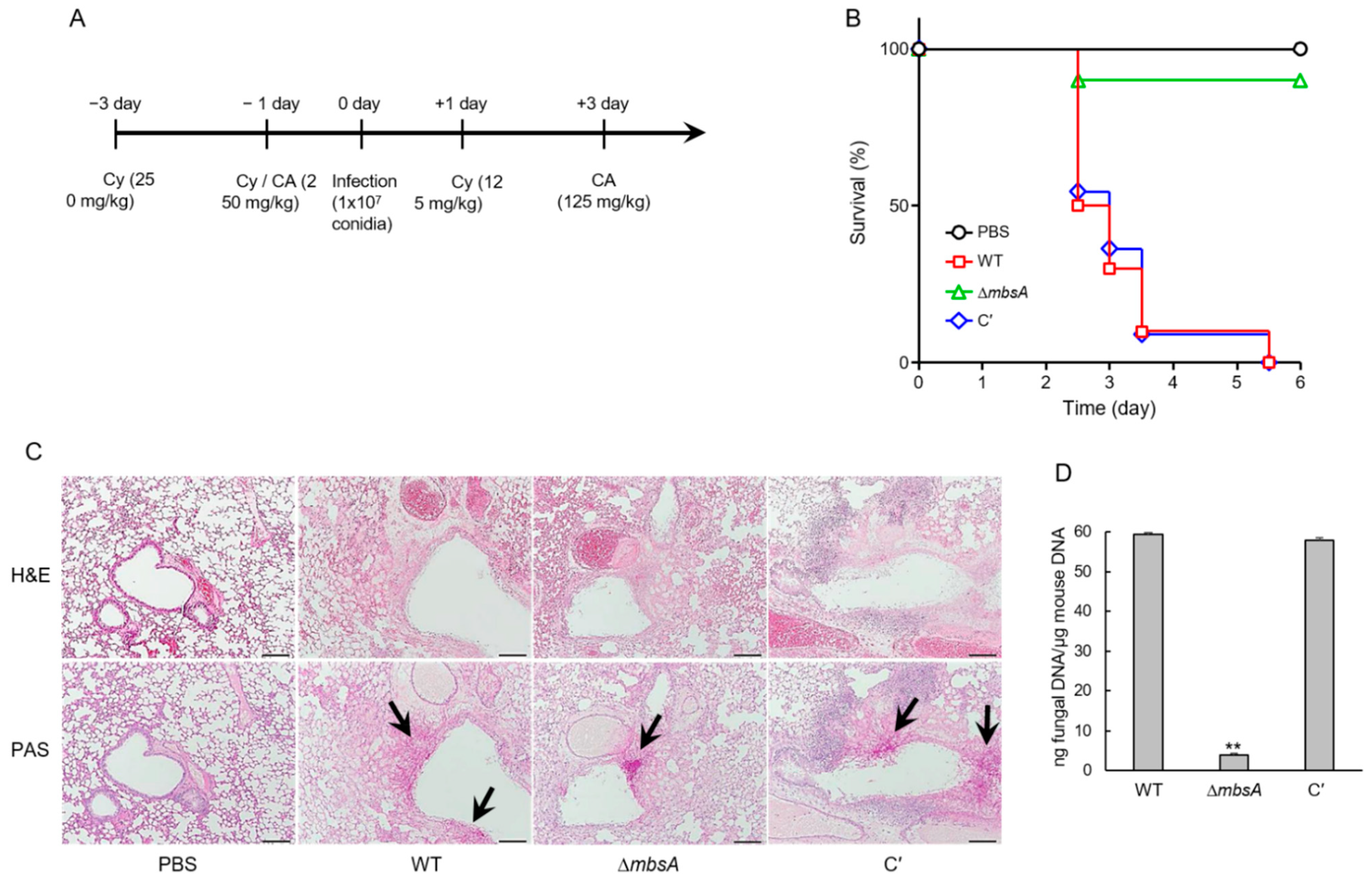
2.8. MbsA Plays an Important Role in Virulence
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Generation of the ΔmbsA Mutant in A. fumigatus
4.3. Nucleic Acid Isolation and Manipulation
4.4. Measurement of Germination Rate and Yield of Conidia
4.5. Determination of the Conidial Hydrophobicity
4.6. Microscopy
4.7. Transcriptome Analysis
4.8. Detection of Gliotoxin (GT)
4.9. Murine Virulence Assay
4.10. Immunoblotting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Y.; Su, H.; Zhou, J.; Feng, H.; Zhang, K.Q.; Yang, J. The APSES family proteins in fungi: Characterizations, evolution and functions. Fungal Genet. Biol. 2015, 81, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Dutton, J.R.; Johns, S.; Miller, B.L. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 1997, 16, 5710–5721. [Google Scholar] [CrossRef]
- Stoldt, V.R.; Sonneborn, A.; Leuker, C.E.; Ernst, J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997, 16, 1982–1991. [Google Scholar] [CrossRef] [PubMed]
- Sonneborn, A.; Bockmuhl, D.P.; Ernst, J.F. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 1999, 67, 5514–5517. [Google Scholar] [CrossRef]
- Doedt, T.; Krishnamurthy, S.; Bockmuhl, D.P.; Tebarth, B.; Stempel, C.; Russell, C.L.; Brown, A.J.; Ernst, J.F. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 2004, 15, 3167–3180. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Doedt, T.; Chiang, L.Y.; Kim, H.S.; Chen, D.; Nierman, W.C.; Filler, S.G. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 2005, 16, 5866–5879. [Google Scholar] [CrossRef]
- Twumasi-Boateng, K.; Yu, Y.; Chen, D.; Gravelat, F.N.; Nierman, W.C.; Sheppard, D.C. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 2009, 8, 104–115. [Google Scholar] [CrossRef]
- Jun, S.C.; Choi, Y.H.; Lee, M.W.; Yu, J.H.; Shin, K.S. The Putative APSES Transcription Factor RgdA Governs Growth, Development, Toxigenesis, and Virulence in Aspergillus fumigatus. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Song, M.H.; Lee, J.W.; Kim, M.S.; Yoon, J.K.; White, T.C.; Floyd, A.; Heitman, J.; Strain, A.K.; Nielsen, J.N.; Nielsen, K.; et al. A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans. Eukaryot. Cell 2012, 11, 53–67. [Google Scholar] [CrossRef]
- Miller, K.Y.; Wu, J.; Miller, B.L. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992, 6, 1770–1782. [Google Scholar] [CrossRef]
- Miller, K.Y.; Toennis, T.M.; Adams, T.H.; Miller, B.L. Isolation and transcriptional characterization of a morphological modifier: The Aspergillus nidulans stunted (stuA) gene. Mol. Gen. Genet. 1991, 227, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, L.H.; Kim, H.E.; Park, J.S.; Han, K.H.; Han, D.M. A putative APSES transcription factor is necessary for normal growth and development of Aspergillus nidulans. J. Microbiol. 2013, 51, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Wong Sak Hoi, J.; Lamarre, C.; Beau, R.; Meneau, I.; Berepiki, A.; Barre, A.; Mellado, E.; Read, N.D.; Latge, J.P. A novel family of dehydrin-like proteins is involved in stress response in the human fungal pathogen Aspergillus fumigatus. Mol. Biol. Cell 2011, 22, 1896–1906. [Google Scholar] [PubMed]
- Wezensky, S.J.; Cramer, R.A., Jr. Implications of hypoxic microenvironments during invasive aspergillosis. Med. Mycol. 2011, 49 (Suppl. 1), S120–S124. [Google Scholar] [CrossRef] [PubMed]
- Rogg, L.E.; Fortwendel, J.R.; Juvvadi, P.R.; Steinbach, W.J. Regulation of expression, activity and localization of fungal chitin synthases. Med. Mycol. 2012, 50, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Borgia, P.T.; Iartchouk, N.; Riggle, P.J.; Winter, K.R.; Koltin, Y.; Bulawa, C.E. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 1996, 20, 193–203. [Google Scholar] [CrossRef]
- Fukuda, K.; Yamada, K.; Deoka, K.; Yamashita, S.; Ohta, A.; Horiuchi, H. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot. Cell 2009, 8, 945–956. [Google Scholar] [CrossRef]
- Yanai, K.; Kojima, N.; Takaya, N.; Horiuchi, H.; Ohta, A.; Takagi, M. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 1994, 58, 1828–1835. [Google Scholar] [CrossRef]
- Aufauvre-Brown, A.; Mellado, E.; Gow, N.A.; Holden, D.W. Aspergillus fumigatus chsE: A gene related to CHS3 of Saccharomyces cerevisiae and important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet. Biol. 1997, 21, 141–152. [Google Scholar] [CrossRef]
- Bruder Nascimento, A.C.; Dos Reis, T.F.; de Castro, P.A.; Hori, J.I.; Bom, V.L.; de Assis, L.J.; Ramalho, L.N.; Rocha, M.C.; Malavazi, I.; Brown, N.A.; et al. Mitogen activated protein kinases SakA(HOG1) and MpkC collaborate for Aspergillus fumigatus virulence. Mol. Microbiol. 2016, 100, 841–859. [Google Scholar] [CrossRef]
- Rocha, M.C.; Fabri, J.H.; Franco de Godoy, K.; Alves de Castro, P.; Hori, J.I.; Ferreira da Cunha, A.; Arentshorst, M.; Ram, A.F.; van den Hondel, C.A.; Goldman, G.H.; et al. Aspergillus fumigatus MADS-Box Transcription Factor rlmA Is Required for Regulation of the Cell Wall Integrity and Virulence. G3 Genes Genomes Genet. 2016, 6, 2983–3002. [Google Scholar]
- Kovacs, Z.; Szarka, M.; Kovacs, S.; Boczonadi, I.; Emri, T.; Abe, K.; Pocsi, I.; Pusztahelyi, T. Effect of cell wall integrity stress and RlmA transcription factor on asexual development and autolysis in Aspergillus nidulans. Fungal Genet. Biol. 2013, 54, 1–14. [Google Scholar] [CrossRef]
- Fujioka, T.; Mizutani, O.; Furukawa, K.; Sato, N.; Yoshimi, A.; Yamagata, Y.; Nakajima, T.; Abe, K. MpkA-Dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 2007, 6, 1497–1510. [Google Scholar] [CrossRef]
- Paris, S.; Debeaupuis, J.P.; Crameri, R.; Carey, M.; Charles, F.; Prevost, M.C.; Schmitt, C.; Philippe, B.; Latge, J.P. Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 2003, 69, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Aimanianda, V.; Bayry, J.; Bozza, S.; Kniemeyer, O.; Perruccio, K.; Elluru, S.R.; Clavaud, C.; Paris, S.; Brakhage, A.A.; Kaveri, S.V.; et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009, 460, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, I.; Dupres, V.; Stephen-Victor, E.; Guijarro, J.I.; Gibbons, J.; Beau, R.; Bayry, J.; Coppee, J.Y.; Lafont, F.; Latge, J.P.; et al. Role of Hydrophobins in Aspergillus fumigatus. J. Fungi 2017, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Zhang, F.; Nie, X.; Wang, X.; Yuan, J.; Zhuang, Z.; Wang, S. Essential APSES Transcription Factors for Mycotoxin Synthesis, Fungal Development, and Pathogenicity in Aspergillus flavus. Front. Microbiol. 2017, 8, 2277. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.V.G.; Ray, S.C.; Puccia, R.; Rappleye, C.A. Characterization of the APSES-family transcriptional regulators of Histoplasma capsulatum. FEMS Yeast Res. 2018, 18, foy087. [Google Scholar] [CrossRef]
- Kafer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977, 19, 33–131. [Google Scholar]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Dominguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006, 1, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.S.; Kim, Y.H.; Yu, J.H. Proteomic analyses reveal the key roles of BrlA and AbaA in biogenesis of gliotoxin in Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2015, 463, 428–433. [Google Scholar] [CrossRef]
- Mah, J.H.; Yu, J.H. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Seo, J.A.; Yu, J.H. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 2004, 51, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Dang, X.; He, Y.Q.; Zhang, T.; Wang, H.Y. Selection of housekeeping genes as internal controls for quantitative RT-PCR analysis of the veined rapa whelk (Rapana venosa). PeerJ 2017, 5, e3398. [Google Scholar] [CrossRef][Green Version]
- Huan, P.; Wang, H.; Liu, B. Assessment of housekeeping genes as internal references in quantitative expression analysis during early development of oyster. Genes Genet. Syst. 2017, 91, 257–265. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Mortensen, H.D.; Gori, K.; Jespersen, L.; Arneborg, N. Debaryomyces hansenii strains with different cell sizes and surface physicochemical properties adhere differently to a solid agarose surface. FEMS Microbiol. Lett. 2005, 249, 165–170. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.-H.; Jun, S.-C.; Lee, M.-W.; Yu, J.-H.; Shin, K.-S. Characterization of the mbsA Gene Encoding a Putative APSES Transcription Factor in Aspergillus fumigatus. Int. J. Mol. Sci. 2021, 22, 3777. https://doi.org/10.3390/ijms22073777
Choi Y-H, Jun S-C, Lee M-W, Yu J-H, Shin K-S. Characterization of the mbsA Gene Encoding a Putative APSES Transcription Factor in Aspergillus fumigatus. International Journal of Molecular Sciences. 2021; 22(7):3777. https://doi.org/10.3390/ijms22073777
Chicago/Turabian StyleChoi, Yong-Ho, Sang-Cheol Jun, Min-Woo Lee, Jae-Hyuk Yu, and Kwang-Soo Shin. 2021. "Characterization of the mbsA Gene Encoding a Putative APSES Transcription Factor in Aspergillus fumigatus" International Journal of Molecular Sciences 22, no. 7: 3777. https://doi.org/10.3390/ijms22073777
APA StyleChoi, Y.-H., Jun, S.-C., Lee, M.-W., Yu, J.-H., & Shin, K.-S. (2021). Characterization of the mbsA Gene Encoding a Putative APSES Transcription Factor in Aspergillus fumigatus. International Journal of Molecular Sciences, 22(7), 3777. https://doi.org/10.3390/ijms22073777







